Abstract
The recently discovered 150-cavity (formed by loop residues 147–152, N2 numbering) adjacent to the enzymatic active site of group 1 influenza A neuraminidase (NA) has introduced a novel target for the design of next-generation NA inhibitors. However, only group 1 NAs, with the exception of the 2009 pandemic H1N1 NA, possess a 150-cavity, and no 150-cavity has been observed in group 2 NAs. The role of the 150-cavity played in enzymatic activity and inhibitor binding is not well understood. Here, we demonstrate for the first time that oseltamivir carboxylate can induce opening of the rigid closed N2 150-loop and provide a novel mechanism for 150-loop movement using molecular dynamics simulations. Our results provide the structural and biophysical basis of the open form of 150-loop and illustrates that the inherent flexibility and the ligand induced flexibility of the 150-loop should be taken into consideration for future drug design.
Influenza virus causes epidemics and pandemics, which severely impair public health1,2,3,4,5. Two of the major pandemics of the last century were caused by N2 containing influenza viruses: H2N2 (Asian flu) and H3N2 (Hong Kong flu)1. Recent outbreaks of H3N2 and H1N2 swine influenza viruses-caused human febrile respiratory illness in the United States highlight the importance of this subtype in influenza epidemics6,7,8. Besides H1N1, the H3N2 subtype is the major causative agent of severe epidemics and is critical for vaccine development9. Therefore a comprehensive understanding of N2 containing viruses is important for preparedness against highly transmissible influenza viruses.
Hemagglutinin (HA) and neuraminidase (NA) are the two major surface glycoproteins responsible for initiating influenza virus infection10,11,12,13 and virus release14,15,16, respectively. HA and NA of influenza A viruses are divided into subtypes based upon their distinct antigenic properties: seventeen for HA (H1–H17) and ten for NA (N1–N10)17,18,19. Among the influenza A viruses, only N1 and N2 have been found in human isolates responsible for pandemics and recurrent annual epidemics. With the exception of N10, recently identified in a bat influenza A virus genome, the nine NA alleles are classified into two groups according to phylogenetic analysis and structure. Group 1 NA comprises N1, N4, N5 and N8, whereas group 2 comprises N2, N3, N6, N7 and N920. 3-D structures reveal the distinct conformations of the areas adjacent to the enzymatic active site between group 1 and group 2 members though the active site structures are virtually identical among all the NAs20. For typical N1 subtypes (but not for the 2009 H1N1 pandemic N1), the crystal structures reveal a 150-loop (formed by amino acids 147–152, N2 numbering) that adopts an open conformation forming an additional 150-cavity adjacent to the active site. Previously, no group 2 NAs have been crystallographically shown to have a 150-loop in an open conformation, although all-atom molecular dynamics simulations indicate that N2 may be able to adopt such a configuration in solution21. The recently discovered 150-cavity is currently being explored as a novel target for group 1 specific influenza NA inhibitors20,22.
Detailed structural analyses demonstrate that residue 147 plays an essential role in the conformation of the 150-loop. Recently, our group has shown that the N5 structure contains an extended 150-cavity resulting from the unique residue N14723. In the solved N2 structures, a salt bridge between D147 and H150 contributes to a rigid closed 150-loop24. Previous molecular dynamics simulations show that the D147-H150 salt bridge greatly stabilizes the closed 150-loop conformation and that loss of this salt bridge reduces the rigidity of the 150-loop21. D147 is prevalent in N2, but rarely found in other types of influenza NA23. Compared with N2 containing D147, the flexibility of the 150-loop of 2009 H1N1 NA (G147) is much higher21, although the structure of 2009 H1N1 NA also presents a deficient 150-cavity in its crystal structure25.
Since NA plays an essential role in the release of new influenza virions from host cells, inhibition of NA compromises the ability of progeny virions to spread to uninfected cells. Oseltamivir (Tamiflu) and zanamivir (Relenza) are two commercially available NA-targeted competitive inhibitors, which act against both group 1 and group 2 enzymes as well as influenza B NA16. The open 150-loop of group 1 NAs has been found to adopt a closed conformation upon binding of zanamivir, however for oseltamivir carboxylate this effect depends on both inhibitor concentration and soaking time dependent20. The crystal structures of typical group 1 NAs in complex with oseltamivir carboxylate display two 150-loop conformations, indicating a two-step process of oseltamivir carboxylate binding. Molecular dynamics simulations of the free and oseltamivir carboxylate-bound forms of tetrameric N1 suggest a rapid loop switching motion, which demonstrates the flexibility of the 150-loop26. In contrast, in the crystal structures of typical group 2 NA-inhibitor bound complexes, the 150-loops always adopt closed conformation20,27,28,29. Also, it is noteworthy that zanamivir always induces the closed conformation20,27, whether the target is a group-1 or group-2 NA. Moreover, previous reports showed that R152K mutant in flu B lead to zanamivir and oseltamivir resistance30,31, which suggests that 150-loop may also play a role in inhibitor binding.
Here we report a novel half open 150-loop in the crystal structure of a typical group 2 NA for the first time. This suggests that inhibitors targeting the 150-cavity may also effective target group 2 influenza NAs. Furthermore, we demonstrate with molecular dynamics simulations that the protonation state of inhibitors and N2 H150 play an important role on the movement of the 150-loop. Comparison of the differences between oseltamivir carboxylate and zanamivir binding to N2 illustrates the importance of the inhibitor C4 group (zanamivir numbering), providing considerable value for influenza inhibitor design.
