Abstract
Age-onset cataracts are believed to be expedited by the accumulation of UV-damaged human γD-crystallins in the eye lens. Here we show with molecular dynamics simulations that the stability of γD-crystallin is greatly reduced by the conversion of tryptophan to kynurenine due to UV-radiation, consistent with previous experimental evidences. Furthermore, our atomic-detailed results reveal that kynurenine attracts more waters and other polar sidechains due to its additional amino and carbonyl groups on the damaged tryptophan sidechain, thus breaching the integrity of nearby dry center regions formed by the two Greek key motifs in each domain. The damaged tryptophan residues cause large fluctuations in the Tyr-Trp-Tyr sandwich-like hydrophobic clusters, which in turn break crucial hydrogen-bonds bridging two β-strands in the Greek key motifs at the “tyrosine corner”. Our findings may provide new insights for understanding of the molecular mechanism of the initial stages of UV-induced cataractogenesis.
Cataracts are the leading cause of impairment and loss of vision in older adults, affecting nearly 22 million Americans over the age of 40, and 51% of world population over the age of 65 (National Eye Institute: www.nei.nih.gov and Prevent Blindness America www.preventblindness.org). Pathological studies have revealed that cataracts are associated with the aggregation of major lens proteins, the βγ-crystallin family1. Because no protein turnover occurs in the lens core, the accumulation of insoluble aggregates of crystallin scatters incident light, reducing the light focused on the photoreceptors in the retina2,3,4,5,6.
One of the possible mechanisms of protein aggregation in age-related cataract is associated with the ultraviolet (UV)-induced thermal and photochemical reactions to the aromatic residues in crystallin7,8,9,10,11. It has been suggested that UV damage to these aromatic residues may impair the folding and stability of lens crystallins and contribute to cataractogenesis9,12. Previous studies have revealed that tryptophan (Trp) residues in crystallin can absorb and filter most of the UV radiation with wave length of 240–310 nm13 due to its low sensitizing activity and high physical quenching activity14,15,16,17. However, Trp can gradually convert to kynurenine (KN) and its derivatives (see Fig. 1a) during the long-time exposure to UV radiation from the Sun18,19,20. Actually, the content of KN is found to be much higher in cataractous lenses than in normal lenses of the same age21,22,23. Thus, protein side-chain modification of Trp-to-KN may play an important role in cataractogenesis, as indicated by epidemiological studies of humans and laboratory experiments on animals9,24,25. However, the detailed molecular mechanism leading to crystallin misfolding and subsequent aggregation is not yet well understood.
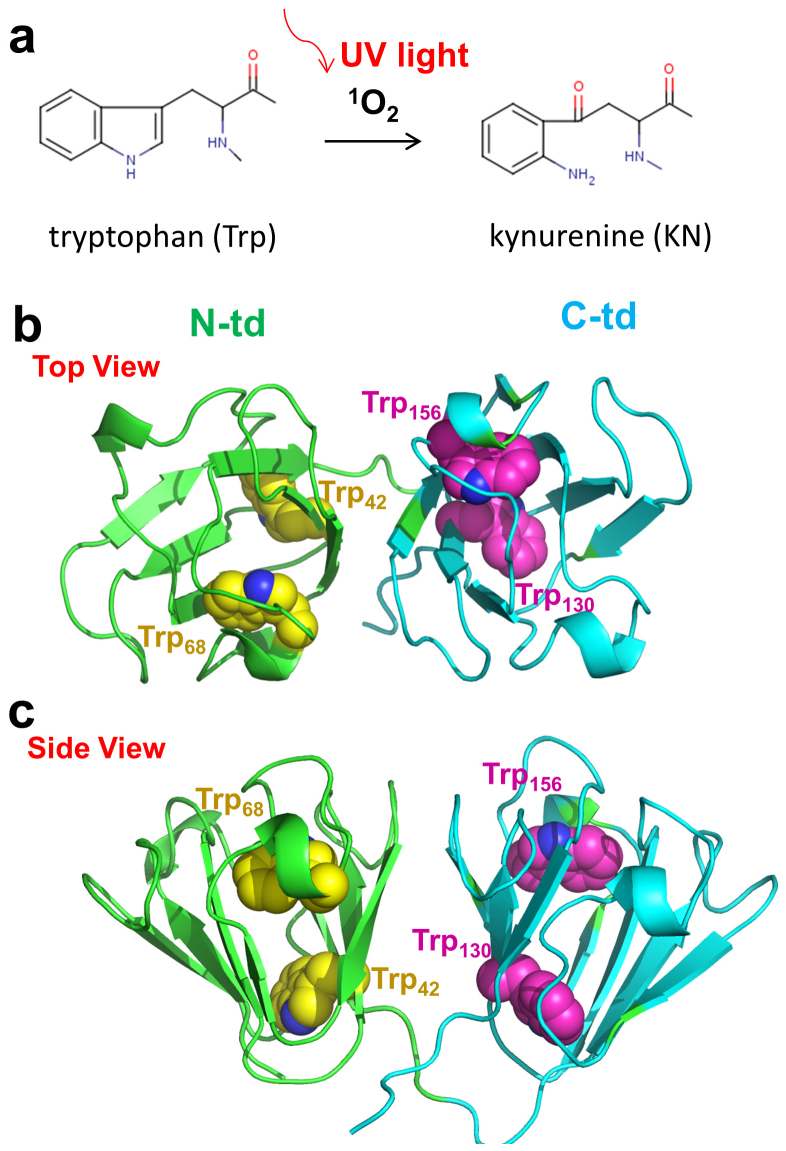
Figure 1.
