Abstract
There is growing interest in identifying regulators of autophagy. The molecular mechanism underlying transforming growth factor-β activated kinase 1 (TAK1)-induced autophagy is poorly understood. We found that TAK1 inhibits p70 S6 kinase1 (S6K1) phosphorylation by interfering interaction of raptor with S6K1, thus inducing autophagy. The factors that determine whether autophagy is cytoprotective or cytotoxic have not been fully elucidated. In Drosophila, TAK1 overexpression leads to an impaired eye phenotype despite inhibition of apoptosis, indicating that the phenotype was mainly due to autophagy. Also, TAK1 overexpression increases lactate dehydrogenase (LDH) level in mammalian cells. When treated with autophagy inhibitors, the level of TAK1-induced cytotoxicity or cell death was significantly attenuated, indicating that TAK1 induces cytotoxic autophagic cell death. This study provides the first in vitro and in vivo evidence of TAK1-induced autophagy and we believe that our findings significantly contribute to the understanding of the mechanisms underlying the induction of autophagy.
Autophagy is an evolutionarily conserved mechanism that degrades long-lived proteins and cytoplasmic organelles. During autophagy, portions of the cytoplasm are sequestered by double-membrane structures called autophagosomes. These autophagosomes eventually fuse with lysosomes to form autolysosomes, where degradation occurs. Microtubule-associated protein 1 light chain 3 (LC3) is initially synthesized in an unprocessed form, proLC3, which is converted into a proteolytically processed form, LC3-I, and finally modified into the phosphatidylethanolamine (PE)-conjugated membrane-bound form, LC3-II, and recruited to the autophagosome1.
Autophagy has multiple cellular roles, participating in both cell survival and cell death2,3,4,5,6. When cells lack essential nutrients, autophagic pathways are activated to supply nutritional components. However, under certain circumstances, autophagy can also be a cell death mechanism, called autophagic cell death. Autophagy has diverse biological roles in the regulation of processes such as aging, development, and tumorigenesis7. Tumorigenesis has been linked with decreased autophagy. Autophagy is negatively regulated by PI3K, Akt, and mTOR, which are often activated in cancer cells2. Autophagy inducers or executors such as phosphatase and tensin homolog (PTEN), tuberous sclerosis 1 (TSC1), tuberous sclerosis 2 (TSC2), autophagy-specific gene 4 (Atg4), and beclin 1 are known to be potent tumor suppressors8. Hence, autophagy may be a tumor-suppression mechanism, and the identification of regulators of autophagy is needed.
Drosophila is an attractive model system for studying the molecular mechanisms of autophagy due to short generation times and relatively simple screening methods. In our previous study, genes regulating cell death in Drosophila were identified through modifier screening of death caspase 1 (DCP1)9. As a result, transforming growth factor-β activated kinase 1 (TAK1) was selected as a candidate for inducing cell death.
TAK1 is a serine/threonine kinase in the mitogen-activated protein kinase kinase kinase (MAPKKK) family10,11. TAK1 is a key regulator in the cascades of cellular responses and its activity is regulated by various cytokines including interleukin-1 (IL-1), transforming growth factor-β (TGF-β) and by toll-like receptors (TLR), CD40 and B cell receptors. Once activated, TAK1 will in turn, activate crucial intra-cellular kinases; the p38 MAPK, c-jun N-terminal kinase (JNK) and I-kappa B kinase complex (IKK). p38 MAPK and JNK control the transcription factors activator protein-1 (AP-1), while nuclear factor-kappa B (NF-kB) is activated by IKK. TAK1 regulates cell survival, differentiation and inflammatory responses via a number of specific transcription factors. Recently, TAK1 has also been implicated in activation of the tumor suppressor proteins, the LKB1 and pVHL12,13,14. TAK1 plays a role in regulating apoptosis. TAK1 promotes or inhibits apoptosis in various types of cells and tissues15,16. However, the role of TAK1 in autophagy has not been completely defined.
Target of rapamycin (TOR) is a highly conserved kinase that exists in two functional complexes, TOR complex 1 (TORC1) and TOR complex 2 (TORC2), that are conserved from yeast to mammals. Mammalian TORC1 (mTORC1) has a primary function in autophagy regulation and contains the regulatory-associated protein of mTOR (raptor), GβL and PRAS40. Raptor is a 150 kDa mTOR-binding protein that also binds S6K1, serves as a scaffold protein of mTOR and facilitates mTOR phosphorylation of S6K117,18,19. It is well known that TOR has a central role in autophagy regulation, and p70 S6 kinase 1 (S6K1) is a direct substrate of TOR20,21,22. S6K1 has been implicated as an important positive regulator of biological processes, such as cell growth, proliferation, and protein synthesis23,24. Previous reports have suggested that S6K1 negatively regulates autophagy25,26. S6K1 has dual functions in autophagy regulation. Phosphorylation of S6K1 is critical for its function and most closely correlates with kinase activity in vivo27,28.
In detail, S6K1 plays a negative role for autophagy in normal conditions. When starvation induces autophagy, S6K1 may act as a positive regulator of autophagy29,30.
Using a combination of several methods, including generation of transgenic flies, we report that TAK1 is a novel regulator of autophagic cell death. To elucidate the TAK1-induced autophagy pathway, we examined the interactions among TAK1, S6K1 and raptor. We provide the first evidence that TAK1 negatively regulates S6K1, thereby inducing cytotoxic autophagic cell death in both mammalian cells and Drosophila.
Results
TAK1 induces autophagy in vitro and in vivo
In order to identify genes regulating cell death in Drosophila, we screened 15,000 enhancer-promoter (EP) lines and identified 72 DCP1-interacting genes. DCP1 overexpression aggravated the adult eye phenotype, but the co-expression of Drosophila TAK1 (one of the 72 DCP1 interacting genes) and DCP1 showed lethality. Therefore, we raised a question if TAK1 contributes to cell death. The mechanisms that can lead to cell death are apoptosis, necrosis, and autophagic cell death. Among these mechanisms, we examined the role of TAK1 in the regulation of autophagy.
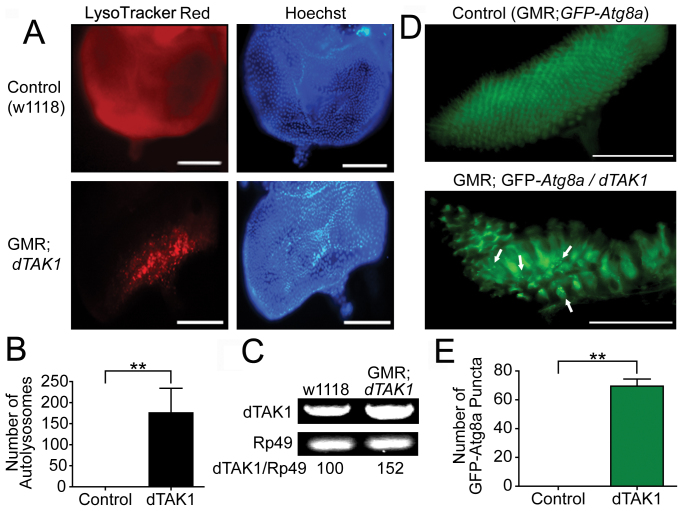
To test whether TAK1 overexpression induces autophagy in vivo, we designed Drosophila model experiments using transgenic lines with an eye-specific glass multimer reporter-GAL4/upstream activation sequence (GMR-GAL4/UAS) system. To investigate the ability of TAK1 to induce autophagy, we quantified the formation of autolysosomes by staining with LysoTracker Red, which is an effective marker of autolysosomes. In the third instar larvae of dTAK1-overexpressing flies, LysoTracker Red-positive puncta were observed in the developing imaginal eye disc posterior to the morphogenetic furrow. The number of autolysosomes in GMR; dTAK1 flies was significantly increased compared with the number of autolysosomes in the eye discs of control flies (Fig. 1A, B).
Figure 1. dTAK1 overexpression induces autophagy in Drosophila eye discs.
(A) Third-instar larval eye discs were stained with 1 μM LysoTracker Red. Hoechst staining of nuclei is shown in blue. Scale bars, 100 μm. (B) Quantification of the number of LysoTracker Red-positive punctate structures. The values represent the means ± s.d. of three independent experiments. * P < 0.05, ** P < 0.01, and *** P < 0.001. (C) The transcript levels of dTAK1 in the third-instar larvae were analyzed by RT-PCR. Rp49 was used as an internal control. (D) The punctate structures containing GFP-Atg8a were used as markers of autophagosome formation in the eye discs of third-instar larvae. (E) Quantitative analysis of GFP-Atg8a puncta. The values represent means ± s.d. of three independent experiments. * P < 0.05, ** P < 0.01, and *** P < 0.001.