Results
Oseltamivir carboxylate induced opening of the rigid N2 150-loop: the first open 150-cavity structure of group 2 NA
Sequence alignment shows that D147 is commonly found in N221,23. D147 can form a rigid salt bridge with H150, which plays an important role in holding the N2 150-loop in a closed conformation (PDB 1NN2)24. Thus, all the N2 crystal structures in the PDB database contain a rigid closed 150-loop. The free N2 observed in our experiment also presents a closed 150-loop (hereafter referred to as “Free N2”) (Fig. 1a).
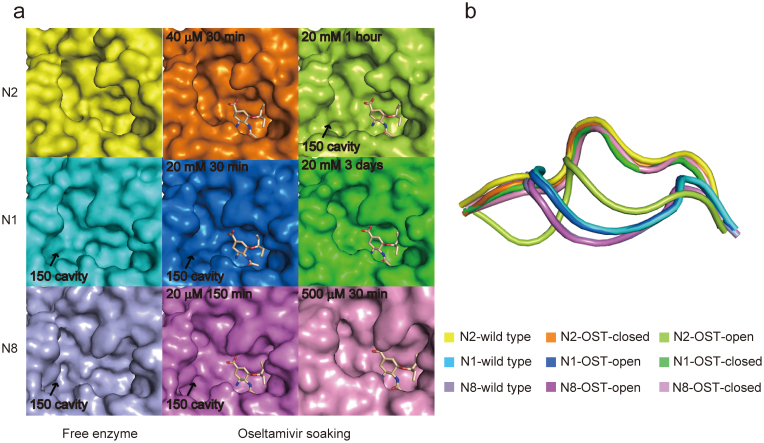
Figure 1. Comparison of oseltamivir carboxylate induced 150-loop conformational change in N2, N1 and N8.
(a) The active sites of free N2 (yellow), N1 (cyan), N8 (lightblue) and oseltamivir carboxylate-bound complex in different soaking conditions are displayed in surface representation. Free N2 (yellow), N2-40 μM-30 min (orange), N1-20 mM-3 days (green) and N8-500 μM-30 min (pink) have no 150-cavity, however free N1, N8, N2-20 mM-1 hour (limon), N1-20 mM-30 min (marine) and N8-20 μM-150 min (magenta) contain a 150-cavity. Oseltamivir carboxylate is shown in wheat stick. (b) Superposition of the 150-loop of free N1, N2, N8 and N1, N2, N8 complexed with oseltamivir carboxylate under different soaking conditions. The 150-loops are shown in cartoon representation, and colors are corresponding to Fig. 1a.
However, upon soaking with oseltamivir carboxylate, we found that the rigid N2 150-loop can actually be opened (Fig. 1a). Our crystal structure data indicates that the conformation of the N2 150-loop is dependent on the concentration of oseltamivir carboxylate (Fig. 1a). Specifically, N2 presents an open 150-cavity when soaking crystals in 20 mM oseltamivir carboxylate for 1 hour (hereafter referred to as “N2-oseltamivir open form”), while it shows the same conformation as in the free N2 when soaking into 40 μM oseltamivir carboxylate for 30 min (hereafter referred to as "N2-oseltamivir closed form") (Fig. 1a).
An influence of oseltamivir carboxylate concentration and soaking-time on the 150-loop conformation has also been observed in both N1 and N8 (Fig. 1a), indicating that oseltamivir carboxylate has great influence on the conformation of the 150-loop. When using typical group 1 NA N1 (PDB 2HTY) and N8 (PDB 2HT5)20 and typical group 2 N2 as open and closed references, it is clear that the 150-loop of N2-oseltamivir open form adopts a half-open conformation (Fig. 1b), which is distinct from other types of NA. Superposition of the free N2 Cα atoms with those of N2-oseltamivir open form and N2-oseltamivir closed form reveals root-mean-square deviations (RMSD) of 3.0 Å and 0.1 Å in the 150-loop, respectively (only Cα atoms were used to calculate RMSD). The main chain of the 150-loop in N2-oseltamivir closed form is the same as free N2. The 150-loop conformation in the oseltamivir carboxylate complexes with N1 and N8 is similar to free N2 with a RMSD of 0.5 Å and 0.6 Å in the 150-loop, respectively.
Intermolecular interaction of the tetrameric NA plays an essential role in stabilizing the open N2 150-cavity
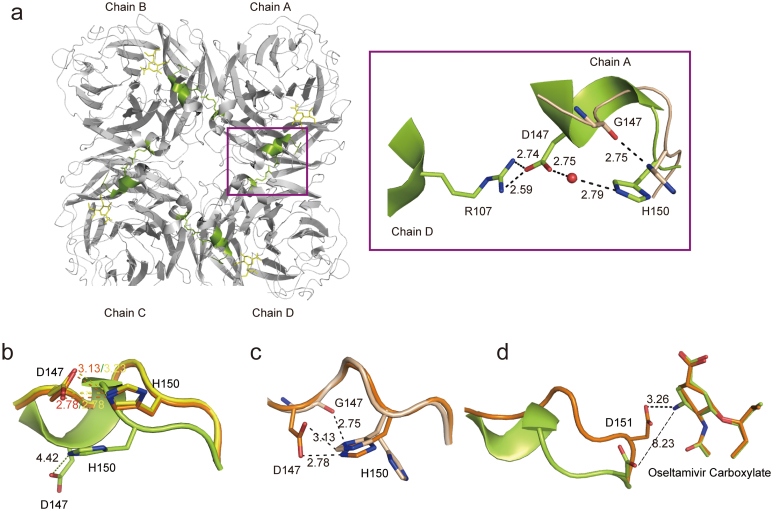
In the N2-oseltamivir open form crystal structure the novel half-open form of the 150-cavity appears to be quite stable. The side chain of D147 is fixed by the side chain of R107 in the neighboring NA molecule and a water molecule forms hydrogen bonds with D147 and H150 with distances of 2.75 Å and 2.79 Å, respectively (Fig. 2a). Additionally, the 150-loop turns into a 310 helix, further stabilizing an open 150-loop conformation (Fig. 2b). To investigate the importance of the D147-R107 intermolecular interaction in maintaining the open conformation of the N2 150-loop, D147 was replaced by glycine. Although the crystal of N2-G147 was soaked in the same condition as N2-oseltamivir open form, the 150-loop of the N2-G147-oseltamivir carboxylate complex still adopts a closed conformation identical to that of free N2 with D147 (Fig. 2c). Specifically, the glycine does not have a side chain, so it cannot form hydrogen bond with R107 in neighboring molecule to further stabilize the 150-loop in an open conformation. Still, the side chain of N2-G147 H150 is flexible and adopts two conformations, in one of these conformations the H150 ND1 forms hydrogen bond with the oxygen atom of the G147 main chain, stabilizing a closed 150-loop conformation (Fig. 2c). Additionally, the interaction between the side chain of D151 and the amino group of oseltamivir carboxylate is another factor that stabilizes the 150-loop in a closed conformation. In the N2-oseltamivir closed form structure, the amino group of oseltamivir carboxylate forms a hydrogen bond with the side chain of D151, at a distance of 3.26 Å (Fig. 2d). While in the N2-oseltamivir-open structure, there is no interaction between the OD1 on the side chain of D151 and the amino group of oseltamivir carboxylate, with a distance of 8.23 Å (Fig. 2d).