(a) An oxidative chemical reaction breaks the aromatic ring of Trp during absorption of UV light. (b) and (c) The overall structure of HγD-Crys protein. The backbone of each domain is represented as cartoon (green, N-terminal domain; Cyan, C-terminal domain). The Trp residues in the protein are presented as spheres (Yellow, N-terminal domain; Magenta, C-terminal domain).
Human γD crystallin (HγD-Crys) is one of the most abundant and longest-lived γ-crystallins in the lens nucleus. HγD-Crys contains four intercalated anti-parallel β-sheet Greek key motifs in two homologous domains (Fig. 1b and c)26. Four tryptophan residues, Trp42 and Trp68 in the N-terminal domain (N-td) plus Trp130 and Trp156 in the C-terminal domain (C-td), are found conserved in all the vertebrate β- and γ-crystallins (Fig. 1b and c). Our previous tryptophan fluorescence quenching studies with triple Trp-to-Phe substitutions have shown that those Trp residues contribute significantly to the overall stability of HγD-Crys16,17,27. Furthermore, both our recent experimental and computational studies on the wild-type HγD-Crys have revealed a sequential unfolding pathway of HγD-Crys (N-td unfolds first, followed by the C-td) and a “domain swapping” mechanism for the protein polymerization28,29,30,31,32,33.
In this study, we used extensive atomistic molecular dynamics simulations to investigate the effect of the conversion from Trp to KN on structural stability and unfolding dynamics of HγD-Crys. We found Trp-to-KN substitutions, expected to be caused by UV-radiation, could significantly increase the unfolding speed of the long-lived HγD-Crys protein. Unlike the wild-type, we observed similar unfolding speed for the N-td and C-td in the quadruple KN mutant. Our simulations indicate that the extra amino and carbonyl groups in KN's sidechain attract more water and other polar sidechains, which essentially introduces “leakage” to the hydrophobic central region of each domain (more profound in C-td), and thus breaks the integrity of the dry cavities. In addition, we found that the original Tyr-Trp-Tyr sandwich-like aromatic cluster quickly loses the stability at the beginning of unfolding when the middle residue Trp is substituted by KN, consistent with our Trp-to-Phe mutagenesis studie16,17,27. The large fluctuation then propagates to the “tyrosine corner” and thus breaks the crucial hydrogen bond that bridges two β-strands in the Greek key motifs. Individual single position substitution indicates that the stability of C-td is more sensitive to the Trp-to-KN substitutions than the N-td. These findings, for the first time, provide a detailed unfolding pathway to the KN substituted HγD-Crys and molecular insights into the UV-induced age-onset cataractogenesis.
Results
The stability of human γD-crystallin with Trp-to-KN substitutions was analyzed with the molecular dynamics (MD) unfolding simulations. Because of the high physical integrity of the HγD-Crys, it has been difficult to study the structural stability in native environments in either experiments or theoretical simulations. In fact, HγD-Crys is hard to be denatured even in high concentration urea30, and the thermal denaturation midpoint is as high as 358.0 K33 with half life time of ~19 years from the initial unfolding step34. As control run, in our 200+ ns MD simulations, we found the wild-type (and the quadruple Trp-to-KN mutant too) was very stable in pure water at 310K (see RMSD plot in Figure S1). Following our previous chemical/thermal unfolding studies for the wild-type HγD-Crys28,29, we tried to unfold both the wild-type and quadruple KN mutant at two different temperatures, 380 K and 425 K, in 8 M urea (see Method section for more details). For MD simulations at 380 K, we did not observe significant unfolding of HγD-Crys for the wild-type even after 500 ns. Therefore, we mainly discuss the results from simulations at 425 K here. In the following sections, we first present the unfolding simulation results derived from the wild-type and the quadruple Trp-to-KN mutant, and then illustrate the influence of the single position KN substitutions in both N-td and C-td.
Faster unfolding speed in quadruple kynurenine mutant than the wild-type
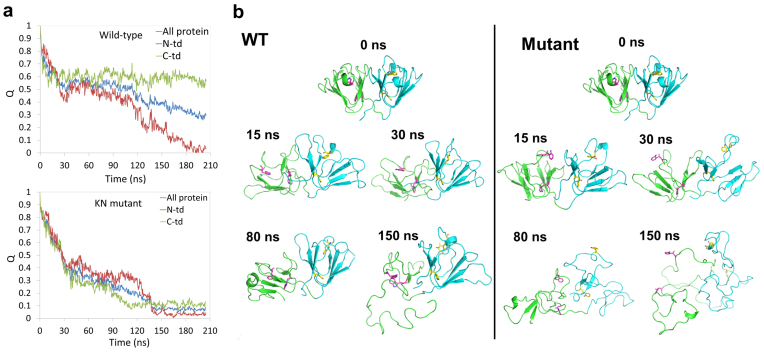
In general, we found faster unfolding speed in the quadruple KN mutant than in the wild-type. We compared the time dependence of the fraction of native contacts Q and RMSDs for both the wild-type and quadruple KN mutant. The results of a representative trajectory are shown in Fig. 2a, 2b, and Supp. Fig. S2 (similar trends were also found in the other four independent trajectories; see Fig. 3a and 3b later). Here, a native contact between residues is counted if any heavy atom of one residue i is within 6.5 Å of any heavy atom of another residue j (j-i > 3) in the crystal structure of the protein. The quadruple KN mutant trigged a complete unfolding of entire protein, where the full protein Q (QKN) quickly drops to < 0.4 after ~30 ns and further decreases to < 0.1 after ~150 ns in the simulation (Fig. 2a). In comparison, the wild-type still keeps the partially folded structure till the end of the simulations, where the full protein Q (QWT) mainly remains at around 0.5 for ~100 ns and slowly drops to 0.3 after 200 ns simulations (Fig. 2a). The quick loss of tertiary and secondary structures is even clearer in quadruple KN mutant from the detailed trajectory comparisons. Representative conformations during the unfolding simulations are shown in Fig. 2b. Four ab folded Greek Key motifs in HγD-Crys gradually lost their β-sheet packs in the quadruple KN mutant during the simulation. While in the wild-type, a clear “two-stage” unfolding process was observed, with the N-td unfolding before the C-td (the C-td remained mostly folded even after the N-td completely unfolded). In the wild-type HγD-Crys, the native structure of the C-td was well kept even after 150 ns MD simulations in 8 M urea at 425 K (Fig. 2b). Our unfolding simulations indicate that the Trp to KN substitution greatly impairs the stability of HγD-Crys, particularly the C-td, which is consistent with our previous tryptophan fluorescence quenching experiments that those Trp residues contribute significantly to the overall stability of HγD-Crys16,17,27 (more discussions below).