The efficient overexpression of dTAK1 in the GMR; dTAK1 flies was confirmed by reverse transcription-polymerase chain reaction (RT-PCR) (Fig. 1C). The detection of green fluorescence protein-tagged autophagy-specific gene 8a (GFP-Atg8a), a homolog of GFP-tagged microtubule-associated protein 1 light chain 3 (GFP-LC3), using fluorescence microscopy is one of the most useful methods for monitoring autophagic activity in Drosophila. The overexpression of dTAK1 in the developing eye discs resulted in a significant accumulation of GFP-Atg8a punctate structures (which represent autophagosomes). In contrast, no punctate structures were detected in control eye discs (Fig. 1D, E).
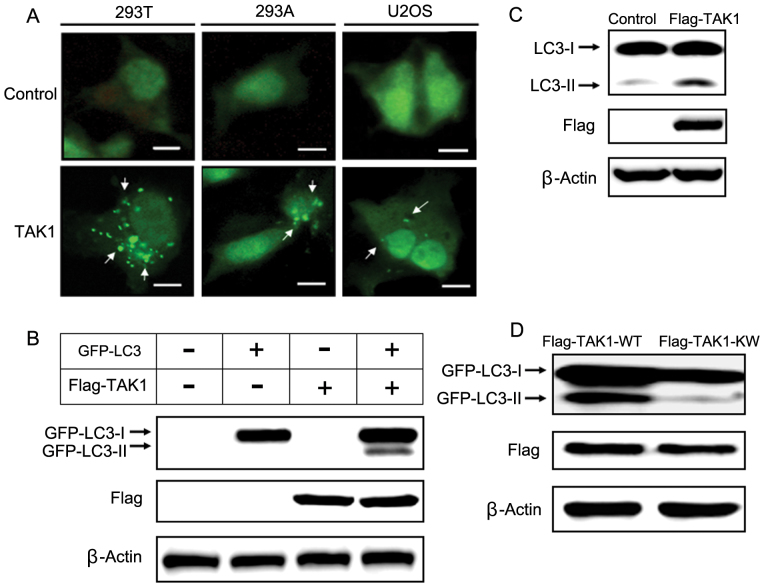
To investigate whether TAK1 induces autophagy in vitro, we co-transfected GFP-LC3 with TAK1 or a control vector in several mammalian cell lines and examined the accumulation of GFP-LC3 punctate structures using fluorescence microscopy. During autophagy, LC3-II relocalizes to the autophagosomal membranes. Thus, the accumulation of GFP-LC3 puncta provides an effective way to detect autophagosomes. In control cells, GFP-LC3 punctate structures were not detected, but GFP-LC3 punctate structures were markedly increased in cells overexpressing TAK1 (Fig. 2A).
Figure 2. TAK1 overexpression induces autophagy in several cell types.
(A) GFP-LC3 was co-transfected with a control plasmid or a TAK1 plasmid. After transfection, the cells were incubated for 48 h, and GFP-LC3 puncta were then detected using fluorescence microscopy. Scale bars, 10 μm. (B) Cells were co-transfected with GFP-LC3 along with the indicated constructs for 48 h and then harvested for immunoblot analysis. β-actin was used as a loading control. GFP-LC3-II was detected as the faster migrating form of LC3 by immunoblot analysis on a 12% SDS-PAGE gel. The expression of TAK1, which was Flag-tagged, was measured using an anti-Flag antibody. (C) Cells were transfected with a control plasmid or a TAK1 plasmid for 48 h and then harvested for immunoblot analysis. Endogenous LC3 processing was evaluated by immunoblotting. (D) HEK 293T cells were co-transfected with GFP-LC3 and either TAK1-WT or TAK1-KW plasmid for 48 h. After transfection, cells were harvested and analyzed by immunoblottng.
GFP-LC3-II can be detected by immunoblotting with antibodies against GFP, and the LC3-II level correlates with the number of autophagosomes. The LC3-II level was increased markedly in TAK1-overexpressing cells compared with controls (Fig. 2B). In addition to exogenous GFP-LC3-II, we examined the level of endogenous autophagy with an anti-LC3 antibody. TAK1-overexpressing cells showed higher LC3-II levels compared with control cells (Fig. 2C). Flag-tagged wild type TAK1 (TAK1-WT) and kinase inactive TAK1 (TAK1-KW) were used in immunoblot analyses. The LC3-II level was decreased markedly in TAK1-KW overexpressing cells when compared with that of TAK1-WT overexpressing cells (Fig. 2D).
Taken together, our study has demonstrated that TAK1 can induce autophagy in line with the recent report which mentioned that dominant-negative TAK1 or knockdown of TAK1 inhibits autophagy31. Consistent with our in vivo data (see Figure 1), the overexpression of TAK1 could induce autophagy in vitro.
Autophagy-related proteins are involved in TAK1-induced autophagy
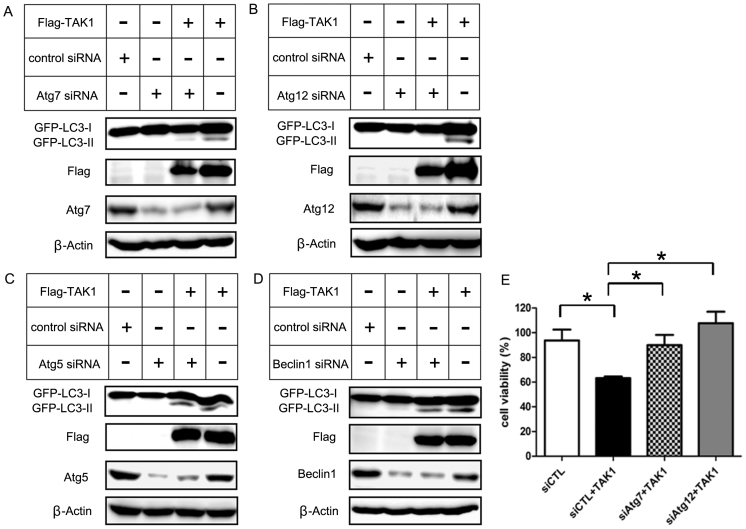
To further examine whether several autophagy-related proteins were involved in TAK1-induced autophagy, we transfected siRNAs against several Atg proteins (Atg5, Atg7, Atg12, and beclin 1) into HEK 293T cells and determined their influence on TAK1-induced autophagy. We assessed the alteration of autophagic activity by Atg proteins using GFP-LC3 co-transfection. In the case of Atg7 or Atg12 knockdown in combination with TAK1 overexpression, GFP-LC3-II was decreased significantly compared with the TAK1 overexpression alone condition (Fig. 3A, B). In contrast, the GFP-LC3-II conversion induced by TAK1 overexpression was only slightly inhibited by Atg5 or beclin 1 siRNA (Fig. 3C, D). In addition, we examined the influence of Atg7 and Atg12 on TAK1-induced cell death using trypan blue exclusion assays. Transfection with siRNA against Atg7 or Atg12 significantly diminished the cytotoxic effect of TAK1 relative to cells overexpressing TAK1 alone (Fig. 3E). Interestingly, the GFP-LC3-II level was inversely proportional to cell viability. Atg12 is activated by Atg7 and is involved in the elongation of isolation membranes. Perhaps, in TAK1-induced autophagy, Atg7- and Atg12-related steps contribute to autophagic cell death and TAK1 induces cytotoxic autophagic cell death through regulation of Atg7 and Atg12. Therefore, knockdown of Atg7 and Atg12 reduced GFP-LC3 II level and restored cell viability. Similarly, Funasaka et al. reported that Tpr depletion inhibits cell growth and promotes autophagy32. They also examined the effects of Tpr depletion on several autophagic factors (LC3, Beclin1, Atg3, Atg5, Atg7 and Atg12). These authors reported that Atg7 and Atg12 are involved in Tpr depletion-induced autophagy.
Figure 3. Autophagy-related proteins are involved in TAK1-induced GFP-LC3-II conversion and cell death.
(A–D) HEK 293T cells were transfected with 100 nM of the indicated siRNAs for 16 h and transfected with TAK1 for another 24 h. The expression levels of TAK1, GFP-LC3-I, GFP-LC3-II, Atg5, Atg7, Atg12, beclin1 and β-actin were analyzed by immunoblotting. (E) Cell viability of HEK 293T cells transfected with indicating siRNAs and TAK1 was measured by trypan blue assay. The values represent the means ± s.d. of three independent experiments. * P < 0.05.
However, knockdown of Atg5 or Beclin1 did not significantly change in GFP-LC3 II levels and cell viability (Supplementary Fig. 1). Similarly, Chang et al. reported that siRNA for Beclin1 and Atg5 minimally affected the LC3 II conversion or cell viability in ConA-induced autophagy33. ConA (concanavalin A) induces cytotoxic autophagic cell death. These findings indicate that Atg7 and Atg12 might be involved in the regulation of TAK1-induced autophagy. We examined the involvement of Atg7 and Atg12 in TAK1-induced autophagy briefly in our study, but it needs to be further investigated.