Figure 2. Structural basis of an open 150-loop in N2 neuraminidase.
(a) Overview of the open form of 150-loop in the N2-oseltamivr complex structure with an emphasis on the intermolecular interaction indicated with a violetpurple box. The residues of N2-wild type are shown in limon, while the residues of N2-G147 are shown in wheat. The water molecule is shown in red sphere. (b) Superposition of the 150-loop conformation between free N2 (yellow), N2-oseltamivir closed form (orange) and N2-oseltamivir open form (limon). Compare the interaction between D147 and H150. (c) Superposition of 150-loop conformation between N2-oseltamivir closed form (orange) and N2-G147 after a 1 hour soak with 20 mM oseltamivir carboxylate (wheat). Compare the interaction between Residue 147 and 150. (d) Superposition of 150-loop conformation between N2-oseltamivir closed form (orange) and N2-oseltamivir open form (limon). Compare the interaction between D151 and amino group of oseltamivir carboxylate. The side chain of the residues is displayed in stick representation with hydrogen bonds and salt bridges indicated by dotted lines. The interaction with a distance greater than 3.5 Å is shown as a broken line. The shared residues are highlighted in black and labeled.
Mechanism for oseltamivir carboxylate induced opening of the N2 150-loop
The crystal structures of N2 in complex with oseltamivir carboxylate and zanamivir were determined at pH 9.0, while the pKa values of the oseltamivr amino group and zanamivir guanidino group are 7.75 and 1332, respectively. Thus, the amino group of oseltamivir carboxylate should be neutral approximately 94.7% of the time while the guanidino moiety in zanamivir should be positively charged almost 99.9% of the time according to the Henderson-Hasselbach equation. The pKa of H150 in the closed 150-loop conformation is 1 pK unit higher than that in the open conformation as predicted by H++33. This indicates that H150 is more likely to be protonated in the closed conformation compared to the open conformation. To explore the mechanism of how inhibitors influence conformational changes of the 150-loop we performed three separated 200 ns molecular dynamics simulations (MDs) for tetrameric N2 with different protonation states of H150 and the amino group of oseltamivir carboxylate. One 200 ns MDs with zanamivir and one 200 ns MDs without any ligand were also performed (Table 1).
Table 1. Information of five simulation systems.
| Systems | Ligand | Charge in C4 group (zanamivir numbering) of inhibitor | Protonation state of H150 |
|---|---|---|---|
| OST-NH3-H150-Pos* | Oseltamivir carboxylate | Positive | Positive |
| OST-NH2-H150-Pos | Oseltamivir carboxylate | Neutral | Positive |
| OST-NH2-H150-Neu | Oseltamivir carboxylate | Neutral | Neutral |
| FREE-H150-Neu | - | - | Neutral |
| ZMR-H150-Pos** | Zanamivir | Positive | Positive |
*OST refers to oseltamivir carboxylate.
**ZMR refers to zanamivir.
In the OST-NH3-H150-Pos system, the C4 amino group of oseltamivir carboxylate and H150 are all positively charged. The 150-loop remains closed for 98.1% of the conformations sampled in the simulation (Table 2, Supplementary Fig. S1). The positively charged oseltamivir carboxylate amino group forms a hydrogen bond with the D151 side chain, which stabilizes the closed 150-loop conformation. The salt bridge between D147 and H150 contributes greatly to a closed 150-loop (Supplementary Fig. S1).
Table 2. Population analysis of 150-cavity.
| Volume rate (%) | ||
|---|---|---|
| System | 0–40 | >40 |
| OST-NH3-H150-Pos | 98.1 | 1.9 |
| OST-NH2-H150-Pos | 59.2 | 40.8 |
| OST-NH2-H150-Neu | 39.9 | 60.1 |
| FREE-H150-Neu | 72 | 28 |
| ZMR-H150-Pos | 97.8 | 2.2 |
In the OST-NH2-H150-Pos system, the oseltamivir carboxylate C4 amino group is neutral and H150 is positively charged. During the simulation, an open 150-cavity can be observed in all four chains and 40.8% of the configurations present a 150-cavity volume larger than 40 Å3 (Table 2). Specifically, in chain A and chain D, the 150-loop started from a closed form, opened for a while, resulting in the formation of a 150-cavity, and then closed again, leading to absence of the 150-cavity. In the other two chains the 150-loop also opened (Supplementary Fig. S2). The open and closed conformation of the 150-loop are directly related to the interaction between D147 and H150 (Supplementary Fig. S2). Compared with the OST-NH3-H150-Pos system, the deprotonated amino group of oseltamivir carboxylate exerts a significant effect on 150-loop opening. The interaction between the amino group of oseltamivir carboxylate and the side chain of D151 is almost completely lost (Fig. 3a) which results in increasing flexibility of the 150-loop. The open and closed conformation of the 150-loop and environmental change of H150. Histidine is sensitive to environmental changes, so when oseltamivir carboxylate influences the rigid closed 150-loop structure, H150 may lose a proton which will reduce the interaction between D147 and H150, and further lead to the 150-loop open.