Figure 2.
(a) The time evolution of the fraction of native contacts Q for both the wild-type and the quadruple KN mutant. The native contact between residues is counted if any heavy atom of one residue i is within 6.5 Å of any heavy atom of another residue j (j-i > 3) in the crystal structure of the protein. (b) Representative conformations of the wild-type and the quadruple KN mutant during unfolding simulations at 425 K.
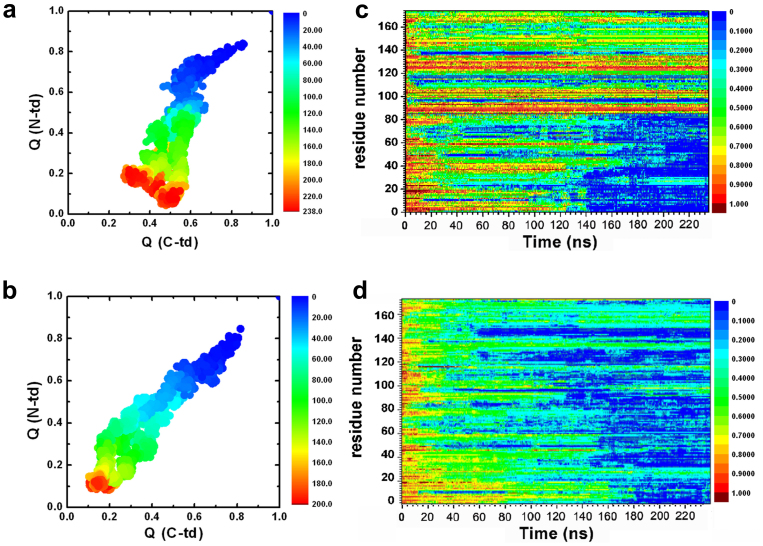
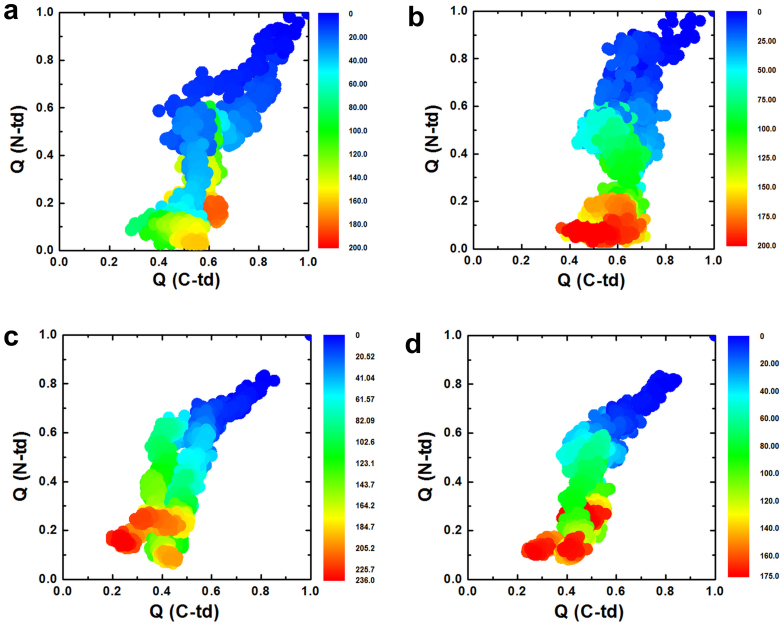
Figure 3. The fractions of native contacts for the two domains are plotted against each other.
(a) wild-type, (b) quadruple KN mutant. The fraction of native contacts formed, Qres, for each residue. (c) wild-type, (d) quadruple KN mutant.
Simultaneous unfolding of N-td and C-td in the kynurenine mutant HγD-Crys
The importance of asynchronous domain unfolding processes in the wild-type HγD-Crys, with a largely folded C-td domain and a mostly unfolded N-td intermediate, indicates that HγD-Crys may aggregate through successive domain swapping of those intermediate conformations. This was found in our previous folding/unfolding experiments in guanidine hydrochloride30,31,32,33,34 and MD simulations in 8 M urea28,29. On the contrary, for quadruple KN mutant, simultaneous unfolding of N-td and C-td was found in HγD-Crys among all five independent trajectories. Fig. 3a and 3b summarize the simulated unfolding reaction of the wild-type and quadruple KN mutant HγD-Crys protein performed at 425 K in 8 M aqueous urea. The time evolution of the fraction of native contacts of the two domains, QN-td and QC-td are plotted against each other, as obtained from five different unfolding trajectories with a total simulation time of 2.3 μs. The plot clearly shows that different unfolding patterns between the wild-type and quadruple KN mutant, indicating a sequential unfolding in the wild-type and simultaneous unfolding in the quadruple KN mutant of two domains (Fig. 3a and 3b).