S6K1 is involved in TAK1-induced autophagy
The pathways that regulate autophagy are evolutionarily conserved. Therefore, we crossed fly lines containing some autophagy regulators (PI3K, Akt, dTOR and dS6K) with dTAK1 to examine the eye phenotype changes (data not shown). Among these, dS6K showed significant phenotype change. Then, we focused on S6K1 to elucidate its involvement in TAK1-induced autophagy. It was reported that autophagy is inhibited by S6K1 in mammalian cells, and the phosphorylation of S6K1 coincides with the inhibition of autophagy25,26,34.
Strictly speaking, S6K1 has an inhibiting effect on autophagy under normal nutritional conditions unlike starvation conditions. In addition to the various autophagy detection methods described previously, we performed FACS analysis to show TAK1-induced autophagic flux (Supplementary Fig. 2). A new method using FACS to allow quantitative analysis of GFP-LC3 turnover has been reported. Through FACS analysis, autophagic flux can be quantified by measuring the delivery of GFP-LC3 into lysosomes.
The disappearance of GFP-LC3 is detectable in cells using FACS. As LC3 is delivered into the lysosomes during autophagy, the disappearance of total GFP-LC3 is a good indicator of autophagic activity35. The GFP-LC3 level was examined in human embryonic kidney (HEK) 293 T cells. The GFP-LC3 level of TAK1 overexpressing cells was reduced compared to mock vector-transfected cells. This reduction was due to the induction of autophagy, indicating that autophagy was induced by TAK1 overexpression. On the other hand, S6K1 overexpression increased GFP levels.
We showed TAK1-induced autophagy in normal culture conditions. In addition, we tested whether TAK1 can induce autophagy in rapamycin-induced or starvation-induced autophagy. In these conditions, TAK1 also promotes autophagy (Supplementary Fig. 3).
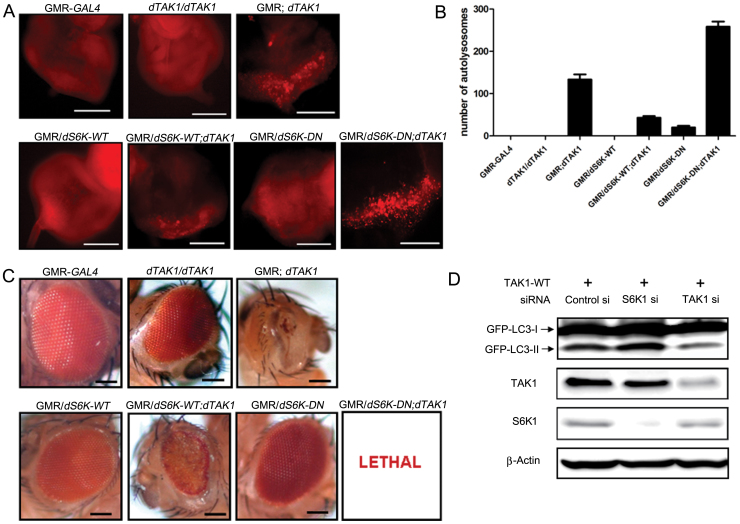
We generated transgenic flies that expressed dTAK1 on several genetic backgrounds to investigate the interaction between S6K1 and TAK1 in autophagy. We compared the abilities of these mutants to induce autophagy using LysoTracker Red staining. As expected, the number of LysoTracker Red-positive puncta was markedly increased in GMR; dTAK1 flies compared with the control files (GMR-GAL4 and dTAK1/dTAK1). In GMR/dS6K-WT flies, few autolysosomes were detected. However, when co-expressed with dTAK1, the number of autolysosomes was increased. In GMR/dS6K-DN flies, a few autolysosomes were detected. Whereas, the number of autolysosomes in the eye discs of flies expressing both Drosophila dominant-negative S6K (dS6K-DN) and dTAK1 was greatly increased (Fig. 4A). The overexpression of dTAK1 alone was sufficient to induce a moderate level of autophagy. However, dTAK1 overexpression in combination with dS6K-WT or dS6K-DN resulted in a significant decrease or increase, respectively, in the number of autolysosomes (Fig. 4B). The use of a GMR-GAL4 driver to overexpress dTAK1 in the developing eye caused a lack of pigmentation and a rough, impaired appearance compared with control flies. The adult eye phenotype of dS6K-WT or dS6K-DN alone was similar to eyes of control flies. Somewhat aggravated eye phenotypes were observed in GMR/dS6K-WT; dTAK1 flies compared with those of GMR/dS6K-WT flies. When co-expressed with dTAK1, pupal lethality was observed on the dS6K-DN background (Fig. 4C). Additionally, the severity of the adult eye phenotype of each fly genotype was correlated with the autolysosome number.
Figure 4. S6K1 is involved in TAK1-induced autophagy in vivo and in vitro.
(A) Eye discs of flies of the indicated genotypes were stained with 1 μM LysoTracker Red to detect autolysosomes. Autolysosomes were visualized as LysoTracker Red-positive punctate structures. Scale bars, 100 μm. (B) The numbers of autolysosomes present in flies of each genotype were quantified. The values represent the means ± s.d. of three independent experiments. (C) Ectopic expression of the indicated genes in Drosophila eyes was induced using the eye-specific driver GMR-GAL4. The adult eye phenotypes of the flies of the indicated genotypes were photographed. Scale bars, 0.1 mm. (D) HEK 293T cells were transfected with TAK1-WT for 24 h and then transfected with the indicated siRNAs for 48 h. Cell lysates were then harvested and analyzed by immunoblotting using the indicated antibodies. A GFP antibody was used to measure the levels of LC3-II, which are correlated with the number of autophagosomes.
To elucidate the role of S6K1 in TAK1-induced autophagy, the effect of S6K1 depletion by gene silencing was examined in TAK1-overexpressed HEK 293T cells. The S6K1 silencing increased GFP-LC3-II level compared with that of TAK1 overexpression alone. However, upon TAK1 silencing, we observed a reduction in GFP-LC3-II, indicating that the autophagic activity was reduced (Fig. 4D). In addition to exogenous GFP-LC3-II levels, TAK1 knockdown decreased endogenous LC3-II levels compared to mock vector-transfected cells. TAK1 overexpression reduced the phosphorylation of S6K1, and the phosphorylation level was somewhat rescued when TAK1 was silenced (Supplementary Fig. 4A).
IL1-β is a well-known TAK1 activator. Therefore, we examined S6K1 phosphorylation in IL1-β treated cells. The S6K1 phosphorylation pattern was similar to Supplementary Fig. 4A, but the phosphorylation level was weaker than normal conditions (Supplementary Fig. 4B, C). Perhaps, this weaker S6K1 phosphorylation is due to TAK1 activation. Activated TAK1 may suppress S6K1 phosphorylation more potently.
Taken together, our results indicate that S6K1 has an inhibitory effect on autophagic activity under normal nutritional conditions, consistent with the findings of several other reports25,26,34.
Phosphorylation of S6K1 correlates with the suppression of autophagy36. Thus, the suppression of S6K1 phosphorylation could promote autophagy. To test whether TAK1-induced autophagy was associated with the inhibition of S6K1 phosphorylation, we measured S6K1 phosphorylation and examined the interaction between TAK1 and S6K1.
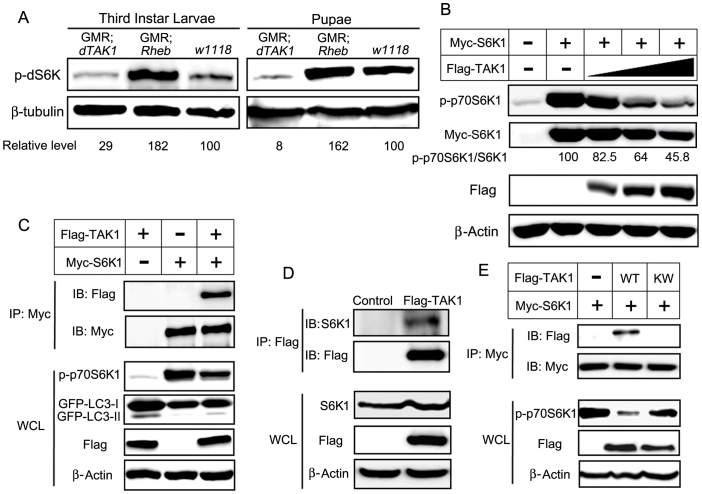
We tested the effects of dTAK1 overexpression on dS6K phosphorylation in Drosophila third-instar larvae and pupae. Immunoblot analyses showed that dS6K phosphorylation was markedly reduced in dTAK1-overexpressing flies. Rheb was used as a positive control37 and w1118 was used as a wild-type control. As expected, Rheb overexpression increased dS6K phosphorylation (Fig. 5A). The overexpression of TAK1 in HEK 293T cells decreased S6K1 phosphorylation in a dose-dependent manner and the quantification of p-p70S6K1/S6K1 level was presented (Fig. 5B).