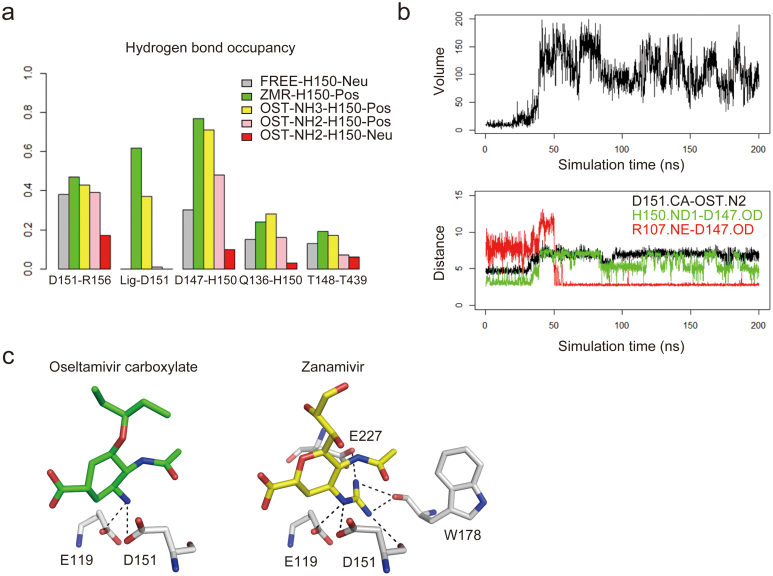
Figure 3. Driving force for the opening of the N2 150-loop.
(a) Hydrogen bond occupancy related to the 150-loop in the five simulation systems. Grey: FREE-H150-Neu; green: ZMR-H150-Pos; yellow: OST-NH3-H150-Pos; pink: OST-NH2-H150-Pos; red: OST-NH2-H150-Neu. (b) The opening process of the 150-loop in OST-NH2-H150-Neu trajectory (Chain D) represents the similar conformational change to the crystal structure. Upper panel: 150-cavity volume; down: the distance between alpha carbon of D151 and nitrogen atom of Oseltamivir carboxylate amino group (black), the nearest distance between OD1 or OD2 of D147 and ND1 of H150 (green), and the nearest distance between OD1 or OD2 of D147 and NE atom of R107 in the neighboring molecule (red). (c) Comparison of the interactions between N2 residues and amino group of oseltamivir carboxylate and guanidino group of zanamivir (within 3 Å).
To validate the above hypothesis, we performed a MD simulation for the OST-NH2-H150-Neu system. In this system, both the amino group of oseltamivir carboxylate and the H150 imidazole side chain are in neutral states. A statistical analysis shows that the 150-cavity with a volume larger than 40 Å3 occurs during 60.1% of the simulation (Table 2), which is 19.3% and 58.2% larger than the OST-NH2-H150-Pos system and OST-NH3-H150-Pos system, respectively. The 150-loop opening is accompanied by the loss of the D147-H150 interaction in all four chains, although different behaviors can be observed in different chains and at specific times. In chain A, the 150-cavity volume decreased due to the orientation of R430. In chain B, the side chain of D147 failed to interact with R107 in the neighboring molecule, and then D147 and H150 formed weak hydrogen bonds again leading to the reformation of a closed 150-loop conformation (Supplementary Fig. S3). In chain D, the 150-loop opening process was observed in a similar manner observed in crystal soaking experiments (Fig. 3b). After 40 ns, the interaction between the oseltamivir carboxylate amino group and D151 was lost (Fig. 3b) enabling the 150-loop to be more flexible. The interaction between D147 and H150 is reduced as a result of deprotonation of H150, thereby increasing the flexibility of D147. Then the side chain of R107 in the neighboring molecule has the opportunity to form salt bridge with D147 enabling the 150-loop to form a stable open conformation.
To validate the effect of the protonation state of H150 on the flexibility of the 150-loop, the FREE-H150-Neu system with H150 in the neutral state was set up. Instantaneous opening of the 150-cavity can be observed in all four of the chains. Specifically, in chain B, C and D, the 150-cavity opened for a short time and then closed again. For chain A, the cavity remains open for a longer time (Supplementary Fig. S4). 72% of the conformations have a 150-cavity smaller than 40 Å3, which is much larger than the closed conformation population in OST-NH2-H150-Neu (39.9%) (Table 2). This indicates that without oseltamivir carboxylate, N2 prefers a closed 150-loop conformation. Although the interaction between D147 and H150 becomes weaker as the deprotonation of H150, hydrogen bonds such as R156-D151, Q136-H150 and T148-T439 also contribute to a closed 150-loop conformation (Fig. 3a).