We further provide a more detailed view of the difference between the wild-type and quadruple KN mutant by calculating the probabilities of native contact formation for each residue, Qres, of HγD-Crys as a function of simulation time during the unfolding process (Fig. 3c and 3d). Overall, the probability of native contact decreases considerably faster in KN mutant than in the wild-type for both N-td and C-td. In addition, more significant change was found in C-td of the mutant, where the probability of native contacts for many residues quickly dropped to < 0.3 within 40 ns during the simulations. Therefore, the quadruple KN substitutions in HγD-Crys largely reduced the stability of the entire protein and severely changed the intrinsic unfolding pathway and its unfolding kinetics of C-td.
Kynurenine mutant introduces “leakages” to the dry central region of each domain
It is of great interest to see how the Trp to KN substitutions result in the above different unfolding behavior for the N-td and C-td from the wild-type. The stability of the folded HγD-Crys is largely related to the hydrophobic central region formed by the two Greek key motifs in each domain. Here we examine the structural difference and the contribution from Trp residues to the hydrophobic central region between N-td and C-td, which has not yet been studied in literature.
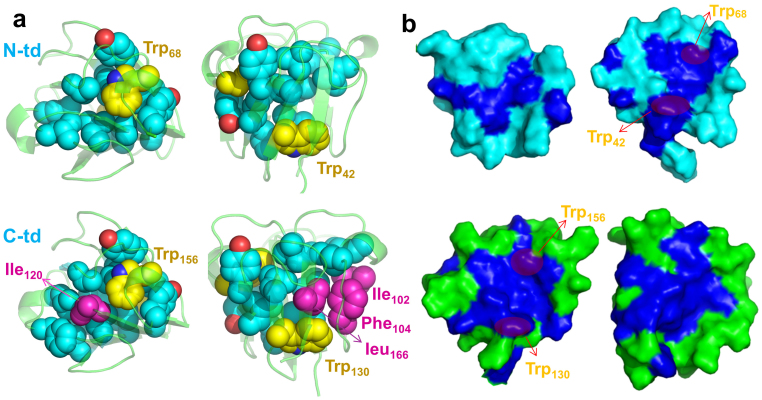
We first compared the number and type of hydrophobic residues which form the dry central region for each domain. We observed that four additional residues, Ile102, Phe104, Ile120, and Leu166, form a more completed/compact hydrophobic central region in C-td (Fig. 4a). The hydrophobicity inside each central region further indicates that C-td is “dryer” than N-td (Fig. 4b). The well packed hydrophobic central region could explain why C-td has higher stability than N-td in the wild-type.
Figure 4.
(a) The hydrophobic central region of each domain. The backbone is represented as cartoon (green) and all the non-polar residues in the dry central region are represented as spheres (cyan). Trp is colored yellow; the additional non-polar residues in C-td are colored in magenta. (b) The inner surface of two domains. The hydrophobic residues are colored in blue and the rest residues are colored in green (C-td) or cyan (N-td).
The four Trp residues in HγD-Crys are important to maintain the stability of the native structures. Trp68 in N-td and Trp156 in C-td are the essential components of the three-residue Tyr-Trp-Tyr clusters linking two Greek key motifs in each domain (Fig. 5d). For example, Trp68 interacts with the aromatic sidechains of Tyr55 and Tyr62 in N-td, and Trp156 interacts with the aromatic sidechains of Tyr143, Tyr150, and Tyr153 in C-td. These aromatic residues block the solvent and other polar residues from reaching the dry central region of each domain. On the other side of each central region, Trp42 and Trp130 tightly interact with the neighboring hydrophobic residues within the central region, which help to keep solvent molecules out. Therefore, these Trp residues are essential and act as the “water blocker” of the dry central region (Fig. 4b). Interestingly, many recent studies have shown that maintaining a dry hydrophobic core is crucial for protein stability and integrity. The nanoscale drying (or dewetting) within the hydrophobic core provides a significant driving force for the protein stability35,36,37,38,39,40,41,42, while the penetration of water (or other solvents) into the hydrophobic core often plays as the most important initial step for protein unfolding43,44,45,46,47,48,49.
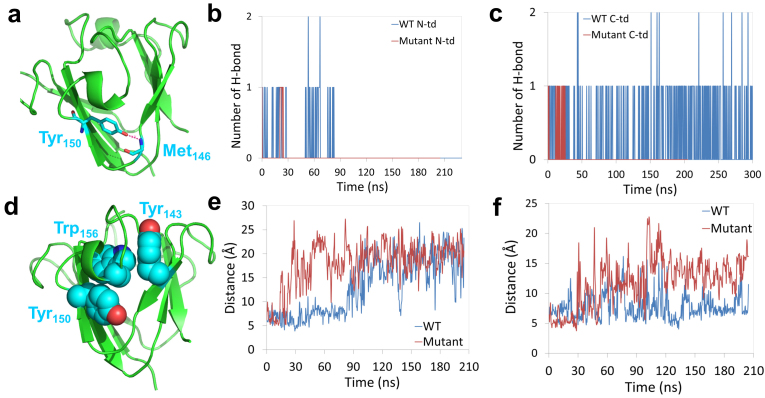
Figure 5.