Figure 5. TAK1 binds to S6K1 and suppresses the phosphorylation of S6K1.
(A) Phosphorylation of dS6K was detected by immunoblotting using a p-dS6K antibody in third-instar larvae and pupae of the indicated genotypes. Protein extracts from pupae and third-instar larvae were used. β-tubulin was used as a loading control, and Rheb-overexpressing flies were used as positive controls. dS6K phosphorylation in each genotype was quantified using Adobe Photoshop and normalized to β-tubulin. (B) Cells were transfected with the indicated constructs and incubated for 48 h. Cell lysates were then collected and immunoblotted with the indicated antibodies. S6K1 phosphorylation decreased in a TAK1 dose-dependent manner. (C) HEK 293T cells were transfected with the indicated constructs, and 48 h after transfection, a Myc immunoprecipitate was obtained and analyzed for the presence of an S6K1-TAK1 complex using a Flag antibody. The blots in the lower four panels were derived from the same cell lysates using the indicated antibodies. (D) Cells were transfected with the denoted constructs, and 48 h after transfection, S6K1-TAK1 complex was detected using an S6K1 antibody. The blots in the lower three panels were derived from the same cell lysates using the indicated antibodies. (E) Myc-S6K1 construct was co-transfected into cells with Flag-tagged TAK1-WT or TAK1-KW and incubated for 48 h. Lysates were collected and immunoblotted with the indicated antibodies.
Immunoprecipitation experiments demonstrated that TAK1 binds to S6K1 (Fig. 5C). Interestingly, GFP-LC3-II level showed an inverse proportion to S6K1 phosphorylation level. TAK1 alone increased the LC3-II level in accordance with Figure 2B. In contrast, S6K1 alone decreased the LC3-II level. When S6K1 was co-expressed with TAK1, S6K1 phosphorylation was decreased, but the LC3-II level was increased compared with that of S6K1 expression alone (Fig. 5C). To identify which part of S6K1 is involved in binding to TAK1, we examined several S6K1 mutants. As a result, the S6K1 C-terminal deletion mutant (Myc-S6K1 delta CT) exhibited significantly reduced binding to TAK1 (Supplementary Fig. 5). Therefore, we propose that the S6K1 C-terminal region is involved in TAK1-S6K1 binding.
These data indicated that S6K1 phosphorylation level is related with autophagy inhibition. Next, we examined whether TAK1 could interact with endogenous S6K1. However, in HEK 293T cells, we could not co-precipitate the endogenous TAK1 and S6K1. We tested the endogenous interaction in NIH 3T3 cells, but we couldn't observe the immune complex, either (data not shown).
Maybe, the commercially available antibodies have relatively low efficacy to detect endogenous interaction. Alternatively, we used Flag-tagged construct to detect endogenous interaction. We examined the interaction of the overexpressed Flag-TAK1 with endogenous S6K1. We found that endogenous interaction was shown after transient expression of Flag-TAK1 (Fig. 5D). In order to examine whether the kinase function of TAK1 is important for S6K1-TAK1 binding, we used TAK1 kinase inactive mutant (TAK1-KW). Importantly, the overexpression of TAK1-KW showed the remarkably reduced binding affinity and considerably restored phosphorylation level of S6K1 compared with TAK1-WT overexpression (Fig. 5E). Additionally, we evaluated the phosphorylation of S6K1 in myc-S6K1 IP. The phosphorylation pattern was similar to the S6K1 phosphorylation pattern in whole cell lysates (Supplementary Fig. 6).
We examined the effect of TAK1-KW on raptor-S6K1 binding. TAK1-WT interfered with the binding of raptor to S6K1, whereas TAK1-KW hardly had an influence on the raptor-S6K1 binding (Supplementary Fig. 7).
TAK1 competes with S6K1 for raptor binding
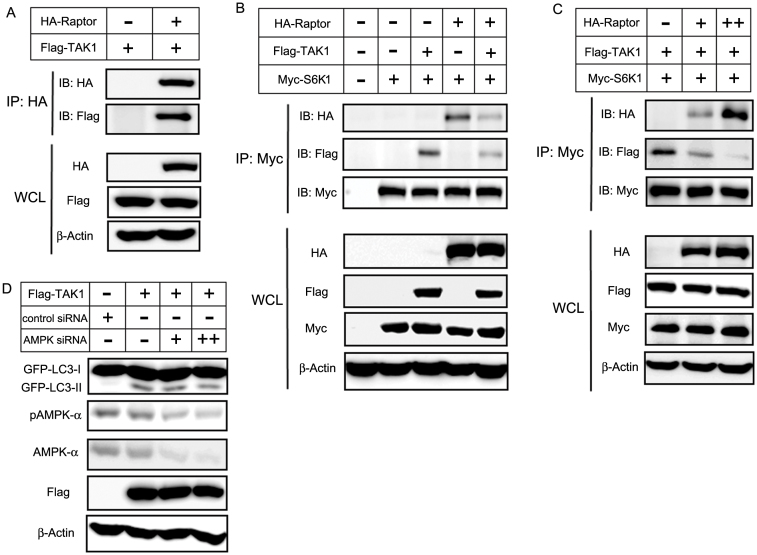
Autophagy is characterized by the inhibition of TORC1 signaling, but the regulation of TORC1 signaling in autophagy is not yet fully understood. Raptor binding to S6K1 is essential for phosphorylation of S6K119,38. Therefore, we targeted raptor to investigate its role in TAK1-induced autophagy. We observed TAK1-raptor interaction by immunoprecipitation (Fig. 6A). Interestingly, TAK1 co-expression resulted in a decrease in raptor-S6K1 binding. (Fig. 6B). Furthermore, TAK1-S6K1 binding decreased in a dose-dependent manner in response to increasing raptor levels. In contrast, raptor-S6K1 binding increased in a raptor dose-dependent manner (Fig. 6C). These results indicate that TAK1 may compete with S6K1 for raptor binding. Therefore, our results suggest that S6K1 and raptor are involved in TAK1-induced autophagy and that TAK1 interferes with the binding of S6K1 to raptor, thereby suppressing S6K1 phosphorylation and activation.
Figure 6. TAK1 competes with S6K1 for binding to raptor.
(A) HA-raptor and Flag-TAK1 were co-transfected into HEK 293T cells. An HA immunoprecipitate was obtained and analyzed for the presence of raptor-TAK1 complexes using a Flag antibody. The blots in the lower three panels were obtained from the same cell lysates using antibodies against Flag, HA or β-actin. (B) HEK 293T cells were transfected with the indicated genes, and 48 h after transfection, cell lysates were immunoprecipitated with an anti-Myc antibody, followed by immunoblotting with antibodies to HA, Flag or Myc. The blots in the lower four panels were obtained from the same cell lysates using the indicated antibodies. (C) Dose-dependent co-immunoprecipitations were performed in HEK 293T cells. Cells were transfected with the indicated constructs, along with increasing amounts of HA-raptor, and incubated for 48 h. Cell lysates were collected and immunoblotted with the indicated antibodies. (D) TAK1 induces AMPK-independent autophagy. HEK 293T cells were transfected with the indicated siRNA and TAK1. The expression of TAK1, GFP-LC3-I, GFP-LC3-II, pAMPK-α, AMPK-α and β-actin were analyzed by immunoblot analysis. (‘+' denotes 50 nM AMPK siRNA, and ‘++' denotes 100 nM AMPK siRNA.).
It was reported that TAK1 activates AMP-activated protein kinase (AMPK) to induce cytoprotective autophagy in TNF-related apoptosis-inducing ligand (TRAIL)-treated epithelial cells39. To study the role of AMPK in TAK1-induced autophagy, the expression of AMPK was downregulated using siRNA. We did not observe AMPK phosphorylation when TAK1 was overexpressed. Furthermore, AMPK down-regulation had little influence on GFP-LC3-II level which was induced by TAK1 overexpression (Fig. 6D). It is possible that wild-type TAK1 overexpression itself does not affect AMPK phosphorylation14. Another possibility is that TRAIL may influence other signals besides TAK1. Therefore, our results indicate that TAK1 can induce autophagy independent of AMPK phosphorylation.