The guanidino group of zanamivir is virtually always positively charged due to its high pKa. Thus, in the ZMR-H150-POS system, the guanidino group and H150 are both set as positively charged. During the simulation, 97.8% conformations sampled in this trajectory has a 150-cavity smaller than 40 Å3. The 150-loop remains closed almost throughout the entire simulation (Supplementary Fig. S5), and has strong interactions with zanamivir. Compared with oseltamivir carboxylate, the hydrogen bond occupancy between zanamivir and D151 is stronger (Fig. 3a), which indicates that zanamivir has a higher avidity with the 150-loop. Moreover, the N2-inhibitor complex structure clearly shows that the interaction between the zanamivir guanidino group and neighboring residues (less than 3.5Å) is stronger than that of the oseltamivir carboxylate amino group (Fig. 3c). Specifically, the zanamivir guanidino can bind to four residues (E119, D151, W178 and E227), while the amino group of oseltamivir carboxylate can only interact with two residues (E119 and D151). The interactions between zanamivir and the main chain of the 150-loop also help to maintain the closed 150-loop conformation. The structure and simulation data suggest that the proper size and charge of the C4 moiety plays a critical role in determining 150-loop dynamics upon ligand binding.
Discussion
The influenza NA 150-cavity was first observed in group 1 NAs, and several group 1 specific inhibitors were subsequently designed to target this 150-cavity22. An open 150-loop has never been observed in X-ray crystallography experiments with group 2 NAs, especially in N2 containing a D147-H150 salt bridge, although simulation work suggested N2 may be able to adopt such a configuration in solution dynamics21. In this report, we demonstrate for the first time that the N2 150-loop can adopt a half-open conformation in its crystal structure. Furthermore we have carried out extensive molecular dynamics simulations in order to elucidate mechanisms of 150-loop motion.
The free N2 crystal structure possesses a salt bridge between D147 and H150, which is one of the key factors to determine the rigidity of its 150-loop. However, this rigid interaction can be broken under the condition of high concentrations oseltamivir carboxylate. With the exception of N5, which contains N147, all other types of NA possess G147. It can be inferred that the 150-loop should be more flexible with G147 in place of D147. Alternatively, the conformation of the 150-loop in these NAs should be more easily influenced by extrinsic factors. It seems that both group 1 and group 2 NAs are able to adopt an open 150-cavity under certain conditions. From this perspective, the conformation of the 150-loop may not be a true group specific feature which is consistent with previous molecular dynamics simulation results21. This notion can also be supported by the 150-cavity deficiency in the atypical group 1 member, 2009 pandemic N125.
It is worthy to note that an open 150-cavity in all types of NA has never been observed in the presence of zanamivir and it has been confirmed that zanamivir remains effective against oseltamivir-resistant viruses. Based on analysis of the complex structures of N2 and inhibitors, we note that the 4-guanidino group of zanamivir provides stronger interactions with the NA active site to facilitate high affinity binding. This might compensate for lost interactions resulting from drug resistant mutations. A previous study illustrates that I223R (IR) and I223R/H275Y (IRHY) mutations of H1N1-2009 NA lead to reduce binding efficacy of oseltamivir carboxylate. The 150-loop of IRHY:osel shows a marked conformational change to the open form, resulting in a loss of the hydrogen bonds between the inhibitor and the 150-loop, contributing to resistance to oseltamivir34. Whearas the avidity of zanamivir is affected less by the IR and IRHY mutations as it is able to maintain hydrogen bonds to the 150-loop. Moreover, a R152K mutation in flu B results in oseltamivir and zanamivir resistance30,31 demonstrating that the interactions between key 150-loop residues in the 150-loop and inhibitors play an important role in high avidity binding. Therefore, increasing the hydrogen bonds between NA inhibitors and residues in the 150-loop may help to overcome drug resistance. Additionally, it is difficult for the zanamivir guanidino group to lose its charge due to its high pKa, and the positively charged guanidino group has a high affinity for the 150-loop.
In conclusion, this study demonstrates the inherent flexibility of the 150-loop and the importance of inhibitor protonation state on influencing active site conformations. The open 150-cavity discovered in group 2 NA suggests that protein active site conformations can be greatly influenced by introduction of a ligand. Thus, both intrinsic protein dynamics and ligand induced protein dynamics should be considered in the future antiviral drug design.
Methods
Recombinant NA production
NA was prepared in baculovirus expression system as previous reported1425. cDNA encoding amino acid residues 83–469 of p57N2 (1957 Asian pandemic H2N2 NA, Sequence GenBank accession No. M18648.1) were cloned into the baculovirus transfer vector pFastBac1 (Invitrogen), with a GP67 signal peptide, a 6X histag, a tetrameric sequence and a thrombin cleavage site at the N terminus. A point mutation corresponding to G147 was introduced by site directed mutagenesis based on the p57N2 sequence. Recombinant baculovirus was prepared based on the manufacturer's protocol (Invitrogen). Sf9 cells were used to prepare recombinant baculovirus.
Protein crystallization and inhibitor soaking assays
Protein concentrations were determined using a BCA protein assay according to the manufacturer's instructions (Pierce). Crystallization screens were initiated using commercial kits (Hampton Research). All crystallizations were performed using the hanging drop vapor diffusion technique. The N2 wild type and N2-G147 crystals were observed in PEGRx I 27 (0.1 M BIS-TRIS propane pH9.0, 10% v/v Jeffamine ED-2001 pH7.0) and PEGRx I 26 (0.1 M Imidazole pH7.0, 20% v/v Jeffamine ED-2001 pH7.0), repectively. The N2 crystals were incubated in mother liquor containing 20 mM and 40 μM oseltamivir carboxylate, and 20 mM Zanamivir at 291 K for 1 hour, 30 min and 1 hour, respectively. The N2-G147 crystal was incubated in mother liquor containing 20 mM oseltamivir carboxylate for 1 hour at 291 K.