(a) Structural view of the hydrogen bond formed at the “tyrosine corner” in C-td. (b) and (c) Number of hydrogen bonds at the “tyrosine corner” during the unfolding simulation for both the wild-type and quadruple KN mutant. (d) Structural view of the Tyr-Trp-Tyr sandwich-like cluster. (e) and (f) sidechain-sidechain distance between the Trp/KN and Tyr around the mutation site in N-td and C-td, respectively. The mutation destabilized the ‘‘tyrosine corner’’ and broke the Tyr-Trp-Tyr sandwich-like cluster.
The hydrophobicity of the central region is largely reduced, however, when the four Trp are substituted to KN. As shown in Figure 1a, the UV-induced reaction breaks the five-member nitrogen-containing pyrrole ring in Trp and introduces additional amino (–NH2) and carbonyl (C = 0) groups to form KN. Therefore, the KN mutant is more favorable to the solvent (e.g., water) and polar residues, resulting in a destabilizing factor to the dry central region in each domain. Trp/KN130 stays inside the dry central region and is completely buried by hydrophobic residues in C-td. We observed significant solubility changes of Trp/KN130 by calculating the water contact number and the non-polar residue number during the simulations (Supp. Fig.S3 and S4). At both 380 K and 425 K simulations, the water contact number in KN130 is much higher than that in Trp130, while the non-polar residue contact number shows a totally opposite trend (Supp. Fig. S3 and S4). Consequently, the KN substitutions break the integrity of the hydrophobic core and introduce two “leakages” to the dry central region in the C-td, which could be a crucial reason why the quadruple KN mutant is easier and faster to unfold than the wild-type.
The tryptophan to kynurenine substitutions trigger an early stage unfolding of crystallin
From Figure 3d, we found residues ranging from 140 to 150 lost their native contacts at the earliest stage in C-td during the simulations (Qres < 0.2 within 60 ns), indicating residues 140–150 being the possible starting point of the unfolding process. Detailed structural analysis shows that the segment of residues 140–150 contains the three-residue Tyr143-Trp156-Tyr150 sandwich-like clusters (Fig. 5d) and the important ‘‘tyrosine corner’’ (Trp150 in C-td, Fig. 5a) which links two Greek key motifs in each domain. Similar to the aforementioned three-residue Tyr-Trp-Tyr clusters, the “tyrosine corner” is a classic feature of the Greek key motifs, bridging two β-strands by a hydrogen bond between the tyrosine hydroxyl group and a backbone carboxyl group. Loss of the hydrogen bond in the ‘‘tyrosine corner’’ will cause the two Greek key motifs to decouple from each other and thus introduce solvent molecules into the dry central region in each domain. We plot the dynamics of the hydrogen bond formation in both N-td and C-td in the wild-type and quadruple KN mutant (Fig. 5b and 5C). We found that the hydrogen bond in the “tyrosine corner” was totally lost at the very early stage of the unfolding in quadruple KN mutant, about 30 ns in both N-td and C-td. However, for the wild-type, the hydrogen bond lasted for ~90 ns in N-td and was well-maintained in C-td even after 300 ns MD simulations. As a result, the Trp-to-KN substitution greatly interrupts the stability of the “tyrosine corner” and accelerates the unfolding in C-td, again consistent with the Trp → Phe mutagenesis studies16,17,27 where the five-member nitrogen-containing pyrrole ring was lost in both cases.
The stability of the ‘‘tyrosine corner’’ may be interrupted by the Tyr-Trp-Tyr sandwich-like clusters because they share a same aromatic residue Tyr in each domain. The sandwich-like packing is thus loosed by the Trp to KN substitutions. To confirm our assumption, we investigated the interaction between Tyr and Trp/KN in the clusters by calculating the distance between the sidechains in both the wild-type and the quadruple KN mutant (Tyr62 and Trp/KN68 in N-td, Tyr 150 and Trp/KN156 in C-td). We found a much larger fluctuation of the sidechain distance for the quadruple KN mutant than the wild-type in both N-td and C-td: the sidechain-sidechain distances were mostly larger than 10 Å for N-td and C-td after ~30 ns simulation for the quadruple KN mutant (Fig. 5e and 5f). This fluctuation was quickly (within a few of nanoseconds) propagated to the ‘‘tyrosine corner’’ by the amino acid Tyr62/150, and destabilized the hydrogen bond between two Greek key motifs. Thus, no direct interactions remained between the two Greek key motifs at the end, i.e., after the loss of hydrogen bond and the decoupling of the sandwich-like aromatic packing, which essentially led to the exposure of the hydrophobic residues in the central region to the solvent.
Trp130-Phe104 is an additional aromatic pair bridging the two Greek key motifs at C-td (Supp. Fig. S5a). Larger fluctuations were also found in these aromatic pair in the quadruple KN mutant than in the wild-type, in which the two sidechains were separated from each other after ~30 ns and then allowed the solvent molecules to enter the dry central region (Supp. Fig. S5b). Therefore, the KN substitutions destroyed one extra aromatic pair between the two Greek key motifs in C-td, which could explain why the stability of C-td is more sensitive to the Trp-to-KN substitutions.