TAK1 induces cytotoxic autophagic cell death
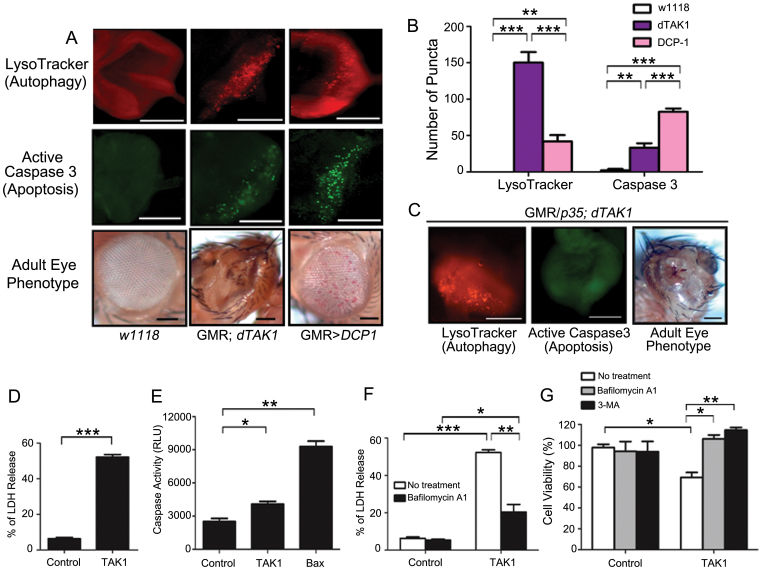
In our previous study, the co-expression of dTAK1 with DCP1 showed lethality9. Thus, we examined the effect of TAK1 and DCP1 on apoptosis and autophagy, respectively. Considering the disrupted eye phenotype of GMR; dTAK1 flies, we investigated whether this phenotype is due to autophagy or not. We used LysoTracker Red staining to detect autophagy and immunostaining with an active caspase 3 antibody to detect apoptosis. The number of autolysosomes in dTAK1-overexpressing flies was significantly higher than the number of autolysosomes in DCP1-overexpressing flies. When overexpressed, DCP1 induced a marked increase in the number of caspase 3-positive puncta compared with wild-type eye discs and dTAK1-overexpressing eye discs. In the GMR; dTAK1 eye discs, a relatively low number of caspase 3-positive puncta were observed compared with the eye discs of GMR > DCP1 flies (Fig. 7A, B). Despite suppression of apoptosis using p35 (apoptosis inhibitor), the rough, impaired adult eye phenotype was still observed in GMR/p35; dATK1 flies (similar to that of dTAK1-overexpressing flies), and there were many LysoTracker Red-positive puncta, i.e., autolysosomes (Fig. 7C). These suggest that TAK1-induced autophagy may contribute to cytotoxic effect, not cytoprotective function. To confirm the efficient expression of p35 in the transgenic flies, we used immunostaining with an active caspase 3 antibody to detect apoptosis. Few caspase 3-positive punctate structures were observed. Therefore, these results indicate that the dTAK1-induced impaired eye phenotype is mainly due to autophagy.
Figure 7. TAK1 induces cytotoxic autophagic cell death.
(A) Eye discs of third-instar larvae of the indicated genotypes were stained with LysoTracker Red to detect autophagic activity and with an active caspase-3 antibody to detect apoptosis. Adult eye phenotypes of the indicated genotypes were visualized. Scale bars indicate 100 μm. (B) The number of puncta in each immunofluorescent image was quantified. Bars indicate means ± s.d. of three independent experiments. (C) The apoptosis inhibitor p35 had little influence on the dTAK1-induced eye phenotype. Immunostaining of third-instar larval eye discs with 1 μM LysoTracker Red shows many punctate structures. The eye discs were stained with active caspase 3 to allow detection of apoptosis. The adult eye phenotype of flies co-expressing dTAK1 and the effector caspase inhibitor p35 was visualized. Scale bars, 100 μm. (D) HEK 293T cells were transfected with the indicated constructs for 24 h, and cytotoxicity was then measured using a LDH release assay. The error bars represent the means ± s.d. of three independent experiments. (E) HEK 293T cells were transfected with the indicated constructs for 48 h. Luminogenic caspase 3/7 substrates were added, and the cells were then incubated for 1 h at room temperature. Caspase activity was analyzed using a luminometer. Bars represent means ± s.d. of three independent experiments. (F) HEK 293T cells were transfected with the indicated constructs and left untreated or treated with 100 nM bafilomycin A1. The cells were analyzed for cytotoxicity using a LDH release assay. The values represent the means ± s.d. of three independent experiments. (G) The level of TAK1-induced cell death is restored by treatment with the autophagy inhibitors bafilomycin A1 and 3-MA. The cells were pre-treated with 20 nM bafilomycin A1 or 2 mM 3-MA for 1 h. After drug treatment, the cells were transfected with either a control plasmid or a TAK1 plasmid. 48 hours after transfection, cells were harvested, and cell viability was assessed using a trypan blue exclusion assay. The values represent the means ± s.d. of three independent experiments. * P < 0.05, ** P < 0.01, and *** P < 0.001.
In addition to its cytoprotective role, autophagy also participates in cell death. However, it is not fully understood what factors determine whether autophagy is cytoprotective or cytotoxic. Considering the deleterious effects of dTAK1 overexpression on the adult eye (Fig. 4C), we hypothesized that TAK1 overexpression may cause cytotoxic cell death. Therefore, we tested the potential role of TAK1-induced autophagy in promoting cell death by measuring cytotoxicity with a lactate dehydrogenase (LDH) release assay in HEK 293T cells. TAK1 overexpression was significantly more cytotoxic than control (Fig. 7D). We evaluated the LDH levels with S6K1 overexpression and co-expression of TAK1 and S6K1 decreased LDH levels (Supplementary Fig. 8).
However, the caspase 3/7 activities resulting from the overexpression of TAK1 and control showed slight difference (Fig. 7E). Bax used as a positive control to confirm the efficient caspase 3/7 activity. To further confirm the role of TAK1-induced autophagy in regulating cell death, bafilomycin A1, an autophagy inhibitor, was used. When autophagy was blocked using bafilomycin A1, the cytotoxicity of TAK1 was significantly reduced (Fig. 7F).
To determine whether autophagy induced by TAK1 was responsible for the reduced cell viability, we used a trypan blue exclusion assay and measured the viability of cells treated with 3-methyladenine (3-MA) (an inhibitor of class III phosphatidylinositol 3-kinase that blocks autophagy) or bafilomycin A1 to inhibit autophagy. The treatment of cells with 3-MA or bafilomycin A1 prior to the transfection of TAK1 significantly rescued the cell viability defect, indicating that TAK1-induced autophagy contributes to cell death, not cell survival (Fig. 7G).
Discussion
In this study, we addressed the mechanism of TAK1-induced autophagy by combining in vitro and in vivo approaches. We identified TAK1 as a positive mediator of autophagy via the suppression of S6K1 phosphorylation and showed that the overexpression of TAK1 induces autophagy by measuring GFP-LC3 punctate structures and GFP-LC3-II level in HEK 293T cells and using the GMR-GAL4 system in Drosophila eyes. Furthermore, the cytotoxicity of TAK1-induced autophagy can be attenuated by pretreatment with 3-MA or bafilomycin A1, both of which are autophagy inhibitors. We observed that TAK1 overexpression induces autophagy and provided evidence that TAK1-induced autophagy is cytotoxic, not cytoprotective.
S6K1 is an important regulator of cell proliferation and growth. In addition to cell size regulation, S6K1 is involved in the inhibition of autophagy26,34. In detail, S6K1 has dual functions in autophagy regulation. S6K1 has a positive role in autophagy induction under starvation conditions29,30. However, S6K1 inhibits autophagy under normal nutritional conditions40,41.
In particular, it appears that downregulation of S6K1 phosphorylation is correlated in autophagy induction when autophagy acts as a cell death mechanism. For example, ionising radiation (IR) induces autophagic cell death and reduces the phosphorylation level of S6K1. S6K1 was suppressed by IR and autophagy was induced40. Pentagalloylglucose (PGG) induces caspase-independent autophagic cell death and decreases S6K1 phosphorylation41. Similarly, TAK1-induced autophagy act as an autophagic cell death, so S6K1 has an inhibitory effect on autophagy induction.
It is possible that S6K1 has a positive or negative role for autophagy according to nutritional conditions. S6K1 may enhance autophagy (cytoprotective autophagy) under starvation conditions whereas it may suppress autophagy (autophagic cell death) under normal conditions. We suggest that downregulation of S6K1 may promote autophagy (particularly cytotoxic autophagic cell death) under normal nutritional conditions.
The mTOR/S6K1 pathway plays important roles in tumorigenesis42,43. In this regard, regulation of S6K1 could be helpful for the treatment of cancer. Therefore, we investigated how S6K1 phosphorylation is regulated during TAK1-induced autophagy. Furthermore, we examined the interaction of TAK1 and S6K1. We found that TAK1 decreases S6K1 phosphorylation and binds to S6K1.
Upon observing that TAK1 negatively regulates S6K1, we performed additional experiments to further define the molecular mechanism behind this inhibition. Autophagy is characterized by inhibition of mTORC1, so we targeted raptor, which is a major component of mTORC1 and may be another regulator of TAK1-induced autophagy. Our results show that TAK1 negatively regulates S6K1 by interfering with S6K1's binding to raptor.