Data collection, structure determination, refinement and analysis
All crystals were soaked in cryoprotectant (reservoir solution supplemented with 20% glycerol), then flash-cooled and maintained at 100 K in a cryostream. Diffraction data for the free p57N2 native structure were collected at KEK beamline NE3A, while the data for N2 soaked with oseltamivr carboxylate and zanamivir were collected at SSRF beamline BL17U. Diffraction data were processed and scaled using HKL200035. Data collection and processing statistics are summarized in Table 3. The structure of p57N2 was solved by molecular replacement using the A/TOKYO/3/1967 H2N2 N2 molecule (PDB 1IVG)36 as the search model with Phaser37 in the CCP4 program suite38. The other datasets were determined by molecular replacement using the solved N2 structure. Extensive model building was performed by hand with COOT39, and restrained refinement was performed using REFMAC540. Further rounds of refinement were performed using the PHENIX package41. The final structures displayed good stereochemistry (Table 3), as assessed by the program PROCHECK42. Structural figures were prepared with PyMOL (http://www.pymol.org/).
Table 3. Data collection and refinement statistics.
| Parameter | FreeN2 | N2-oseltamivir open form | N2-oseltamivir closed form | N2-G147-oseltamivir |
|---|---|---|---|---|
| Data collecting | ||||
| Space group | C2221 | C2221 | C2221 | C2221 |
| Unit cell dimensions (a, b, c) | 114.83, 39.47, 140.05 | 114.91, 139.39, 140.24 | 115.43, 139.11, 140.00 | 114.33, 139.31, 140.05 |
| Unit cell dimensions (α, β, γ) | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 |
| Resolution range (Å)a | 50.00–1.80 (1.86–1.80) | 50.00–1.60 (1.65–1.60) | 50.00–1.80 (1.86–1.80) | 50.00–2.20 (2.28–2.20) |
| Rmerge (%)b | 10.2 (54.7) | 8.1 (53.8) | 9.7 (49.2) | 17.3 (55.4) |
| I/σ | 17.59 (2.54) | 19.1 (2.29) | 20.19 (4.3) | 11.17 (3.53) |
| Redundancy | 6.2 (5.9) | 4.9 (4.7) | 6.6 (6.5) | 7.2 (7.2) |
| Completeness (%) | 96.0 (98.1) | 98.4 (93.9) | 100 (100) | 99.7 (100) |
| Refinement | ||||
| Resolution (Å) | 37.45–1.80 | 30.00–1.60 | 38.75–1.80 | 37.40–2.20 |
| Rwork (%)c | 16.0 | 13.7 | 14.9 | 19.9 |
| Rfree (%)d | 17.8 | 17.0 | 16.9 | 23.8 |
| RMSDs | ||||
| Bond lengths (Å) | 0.006 | 0.007 | 0.007 | 0.006 |
| Bond angles (°) | 1.199 | 1.231 | 1.219 | 1.245 |
| Ramachandran plot Quality | ||||
| Favored (%) | 96.5 | 96.6 | 96.5 | 95.9 |
| Allowed (%) | 3.5 | 3.4 | 3.4 | 4.1 |
| Outlier (%) | 0 | 0 | 0 | 0 |
aValue in parentheses refer to the highest resolution shell of data.
bRmerge = Σhkl |I-<I>|/ΣhklI where I is the intensity of unique relfection hkl and <I> is the average over symmetry-related observations of unique reflection hkl.
cRwork = Σ|Fobs - Fcalc|/ΣFobs where Fobs and Fcalc are the observed and calculated structure factors, respectively.
dRfree is calculated as for Rcryst but using 5% of reflections sequestered before refinement.
Molecular dynamics simulations
Five simulation systems were built up and designated as OST-NH3-H150-Pos, OST-NH2-H150-Pos, OST-NH2-H150-Neu, ZMR-H150-Pos, and FREE-H150-Neu (Table 1). Atomic coordinates of systems OST-NH3-H150-Pos, OST-NH2-H150-Pos, and OST-NH2-H150-Neu were taken from the crystal structure of the N2-oseltamivir closed form complex; FREE-H150-Neu were taken from the crystal structure of the free N2 and ZMR-H150-Pos were taken from the crystal structure of the N2-zanamivir complex (PDB 3TIC)27. NA tetramers were used in each system. Protonation states of H150 and states of ligand were assigned as shown in Table 1. Water molecules and calcium ions in crystal structures were retained.
RESP43 charge of OST and ZMR were generated using antechamber44 after calculation by Gaussian0945 (HF/6-311G**). Force fields for OST and ZMR were generated based on general AMBER force field in amber package43. Then the system was setup using AmberTools. Amber ff99SB force field was applied to proteins46. The calcium was parameterized in the classical force field. TIP3P waters were added to solvate each system with a boundary of 8 angstroms away from any protein or ligand atoms47. Counterions were added to neutralize each system. The system details can be found in the Supplementary information (Supplementary Table 1). Then the topology of amber format was transformed to GROMACS readable format.
Simulations were performed using the GROMACS 4.5.3 package48. To alleviate any steric clashes, the initial structures were minimized using the steepest descents method with a force tolerance of 10 kJ mol−1 nm−1. Non-hydrogen protein atoms, main chain atoms and protein α-carbon atoms were progressively restrained with a force constant of 1000 kJ mol−1 nm−1. After energy minimization, the systems were heated to 310 K with position restraints on non-water atoms. The temperature was controlled using the V-rescale method with a coupling time of τT = 0.1 ps and the pressure was maintained at 1 bar using Berendsen barostat with τp = 0.1 ps and a compressibility of 4.5 × 10−5 bar. Long range electrostatic interactions were calculated using the particle-mesh Ewad (PME) algorithm49,50. The LINCS51 algorithm was used to constrain hydrogen bond lengths. Further three 500 ps long equilibration runs were position restrained during the whole equilibration process. Finally, 200 ns MD production runs were performed for each of the five tetramer systems. The time step for integration was set to 2 fs. The coordinates of the systems were saved every 10 ps.