In our previous studies, we found that the surface non-native salt-bridge formed between Glu134 and Arg141 is critical in preventing one of the Greek key motifs in C-td from complete unfolding in the wild-type HγD-Crys. In our MD simulations, the non-native salt-bridge was quickly formed in ~20 ns and then kept intact during the rest of the simulations in the wild-type (Supp. Fig. S6), which further confirmed the important role of this salt-bridge in stabilizing the C-td. However, in the quadruple KN mutant, the non-native salt-bridge was broken after ~100 ns, indicating the complete unfolding of C-td (Supp. Fig. S6).
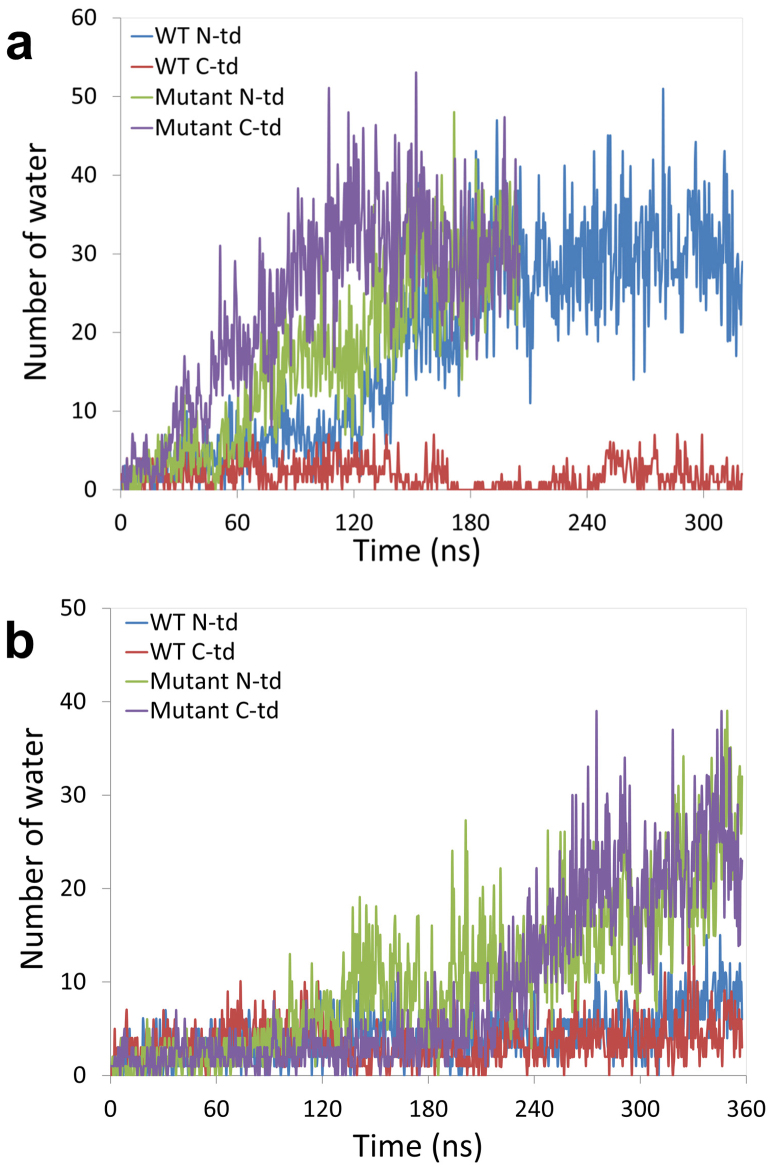
We then looked at the water dynamics in the dry central region in each domain for the wild-type and the KN mutant. The number of water molecules in the hydrophobic central region increased much faster in the quadruple KN mutant than the wild-type in C-td (Fig. 6a). In the quadruple KN mutant, the number of water molecules was gradually increased from 3 at the beginning to ~30 after 100 ns in 425 K simulations. Similar trends were found in 380 K simulations (Fig. 6a) but it took longer simulation time.
Figure 6. The time evolution of water dynamics in each hydrophobic central region for both the wild-type and the quadruple KN mutant.
The number of water inside N-td and C-td hydrophobic central region is counted along the simulation time. MD simulations were performed at two tempratures: (a) 425 K and (b) 380 K.
The stability of the C-td is more sensitive to the Trp-to-KN substitutions than the N-td
In reality, it is unlikely to have all the 4 tryptophan residues substituted to KN in a single HγD-Crys protein. Therefore, we further investigated the influence of individual Trp-to-KN substitutions, in which only one Trp residue was substituted by KN at a time. From our multiple ~200 ns unfolding simulations, the single KN substitution in the N-td (Trp42 or Trp68) resulted in the unfolding of N-td, similar to the wild-type but with slightly accelerated unfolding speed, while the C-td remained largely folded (QC-td > 0.5, similar to the wild-type) (see Fig. 7a and 7b). On the other hand, the single KN substitution in the C-td (Trp130 or Trp156) greatly reduced the overall stability of HγD-Crys, affecting both N-td and C-td (QC-td reduced to 0.2 in some cases; see Figure 7c and 7d).
Figure 7. The fractions of native contacts for the two domains are plotted against each other.
(a) Single KN mutant at position 42 in N-td. (b) Single KN mutant at position 68 in N-td. (c) Single KN mutant at position 130 in C-td, (d) Single KN mutant at position 156 in C-td.