Although previous report mentioned that the relationship between TAK1 and AMPK in TRAIL treated cells39, this is the first study showing that TAK1 binds to S6K1 and TAK1 competes S6K1 for raptor binding in regulating autophagy. Herrero-Martin et al39. reported that TAK1 activates AMPK-dependent cytoprotective autophagy in epithelial cells treated with TRAIL. However, TAK1 did not activate AMPK and induced cytotoxic autophagy in our study. It is possible that TAK1 induces autophagy through several pathways. TAK1 might induce autophagy through not only AMPK-dependent way, but also AMPK-independent pathway. We think that our TAK1-induced autophagy acts through AMPK-independent pathway. Also, it is plausible that TAK1-WT overexpression does not affect AMPK phosphorylation14. Herrero-Martin et al. mentioned that TAK1 is essential, but not sufficient for the effective activation of AMPK. Additionally, it is possible that TRAIL may activate other signals besides TAK1.
Autophagy is a potent tumor suppressive mechanism. Multiple genes that are required for the induction or execution of autophagy are known to be potent tumor suppressors; these include phosphatase and tensin homolog (PTEN), tuberous sclerosis 1 (TSC1), tuberous sclerosis 2 (TSC2), autophagy-specific gene 4 (Atg4), and beclin 18. Moreover, many negative regulators of autophagy, including Akt, class I phosphoinositide 3 kinase (PI3K), mTOR, and S6K1, have oncogenic properties2,44. Autophagy can suppress tumor progression by promoting cell death and inhibiting cell growth. The induction of autophagic cell death has emerged as a new potential therapeutic method to solve the problem of cancer cell resistance to apoptosis or radiotherapy45. Apoptosis-resistant or radio-resistant cancer cells can undergo autophagic cell death when treated with autophagy-inducing agents. These properties of autophagy may play critical roles in tumor suppression.
Autophagy plays contrasting roles in regulating cell death and survival. To resolve these contradictory phenomena, a better understanding of the molecular regulators of autophagy-mediated cellular events is needed. Explaining how a specific pathway may lead to opposite biological effects is important for the development of new therapeutic strategies related to autophagy. Autophagy may participate in either the inhibition or acceleration of cancer cell death, but, under certain circumstances, it may serve as a tumor suppression mechanism before tumors form.
Based on our in vitro and in vivo data that TAK1 overexpression induces autophagic cell death, we can use it as a potential therapy in eliminating apoptosis-resistant or radio-resistant cancer cells. Our study may therefore provide the basis for new anti-cancer therapies. TAK1-induced autophagic cell death will influence drug design and the mechanistic study of cancer cell death. Targeting the autophagic pathway to kill cancer cells has emerged as a promising new approach for drug discovery and cancer therapy. Recent report suggested that cancer cells can undergo death by non-apoptotic pathways such as autophagic cell death46.
In summary, our study demonstrates that TAK1 acts as a novel inducer of autophagic cell death by negatively regulating the S6K1. The regulation of autophagy is important for several aspects of biological processes and medicine, and thus, it is of great importance to find novel regulators of autophagy. Here, we showed the relationship among TAK1, raptor and S6K1. TAK1 competes S6K1 for binding to raptor, thus inducing autophagy. And the S6K1 phosphorylation level was decreased when TAK1 was overexpressed. We also showed that this regulation is evolutionarily conserved between mammalian cells and Drosophila. We believe that our results will be useful in future investigations, including research on anti-cancer therapies. Because TAK1 induces autophagic cell death and negatively regulates S6K1, which is a positive regulator of cell growth, the modulation of TAK1-induced autophagy may be a good candidate for the treatment of cancer.
Methods
Cell culture and transfection
HEK 293T cells, HEK 293A cells, and U2OS human osteosarcoma cells were grown in Dulbecco's Modified Eagle's Medium (DMEM; Gibco-BRL, Carlsbad, CA, USA) supplemented with 10% (v/v) fetal bovine serum (FBS; HyClone, Logan, UT, USA), 100 IU/ml penicillin, and 100 μg/ml streptomycin (Gibco-BRL). All cells were maintained at 37°C in a humidified incubator with 5% CO2. HEK 293T cells were transfected as described in the figure legends using the FuGENE HD Transfection Reagent (Roche, Mannheim, Germany), according to the manufacturer's protocol.
Drosophila stocks
The UAS-dS6K-DN and UAS-Rheb fly lines were obtained from the Bloomington Stock Center (Indiana University, Bloomington, IN, USA). The GMR-GAL4 > UAS-DCP1/CyO mutant fly line was made by Y.I. Kim. The UAS-dTAK1 and eye-specific GMR-GAL4 fly lines were kind gifts from Dr. J. Chung (Seoul National University, Seoul, Republic of Korea). The UAS-GFP-Atg8a fly line was a generous gift from Dr. T.P. Neufeld (University of Minnesota, Minneapolis, MN, USA). The UAS-dS6K-WT fly line was kindly provided by Dr. G. Thomas (University of Cincinnati Genome Research Institute, Cincinnati, OH, USA). The eye phenotypes of adult transgenic flies were photographed using a Carl Zeiss Stemi 2000C microscope with AxioVision AC software.
Immunoprecipitation
The Flag-tagged TAK1-WT construct and Flag-tagged TAK1-KW construct were kindly provided by Dr. H-H Kim (Seoul National University, Seoul, Republic of Korea) and Dr. Seong-Jin Kim (CHA University, Seoul, Republic of Korea). The Myc-tagged S6K1 construct was a kind gift from Dr. J. Chung. The HA-raptor plasmid was purchased from Addgene (Cambridge, MA, USA). Forty-eight hours after transfection, transfected cells were lysed and incubated on ice for 30 min with lysis buffer (20 mM HEPES, pH 7.5, 150 mM NaCl, 10% glycerol, 5 mM EDTA, 1% Triton X-100, a complete protease inhibitor cocktail tablet and a phosphatase inhibitor cocktail tablet from Roche). Cell lysates were centrifuged at 10,000 × g for 15 min at 4°C. Protein lysates from each sample (500 μg) were incubated with 4 μg of specific antibodies overnight at 4°C. After the overnight incubation, 25 μl of Dynabeads Protein G (Invitrogen, Oslo, Norway) was added, followed by an additional 1 h incubation at 4°C. The beads were collected by placing tubes on a magnet for 2 min, washed three times with 200 μl phosphate-buffered saline (PBS) containing 10% lysis buffer, and resuspended in 25 μl of 2× SDS sample buffer. The samples were incubated for 5 min at 95°C, followed by incubation of tubes on the magnet for 2 min. Supernatants were collected and loaded onto a gel. Immunoprecipitated complexes were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 8–12% acrylamide gels and transferred to a nitrocellulose membrane. Following transfer, the membranes were blocked for 1 h at room temperature in blocking buffer (5% skim milk in PBS). The membranes were incubated overnight at 4°C in primary antibodies diluted in 1× 0.1% Tween-20 in PBS (PBST). Phosphospecific S6K1 T389 (1∶1,000 dilution), HA-Tag (for immunoprecipitation, 1∶50 dilution), and Myc-Tag (for immunoprecipitation, 1∶200 dilution) antibodies were purchased from Cell Signaling Technology (Beverly, MA, USA). FLAG M2 (1∶1,000 dilution) and β-actin (1∶10,000 dilution) antibodies were purchased from Sigma (St Louis, MO, USA). HA (for immunoblotting, 1∶1,000 dilution) and Myc (for immunoblotting, 1∶1,000 dilution) antibodies were obtained from Roche (Mannheim, Germany). The membranes were washed three times with PBST. Secondary antibodies were diluted in PBST and were added to the membrane for 1 h 40 min at room temperature. The following secondary antibodies were used: anti-rabbit IgG horseradish peroxidase (HRP)-linked antibody (Cell Signaling Technology, 1∶2,000 dilution) and goat anti-mouse IgG-HRP conjugated antibody (Chemicon, Billerica, MA, USA, 1∶5,000 dilution). The membranes were washed five times with PBST. Chemiluminescent detection was accomplished using the SuperSignal West Pico substrate (Pierce, Rockford, IL, USA). The images were analyzed with an LAS 4000 instrument (Fujifilm, Minato, Tokyo, Japan).