Population analysis based on 150-cavity
Conformations in each chain were extracted by an interval of 100 ps from the trajectories for 150-cavity volume calculation. The volume of 150-cavity was measured using the pocket volume measuring algorithm POVME52. The volume inclusion sphere that defines the 150 cavity was a single sphere that encompassed the 150-cavity in the widely investigated group 1 neuraminidase (PDB 2HU0)20. Each conformation extracted was aligned to 2HU0, and POVME algorithm neglected the volume occupied by atoms within the 150-cavity sphere.
Hydrogen bond statistics and RMSD calculation
VMD53 was used to calculate the hydrogen bonds between two groups or residues. Hydrogen bond distance cutoff was set to 3.5 Å and angle was set to 20 degrees. VMD was also used to calculate the RMSD of 150-loop in crystal structures. All α-carbon atoms were superimposed at first, and then the RMSD of α-carbon atoms in the 150-loop was calculated. Only Cα atoms were used to calculate RMSD values.
Author Contributions
G.F.G. conceived, designed and supervised the project. Y.W. and Y.L. carried out the protein preparation and crystallization. G.Q. and K.Y. did the dynamics simulation. J.Q. collected the data and F.G. solved the structures. Y.W., G.Q., Y.L., K.Y., C.V., H.J. and G.F.G. analyzed the data. Y.W., G.Q., H.J. and G.F.G. wrote the paper.
Supplementary Material
Dataset 1
Acknowledgments
This work is supported by grants from China Ministry of Science and Technology (MOST) 973 Project (Grant No. 2011CB504703 and 2009CB918502) , the National High Technology Research and Development Program of China (863 Program) (No. 2012AA020301 and 2012AA01A305), the China National Grand S&T Special Project (2013ZX10004611) and the National Natural Science Foundation of China (Grant No. 21021063, 81230076 and 21210003). GFG is a leading principal investigator of the Innovative Research Group of the National Natural Science Foundation of China (NSFC, Grant No. 81021003). Assistance by the staff at the Shanghai Synchrotron Radiation Facility (SSRF-beamline 17U) is acknowledged. We thank Yanfang Zhang for help with protein preparation. C.J.V. is a recipient of the Chinese Academy of Sciences Fellowship for Young International Scientists (2011Y2SA01) and the NSFC Research Fund for Young International Scientists (31150110147). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- Cox N. J. & Subbarao K. Global epidemiology of influenza: past and present. Annu Rev Med 51, 407–21 (2000). [DOI] [PubMed] [Google Scholar]
- Guan Y. et al. The emergence of pandemic influenza viruses. Protein Cell 1, 9–13 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y. et al. In silico characterization of the functional and structural modules of the hemagglutinin protein from the swine-origin influenza virus A (H1N1)-2009. Sci China Life Sci 53, 633–42 (2010). [DOI] [PubMed] [Google Scholar]
- Liu D., Liu X., Yan J., Liu W. J. & Gao G. F. Interspecies transmission and host restriction of avian H5N1 influenza virus. Sci China C Life Sci 52, 428–38 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G. F. & Sun Y. It is not just AIV: from avian to swine-origin influenza virus. Sci China Life Sci 53, 151–3 (2010). [DOI] [PubMed] [Google Scholar]
- Update: Influenza A (H3N2)v transmission and guidelines - five states, 2011. MMWR Morb Mortal Wkly Rep 60, 1741–4 (2012). [PubMed] [Google Scholar]
- Swine-origin influenza A (H3N2) virus infection in two children--Indiana and Pennsylvania, July-August 2011. MMWR Morb Mortal Wkly Rep 60, 1213–5 (2011). [PubMed] [Google Scholar]
- Limited human-to-human transmission of novel influenza A (H3N2) virus--Iowa, November 2011. MMWR Morb Mortal Wkly Rep 60, 1615–7 (2011). [PubMed] [Google Scholar]
- World Health Organization Summary of status of development and availability of variant influenza A (H3N2) candidate vaccine viruses. (2012 Mar 8).
- Liu C., Eichelberger M. C., Compans R. W. & Air G. M. Influenza type A virus neuraminidase does not play a role in viral entry, replication, assembly, or budding. J Virol 69, 1099–106 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palese P., Tobita K., Ueda M. & Compans R. W. Characterization of temperature sensitive influenza virus mutants defective in neuraminidase. Virology 61, 397–410 (1974). [DOI] [PubMed] [Google Scholar]
- Skehel J. J. & Wiley D. C. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem 69, 531–69 (2000). [DOI] [PubMed] [Google Scholar]
- Takemoto D. K., Skehel J. J. & Wiley D. C. A surface plasmon resonance assay for the binding of influenza virus hemagglutinin to its sialic acid receptor. Virology 217, 452–8 (1996). [DOI] [PubMed] [Google Scholar]
- von Itzstein M. The war against influenza: discovery and development of sialidase inhibitors. Nat Rev Drug Discov 6, 967–74 (2007). [DOI] [PubMed] [Google Scholar]
- Colman P. M. Neuraminidase: enzyme and antigen. In: The influenza viruses (ed. , ed. ) 175–210 (Plenum Press, New York, 1989). [Google Scholar]
- Colman P. M. New antivirals and drug resistance. Annu Rev Biochem 78, 95–118 (2009). [DOI] [PubMed] [Google Scholar]
- A revision of the system of nomenclature for influenza viruses: a WHO memorandum. Bull World Health Organ 58, 585–91 (1980). [PMC free article] [PubMed] [Google Scholar]
- Li Q. et al. Structural and functional characterization of neuraminidase-like molecule N10 derived from bat influenza A virus. Proc Natl Acad Sci U S A (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X. et al. Crystal structures of two subtype N10 neuraminidase-like proteins from bat influenza A viruses reveal a diverged putative active site. Proc Natl Acad Sci U S A (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell R. J. et al. The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design. Nature 443, 45–9 (2006). [DOI] [PubMed] [Google Scholar]