These results indicate that the C-td is more sensitive to the single position substitution. In fact, the C-td remained largely folded in the wild-type and all the N-td single mutants (native contact faction QC-td ~0.5 at the end of simulations, see Fig. 3a, 7a, and 7b). However, the C-td started to unfold with single mutations in its own domain at either position 130 or 156 (QC-td ~0.2 to 0.4. see Fig. 7c and 7d), and completely unfold in the quadruple KN mutant with two substitutions in C-td (QC-td < 0.1, see Fig. 3b). This clearly shows the accumulation of Trp-to-KN substitutions in the C-td can substantially decrease the stability of the C-td and thus the entire protein, which is also consistent with our previous experimental tryptophan mutagenesis study in GdmCl27. The KN substitutions in the C-td can essentially diminish the unequal structural stability between the N-td and C-td in the wild-type (where C-td is more stable than N-td), and substantially change the unfolding pattern of HγD-Crys protein.
Meanwhile, from our simulation data, it seems unclear which Trp is more important within each domain. For example, in the N-td, similar unfolding processes for N-td (and C-td) were observed for the single KN substitution at position 42 or 68 (representative trajectories shown in Supp. Fig. S7 and S8). Similarly, in the C-td, both single substitutions at position 130 and 156 accelerated the unfolding of C-td (and the entire protein) in a similar manner (see representative trajectories in Supp. Fig. S9 and S10).
Another interesting point to further investigate the importance of these tryptophan residues is to perform a similar but probably more conservative quadruple Trp-to-Phe mutation. Our simulations showed a similar unfolding pattern as compared to the quadruple Trp-to-KN mutant, but to a slightly lesser degree in terms of C-td unfolding (see Supp. Fig. S11). Both the N-td and C-td were unfolded simultaneously in the quadruple Trp-to-Phe mutant as well. This is consistent with our previous experimental Trp-to-Phe mutagenesis studies16,17,27, where the triple Trp-to-Phe mutants were found to be all less stable than the wild-type (denatured under lower concentrations of GdmCl than the wild-type).
Discussion
The stability of Human γD crystallin (HγD-Crys) is critical for retaining the transparency and proper refractive index of the vertebrate eye lens. The UV-induced damage model of cataractogenesis suggests that the aromatic Trp residues could be the key to the folding and stability of lens crystallins9,15,16,17. In this report, we have used large-scale explicit solvent atomistic simulations to examine the stability of HγD-Crys with tryptophan to kynurenine (KN) substitutions by UV damage. Our simulations clearly suggest that the Trp-to-KN substitutions could essentially reduce the stability and increase the unfolding speed of long-lived human γD-crystallin, consistent with our previous tryptophan fluorescence quenching experiments that those Trp residues contribute significantly to the overall stability of HγD-Crys16,17,27. Unlike the sequential unfolding in the wild-type, we found similar unfolding speed in both N-td and C-td of the quadruple KN mutant. Detailed structural analysis shows that the C-td forms more complete hydrophobic central region than the N-td with extra hydrophobic residues (Ile102, Phe104 Ile120, and Leu166) near the Trp130 and Trp156 residues. This could be the main reason why the C-td unfolds much slower than the N-td in the wild-type. However, the extra amino (-NH2) and carbonyl (C = 0) groups in the KN mutant attract more waters and polar sidechains to interact with the hydrophobic central region, thus introducing “water leakages” to the dry central region. Detailed analysis indicates that the unfolding starts from the collapse of Tyr-Trp-Tyr sandwich-like cluster due to the large fluctuations caused by the middle residue substitution (Trp-to-KN). Then the fluctuations rapidly propagate to the “tyrosine corner” and thus break the crucial hydrogen-bond bridging the two β-strands in the Greek key motifs. Single KN substitution at each Trp position indicates that the stability of the C-td is more sensitive to the Trp-to-KN substitutions than the N-td. The single Trp-to-KN substitution in the N-td only affects the N-td, but the C-td remains largely intact; however, the same single Trp-to-KN substitution in the C-td can greatly affect the overall stability of HγD-Crys. Overall, our molecular dynamics simulations combined with previous experimental results provide new insights into the molecular mechanism of the UV-induced cataract formation.
Methods
The x-ray crystal structure of the wild-type human γD-crystallin (PDB entry: 1HK026, 173 residues) was used as the starting structure for molecular dynamics simulations, which have been widely used in studies of biomolecules50,51,52,53,54,55,56. The protein was first solvated in a pre-equilibrated 8 M aqueous urea solution with a box size of 73.1 Å × 73.1 Å × 73.1 Å (1,920 urea and 8,192 water molecules; see Ref 57 for more details). The NAMD258 package was utilized for the MD simulations with the NPT ensemble. The CHARMM (parameter set c32b1) force field59, and the TIP3P water were used as the explicit solvent60,61. The Particle Mesh Ewald (PME) method62 was applied to treat the long-range electrostatic interactions and a 12 Å cutoff was employed for the van der Waals interactions. The force field parameters of kynurenine (KN) were obtained from best matching equivalent amino acid atom types in CHARMM force field (parameters are available upon request). Besides the wild-type, both the quadruple KN substitution (at position of 42, 68, 130, and 156) system and 4 single KN substitution systems were built to investigate the role of overall or individual Trp residue(s) to the stability of human γD-crystallin. In addition, a more conserved quadruple Trp-to-Phe mutant was simulated to further examine the importance of Trp residues and to compare with our previous experimental Trp-to-Phe mutagenesis studies in guanidine hydrochloride27. Both the wild-type and KN mutants of the γD-crystallin systems were equilibrated via a 20,000-step energy minimization, followed by 1-ns NPT equilibrations with 0.5 fs time step at 1 atm and 310 K. First, a control run was performed for the wild-type (and the quadruple Trp-to-KN mutant) at 310 K in pure water. For each system, at least five trajectories were run starting from different initial configurations; the initial structures were randomly picked from the equilibration trajectories. Two alleviated temperatures, 380 K and 425 K, were used to unfold the protein in the wild-type and the quadruple Trp-to-KN mutant due to its unusual stability. The rest single position Trp-to-KN mutants and quadruple Trp-to-Phe mutant were performed only at 425 K to save computational resources. The time step for all production runs was 2.0 fs. The total aggregated simulation time for this study is about 12.0 μs.