Immunoblot analysis
The GFP-LC3 plasmid was a kind gift from Dr. Tamotsu Yoshimori (Osaka University, Osaka, Japan). Forty-eight hours after transfection, whole-cell protein lysates were prepared from transfected cells using 5× sample buffer (250 mM Tris-HCl, pH 6.8, 10% SDS, 30% glycerol, 0.5 M DTT, and 0.02% bromophenol blue). Denatured protein (20 μg) was loaded on 8–12% gels, and electrophoresis was performed. The gels were then transferred onto nitrocellulose membranes for 2 h at 4°C. Following transfer, the membranes were blocked for 1 h at room temperature in blocking buffer (5% skim milk in PBS). The membranes were then incubated overnight at 4°C in primary antibodies diluted in 1× PBST buffer. The following primary antibodies were used: GFP (Santa Cruz Biotechnology, Santa Cruz, CA, USA, 1∶1,000 dilution), FLAG M2 (Sigma-Aldrich, 1∶1,000 dilution), β-actin (Sigma-Aldrich, 1∶10,000 dilution), phosphospecific S6K1 T389 (Cell Signaling Technology, 1∶1,000 dilution), Myc (Roche, 1∶1,000 dilution), pAMPK-α (Cell Signaling Technology, 1∶500 dilution), AMPK-α (Cell Signaling Technology, 1∶1,000 dilution), Atg5 (Cell Signaling Technology, 1∶1,000 dilution), Atg7 (Cell Signaling Technology, 1∶1,000 dilution), Atg12 (Cell Signaling Technology, 1∶1,000 dilution), beclin1 (Cell Signaling Technology, 1∶1,000 dilution) and LC3B (Cell Signaling Technology, 1∶1,000 dilution). The membranes were washed three times with PBST. Secondary antibodies were diluted in PBST and were added for 1 h 40 min at room temperature. The following secondary antibodies were used: anti-rabbit IgG HRP-linked antibody (Cell Signaling Technology, 1∶2,000 dilution) and goat anti-mouse IgG-HRP conjugated antibody (Chemicon, 1∶5,000 dilution). The membranes were washed five times with PBST. Chemiluminescent detection was accomplished using SuperSignal West Pico substrate (Pierce, Rockford, IL, USA), and the images were analyzed with an LAS 4000 instrument (Fujifilm).
Immunoblotting of Drosophila
Fifteen third-instar larvae or pupae of the indicated genotypes were collected and ground in 300 μL of lysis solution (1% Triton X-100, 50 mM Tris pH7.4, 500 mM NaCl, 7.5 mM MgCl2, 0.2 mM EDTA, 1 mM NaVO4, 50 mM β-glycerophosphate, 1 mM DTT, and 25% glycerol). Fifty micrograms of protein were loaded onto a 12% SDS-gel and transferred onto a nitrocellulose membrane. The membrane was incubated overnight at 4°C with a phosphospecific dS6K T398 antibody (Cell Signaling Technology, 1∶500 dilution) or a mouse anti-tubulin antibody (E7, DSHB, University of Iowa, Iowa city, IA, USA, 1∶1,000 dilution).
RT-PCR
For each sample, total RNA from 15 third-instar larvae or pupae was extracted using Trizol reagent (Invitrogen, Carlsbad, CA, USA). Reverse transcription was carried out with the Transcriptor First Strand cDNA synthesis Kit (Roche) using the provided protocol. PCR was then performed as follows: 5 min denaturation at 95°C followed by 30 cycles of amplification (95°C for 1 min, 55°C for 1 min, and 72°C for 2 min) and an additional extension for 10 min at 72°C. The Rp49 primers were as follows: forward (5′-AGA TCG TGA AGA AGC GCA CC-3′) and reverse (5′-CGA TCC GTA ACC GAT GTT GG-3′). The dTAK1 primers were as follows: forward (5′-GAT GAC CAA CAA TCG CGG -3′) and reverse (5′-GCC GAC GAG GTG AGG TTG -3′)47. RT-PCR was performed using a GeneAmp PCR System 9700 (Applied Biosystems, CA, USA).
LysoTracker Red staining of third-instar larvae
Imaginal eye discs were dissected from third-instar larvae in PBS. Eye discs were incubated for 1 min in LysoTracker Red (Molecular Probes, Eugene, OR, USA, 1∶1,000 dilution) and Hoechst 33342 (Sigma-Aldrich, 1∶100 dilution) for 10 s in PBS. The collected discs were washed three times with PBS, mounted on glass slides with coverslips, and immediately photographed using a Leica DM6000B microscope (Leica, Wetzlar, Germany).
Caspase 3 staining of third-instar larvae
Imaginal eye discs were dissected from third-instar larvae in PBS. For caspase 3 staining, the discs were fixed in 4% paraformaldehyde for 20 min at room temperature and were labeled with a cleaved caspase 3 primary antibody (Cell Signaling Technology, 1∶50 dilution) and an Alexa Fluor 488 goat anti-rabbit IgG secondary antibody (Molecular Probes, 1∶200 dilution). The coverslips were mounted on glass slides using 60% glycerol, and images were acquired using a Leica DM6000B microscope.
Caspase activity test
Caspase activity was determined using the Caspase-Glo 3/7 Assay (Promega, Madison, WI, USA). Forty-eight hours after transfection, cells were harvested. Eight thousand cells were counted for each well of a 96-well plate, and the cells were mixed with the Caspase Glo 3/7 reagent (1∶1 ratio). Samples were incubated for 1 h at room temperature. The luminescence of each sample was measured in a Wallac Victor 1420 multilabel counter (PerkinElmer, Waltham, MA, USA).
Autophagy detection using the GFP-Atg8a system in drosophila
The GMR-GAL4/BC; dTAK1/TM6B transgenic fly line was made and then crossed with the UAS-GFP-Atg8a line. The third-instar larvae expressing both dTAK1 and GFP-Atg8a were collected. Eye discs were dissected in PBS and mounted on glass slides with coverslips using 60% glycerol. Eye disc samples were imaged with a Leica DM6000B microscope. At least three fields in each of 10 larvae were imaged, and GFP-positive puncta were counted.
Determination of autophagy using GFP-LC3 and cell death detection
Cells were plated at 1.5 × 105 cells/well in 12-well plates the day before transfection, and 50 ng of GFP-LC3 along with 450 ng of either TAK1 or control vector was co-transfected into the indicated types of cells using FuGENE HD Transfection Reagent (Roche). Forty-eight hours after transfection, GFP-LC3 puncta were visualized using a Leica DM6000B microscope. Cell death was assessed using a LDH release assay (Cytotoxicity Detection KitPLUS LDH, Roche). The mTOR plasmid was purchased from Addgene (Cambridge, MA, USA), and the Bax plasmid was obtained from KRIBB (Daejeon, Republic of Korea). HEK 293T cells were seeded at 7.5 × 104 cells/well in 24-well plates the day before transfection. The cells were treated with 2 mM 3-MA (Sigma-Aldrich) or 20 nM bafilomycin A1 (Upstate, Billerica, MA, USA) for 1 h before transfection. After 48 h of incubation, a trypan blue (Welgene, Daegu, South Korea) exclusion assay was performed to measure cell viability.
RNA interference
The TAK1 and AMPK siRNAs were purchased from Santa Cruz Biotechnology. Control, Atg5, Atg7, Atg12, beclin1 and S6K1 siRNAs were purchased from Cell Signaling Technology. Each siRNA (100 nM) was transfected into the cells41 using FuGENE HD (Roche) according to the manufacturer's protocol.
Statistical analyses
All of the experiments were repeated at least three times. The data are expressed as the mean ± standard deviation (s.d.). Statistical analyses were performed using Student's t test (two-tailed).
Author Contributions
J.H.S., S.H.M., H.K.L. and O.J.Y. designed research; J.H.S. performed experiments; J.H.S., S.H.M., S.J.K., Y.I.K, J.P., H.K.L. and O.J.Y. analyzed data; and J.H.S., H.K.L. and O.J.Y. wrote the manuscript. All authors discussed the results and reviewed the manuscript.
Supplementary Material
Supplementary Information
Acknowledgments
We would like to thank Dr. J. Chung, Dr. G. Thomas, and Dr. T. P. Neufeld for providing the fly stocks and Dr. H. H. Kim for providing the Flag-tagged TAK1 plasmid. We also thank Dr. Tamotsu Yoshimori for providing the GFP-LC3 plasmid and Se-Kyu Choi, Won-Ho Kim, and Dr. Oky Maeng for technological assistance. Especially, we thank Dr. Sujin Park, Dr. Kyung-Min Yang, Dr. Jin Muk Kang, and Staci Jakyong Kim for insightful discussions. Also, I thank Kyung Yun for helpful comments. Ook Joon Yoo was supported by the Research Program for Bio & Medical Technology Development through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2008-2004686). Heung Kyu Lee was supported by the NRF (2012028274) and the National R&D Program for Cancer Control (1020230). Seong-Jin Kim was supported by the NRF (2009-0081756) and MLTM (20046001).