- Amaro R. E. et al. Mechanism of 150-cavity formation in influenza neuraminidase. Nat Commun 2, 388 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudrawar S. et al. Novel sialic acid derivatives lock open the 150-loop of an influenza A virus group-1 sialidase. Nat Commun 1, 113 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M. et al. Influenza A virus N5 neuraminidase has an extended 150-cavity. J Virol 85, 8431–5 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varghese J. N. & Colman P. M. Three-dimensional structure of the neuraminidase of influenza virus A/Tokyo/3/67 at 2.2 A resolution. J Mol Biol 221, 473–86 (1991). [DOI] [PubMed] [Google Scholar]
- Li Q. et al. The 2009 pandemic H1N1 neuraminidase N1 lacks the 150-cavity in its active site. Nat Struct Mol Biol 17, 1266–8 (2010). [DOI] [PubMed] [Google Scholar]
- Amaro R. E. et al. Remarkable loop flexibility in avian influenza N1 and its implications for antiviral drug design. J Am Chem Soc 129, 7764–5 (2007). [DOI] [PubMed] [Google Scholar]
- Vavricka C. J. et al. Structural and functional analysis of laninamivir and its octanoate prodrug reveals group specific mechanisms for influenza NA inhibition. PLoS Pathog 7, e1002249 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins P. J. et al. Crystal structures of oseltamivir-resistant influenza virus neuraminidase mutants. Nature 453, 1258–61 (2008). [DOI] [PubMed] [Google Scholar]
- Xu X., Zhu X., Dwek R. A., Stevens J. & Wilson I. A. Structural characterization of the 1918 influenza virus H1N1 neuraminidase. J Virol 82, 10493–501 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubareva L. V., Webster R. G. & Hayden F. G. Comparison of the activities of zanamivir, oseltamivir, and RWJ-270201 against clinical isolates of influenza virus and neuraminidase inhibitor-resistant variants. Antimicrob Agents Chemother 45, 3403–8 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetherall N. T. et al. Evaluation of neuraminidase enzyme assays using different substrates to measure susceptibility of influenza virus clinical isolates to neuraminidase inhibitors: report of the neuraminidase inhibitor susceptibility network. J Clin Microbiol 41, 742–50 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörwald F. Z. Lead Optimization for Medicinal Chemists, (Wiley-VCH, 2012). [Google Scholar]
- Anandakrishnan R., Aguilar B. & Onufriev A. V. H++ 3.0: automating pK prediction and the preparation of biomolecular structures for atomistic molecular modeling and simulations. Nucleic Acids Res 40, W537–41 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods C. J. et al. Long time scale GPU dynamics reveal the mechanism of drug resistance of the dual mutant I223R/H275Y neuraminidase from H1N1-2009 influenza virus. Biochemistry 51, 4364–75 (2012). [DOI] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. & Charles W. Carter, Jr. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- Jedrzejas M. J. et al. Structures of aromatic inhibitors of influenza virus neuraminidase. Biochemistry 34, 3144–51 (1995). [DOI] [PubMed] [Google Scholar]
- Read R. J. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr D Biol Crystallogr 57, 1373–82 (2001). [DOI] [PubMed] [Google Scholar]
- The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50, 760–3 (1994). [DOI] [PubMed] [Google Scholar]
- Emsley P. & Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, 2126–32 (2004). [DOI] [PubMed] [Google Scholar]
- Murshudov G. N., Vagin A. A. & Dodson E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53, 240–55 (1997). [DOI] [PubMed] [Google Scholar]
- Adams P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R. M. M., Moss D. & Thornton J. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Cryst 26, 283–291 (1993). [Google Scholar]
- Wang J., Wolf R. M., Caldwell J. W., Kollman P. A. & Case D. A. Development and testing of a general amber force field. J Comput Chem 25, 1157–74 (2004). [DOI] [PubMed] [Google Scholar]
- Wang J., Wang W., Kollman P. A. & Case D. A. Automatic atom type and bond type perception in molecular mechanical calculations. J Mol Graph Model 25, 247–60 (2006). [DOI] [PubMed] [Google Scholar]
- Frisch M. J. et al. Gaussian 09, Revision A.1. (Gaussian, Inc.Wallingford CT, 2009).
- Hornak V. et al. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 65, 712–25 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W. & Klein M. L. Comparison of Simple Potential Functions for Simulating Liquid Water. Journal of Chemical Physics 79, 926–935 (1983). [Google Scholar]
- Hess B., Kutzner C., van der Spoel D. & Lindahl E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. Journal of Chemical Theory and Computation 4, 435–447 (2008). [DOI] [PubMed] [Google Scholar]
- Darden T., York D. & Pedersen L. Particle Mesh Ewald - an N. Log(N) Method for Ewald Sums in Large Systems. Journal of Chemical Physics 98, 10089–10092 (1993). [Google Scholar]
- Essmann U. et al. A smooth particle mesh Ewald method. Journal of Chemical Physics 103, 8577–93 (1995). [Google Scholar]
- Hess B., Bekker H., Berendsen H. J. C. & Fraaije J. G. E. M. LINCS: a linear constraint solver for molecular simulations. Journal of Computational Chemistry 18, 1463–1472 (1997). [Google Scholar]
- Durrant J. D., de Oliveira C. A. & McCammon J. A. POVME: an algorithm for measuring binding-pocket volumes. J Mol Graph Model 29, 773–6 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W., Dalke A. & Schulten K. VMD: visual molecular dynamics. J Mol Graph 14, 33–8, 27–8 (1996). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dataset 1