References
- Klionsky D. J. et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 4, 151–175 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo Y., Kanzawa T., Sawaya R. & Kondo S. The role of autophagy in cancer development and response to therapy. Nat Rev Cancer 5, 726–734 (2005). [DOI] [PubMed] [Google Scholar]
- Kourtis N. & Tavernarakis N. Autophagy and cell death in model organisms. Cell Death Differ 16, 21–30 (2009). [DOI] [PubMed] [Google Scholar]
- Scarlatti F., Granata R., Meijer A. J. & Codogno P. Does autophagy have a license to kill mammalian cells? Cell Death Differ 16, 12–20 (2009). [DOI] [PubMed] [Google Scholar]
- Denton D., Nicolson S. & Kumar S. Cell death by autophagy: facts and apparent artefacts. Cell Death Differ 19, 87–95 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. L. et al. A functionalized single-walled carbon nanotube-induced autophagic cell death in human lung cells through Akt-TSC2-mTOR signaling. Cell Death Dis 2, e159 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Yoshimori T. & Levine B. Methods in mammalian autophagy research. Cell 140, 313–326 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiuri M. C. et al. Control of autophagy by oncogenes and tumor suppressor genes. Cell Death Differ 16, 87–93 (2009). [DOI] [PubMed] [Google Scholar]
- Kim Y. I. et al. A genetic screen for modifiers of Drosophila caspase Dcp-1 reveals caspase involvement in autophagy and novel caspase-related genes. BMC Cell Biol 11, 9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landstrom M. The TAK1-TRAF6 signalling pathway. Int J Biochem Cell Biol 42, 585-589 (2010). [DOI] [PubMed] [Google Scholar]
- Delaney J. R. & Mlodzik M. TGF-beta activated kinase-1: new insights into the diverse roles of TAK1 in development and immunity. Cell Cycle 5, 2852–2855 (2006). [DOI] [PubMed] [Google Scholar]
- Tan S. H. et al. Regulation of cell proliferation and migration by TAK1 via transcriptional control of von Hippel-Lindau tumor suppressor. J Biol Chem 284, 18047–18058 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikari A., Xu M. & Chen Z. J. Ubiquitin-mediated activation of TAK1 and IKK. Oncogene 26, 3214–3226 (2007). [DOI] [PubMed] [Google Scholar]
- Xie M. et al. A pivotal role for endogenous TGF-beta-activated kinase-1 in the LKB1/AMP-activated protein kinase energy-sensor pathway. Proc Natl Acad Sci U S A 103, 17378–17383 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrentino A. et al. The type I TGF-beta receptor engages TRAF6 to activate TAK1 in a receptor kinase-independent manner. Nat Cell Biol 10, 1199–1207 (2008). [DOI] [PubMed] [Google Scholar]
- Sayama K. et al. Transforming growth factor-beta-activated kinase 1 is essential for differentiation and the prevention of apoptosis in epidermis. J Biol Chem 281, 22013–22020 (2006). [DOI] [PubMed] [Google Scholar]
- Hara K. et al. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 110, 177–189 (2002). [DOI] [PubMed] [Google Scholar]
- Mehrpour M., Esclatine A., Beau I. & Codogno P. Overview of macroautophagy regulation in mammalian cells. Cell Res 20, 748–762 (2010). [DOI] [PubMed] [Google Scholar]
- Ali S. M. & Sabatini D. M. Structure of S6 kinase 1 determines whether raptor-mTOR or rictor-mTOR phosphorylates its hydrophobic motif site. J Biol Chem 280, 19445–19448 (2005). [DOI] [PubMed] [Google Scholar]
- Hay N. & Sonenberg N. Upstream and downstream of mTOR. Genes Dev 18, 1926–1945 (2004). [DOI] [PubMed] [Google Scholar]
- Jung C. H., Ro S. H., Cao J., Otto N. M. & Kim D. H. mTOR regulation of autophagy. FEBS Lett 584, 1287–1295 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer A. J. & Codogno P. Regulation and role of autophagy in mammalian cells. Int J Biochem Cell Biol 36, 2445–2462 (2004). [DOI] [PubMed] [Google Scholar]
- Ruvinsky I. & Meyuhas O. Ribosomal protein S6 phosphorylation: from protein synthesis to cell size. Trends Biochem Sci 31, 342–348 (2006). [DOI] [PubMed] [Google Scholar]
- Kozma S. C. & Thomas G. Regulation of cell size in growth, development and human disease: PI3K, PKB and S6K1. Bioessays 24, 65–71 (2002). [DOI] [PubMed] [Google Scholar]
- Castedo M., Ferri K. F. & Kroemer G. Mammalian target of rapamycin (mTOR): pro- and anti-apoptotic. Cell Death Differ 9, 99–100 (2002). [DOI] [PubMed] [Google Scholar]
- Blommaart E. F., Luiken J. J., Blommaart P. J., van Woerkom G. M. & Meijer A. J. Phosphorylation of ribosomal protein S6 is inhibitory for autophagy in isolated rat hepatocytes. J Biol Chem 270, 2320–2326 (1995). [DOI] [PubMed] [Google Scholar]
- Pullen N. & Thomas G. The modular phosphorylation and activation of p70S6K1. FEBS Lett 410, 78–82 (1997). [DOI] [PubMed] [Google Scholar]
- Weng Q. P. et al. Regulation of the p70 S6 kinase by phosphorylation in vivo. Analysis using site-specific anti-phosphopeptide antibodies. J Biol Chem 273, 16621–16629 (1998). [DOI] [PubMed] [Google Scholar]
- Scott R. C., Schuldiner O. & Neufeld T. P. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell 7, 167–178 (2004). [DOI] [PubMed] [Google Scholar]
- Armour S. M. et al. Inhibition of mammalian S6 kinase by resveratrol suppresses autophagy. Aging (Albany NY) 1, 515–528 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criollo A. et al. Inhibition of autophagy by TAB2 and TAB3. EMBO J 30, 4908–4920 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funasaka T., Tsuka E. & Wong R. W. Regulation of autophagy by nucleoporin Tpr. Sci Rep 2, 878 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. P., Yang M. C., Liu H. S., Lin Y. S. & Lei H. Y. Concanavalin A induces autophagy in hepatoma cells and has a therapeutic effect in a murine in situ hepatoma model. Hepatology 45, 286–296 (2007). [DOI] [PubMed] [Google Scholar]
- Iwamaru A. et al. Silencing mammalian target of rapamycin signaling by small interfering RNA enhances rapamycin-induced autophagy in malignant glioma cells. Oncogene 26, 1840–1851 (2007). [DOI] [PubMed] [Google Scholar]
- Shvets E. & Elazar Z. Flow cytometric analysis of autophagy in living mammalian cells. Methods Enzymol 452, 131–141 (2009). [DOI] [PubMed] [Google Scholar]
- Codogno P. & Meijer A. J. Autophagy and signaling: their role in cell survival and cell death. Cell Death Differ 12 Suppl 2, 1509–1518 (2005). [DOI] [PubMed] [Google Scholar]
- Lee S. B. et al. ATG1, an autophagy regulator, inhibits cell growth by negatively regulating S6 kinase. EMBO Rep 8, 360–365 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avruch J. et al. Insulin and amino-acid regulation of mTOR signaling and kinase activity through the Rheb GTPase. Oncogene 25, 6361–6372 (2006). [DOI] [PubMed] [Google Scholar]
- Herrero-Martin G. et al. TAK1 activates AMPK-dependent cytoprotective autophagy in TRAIL-treated epithelial cells. EMBO J 28, 677–685 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daido S. et al. Inhibition of the DNA-dependent protein kinase catalytic subunit radiosensitizes malignant glioma cells by inducing autophagy. Cancer Res 65, 4368–4375 (2005). [DOI] [PubMed] [Google Scholar]
- Hu H. et al. Pentagalloylglucose induces autophagy and caspase-independent programmed deaths in human PC-3 and mouse TRAMP-C2 prostate cancer cells. Mol Cancer Ther 8, 2833–2843 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efeyan A. & Sabatini D. M. mTOR and cancer: many loops in one pathway. Curr Opin Cell Biol 22, 169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dann S. G., Selvaraj A. & Thomas G. mTOR Complex1-S6K11 signaling: at the crossroads of obesity, diabetes and cancer. Trends Mol Med 13, 252–259 (2007). [DOI] [PubMed] [Google Scholar]
- Sabatini D. M. mTOR and cancer: insights into a complex relationship. Nat Rev Cancer 6, 729–734 (2006). [DOI] [PubMed] [Google Scholar]
- Kuwahara Y. et al. Enhancement of autophagy is a potential modality for tumors refractory to radiotherapy. Cell Death Dis 2, e177 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara K. et al. Akt inhibitor shows anticancer and radiosensitizing effects in malignant glioma cells by inducing autophagy. Int J Oncol 31, 753–760 (2007). [PubMed] [Google Scholar]
- Chen W., White M. A. & Cobb M. H. Stimulus-specific requirements for MAP3 kinases in activating the JNK pathway. J Biol Chem 277, 49105–49110 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information