Author Contributions
R.Z. and J.A.K. conceived and designed the research. Z.X., Z.Y. and R.Z. co-wrote the manuscript. Z.X., Z.Y. and T.H. carried out the molecular dynamics simulations. Z.X., Z.Y., J.A.K. and R.Z. analyzed the data. All authors discussed the results and commented on the manuscript.
Supplementary Material
Supporting Information
Acknowledgments
We would like to thank Bruce Berne, Payel Das and Seung-gu Kang for helpful discussions. This work was supported by the IBM Blue Gene Program.
References
- Oyster C. W. The Human Eye: Structure and Function (Sinauer Associates, 1999). [Google Scholar]
- Benedek G. B. Cataract as a protein condensation disease - The Proctor Lecture. Invest. Ophthalmol. Vis. Sci. 38, 1911–1921 (1997). [PubMed] [Google Scholar]
- Lerman S. & Borkman R. Spectroscopic evaluation and classification of the normal, aging, and cataractous lens. Ophthalmic. Res. 8, 335–& (1976). [Google Scholar]
- Yu N. T., Barron B. C. & Kuck J. F. R. Distribution of two metabolically related fluorophors in human lens measured by laser microprobe. Exp. Eye. Res. 49, 189–194 (1989). [DOI] [PubMed] [Google Scholar]
- Palme S., Slingsby C. & Jaenicke R. Mutational analysis of hydrophobic domain interactions in gamma B-crystallin from bovine eye lens. Protein Sci. 6, 1529–1536 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. A., Bateman O. A., Jaenicke R. & Slingsby C. Mutation of interfaces in domain-swapped human beta B2-crystallin. Protein Sci. 16, 615–625 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies M. J. & Truscott R. J. W. Photo-oxidation of proteins and its role in cataractogenesis. J. Photoch. Photobio. B 63, 114–125 (2001). [DOI] [PubMed] [Google Scholar]
- Grossweiner L. I. Photochemistry of proteins - A review. Curr. Eye. Res. 3, 137–144 (1984). [DOI] [PubMed] [Google Scholar]
- McCarty C. A. & Taylor H. R. A review of the epidemiologic evidence linking ultraviolet radiation and cataracts. Dev. Ophthalmol. 35, 21–31 (2002). [DOI] [PubMed] [Google Scholar]
- Aarts H. J. M., Lubsen N. H. & Schoenmakers J. G. G. CRYSTALLIN GENE-EXPRESSION DURING RAT LENS DEVELOPMENT. Eur. J. Biochem. 183, 31–36 (1989). [DOI] [PubMed] [Google Scholar]
- Lampi K. J., Shih M., Ueda Y., Shearer T. R. & David L. L. Lens proteomics: Analysis of rat crystallin sequences and two-dimensional electrophoresis map. Invest. Ophthalmol. Vis. Sci. 43, 216–224 (2002). [PubMed] [Google Scholar]
- Sasaki H. et al. High prevalence of nuclear cataract in the population of tropical and subtropical areas. Dev. Ophthalmol. 35, 60–69 (2002). [DOI] [PubMed] [Google Scholar]
- Kolosova N. G., Lebedev P. A., Aidagulova S. V. & Morozkova T. S. OXYS rats as a model of senile cataract. B Exp. Biol. Med. 136, 415–419 (2003). [DOI] [PubMed] [Google Scholar]
- Merriam J. C. et al. An action spectrum for UV-B radiation and the rat lens. Invest. Ophthalmol. Vis. Sci. 41, 2642–2647 (2000). [PubMed] [Google Scholar]
- Chen J., Callis P. R. & King J. Mechanism of the Very Efficient Quenching of Tryptophan Fluorescence in Human gamma D- and gamma S-Crystallins: The gamma-Crystallin Fold May Have Evolved To Protect Tryptophan Residues from Ultraviolet Photodamage. Biochemistry (Mosc). 48, 3708–3716 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Flaugh S. L., Callis P. R. & King J. Mechanism of the highly efficient quenching of tryptophan fluorescence in human gamma D-crystallin. Biochemistry (Mosc). 45, 11552–11563 (2006). [DOI] [PubMed] [Google Scholar]
- Chen J., Toptygin D., Brand L. & King J. Mechanism of the efficient tryptophan fluorescence quenching in human gamma D-crystallin studied by time-resolved fluorescence. Biochemistry (Mosc). 47, 10705–10721 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez S., Aquilina J. A., Jamie J. F., Sheil M. M. & Truscott R. J. W. Novel protein modification by kynurenine in human lenses. J. Biol. Chem. 277, 4867–4873 (2002). [DOI] [PubMed] [Google Scholar]
- Hains P. G. & Truscott R. J. W. Post-translational modifications in the nuclear region of young, aged, and cataract human lenses. J. Proteome Res. 6, 3935–3943 (2007). [DOI] [PubMed] [Google Scholar]
- Hood B. D., Garner B. & Truscott R. J. W. Human lens coloration and aging - Evidence for crystallin modification by the major ultraviolet filter, 3-hydroxy-kynurenine O-beta-D-glucoside. J. Biol. Chem. 274, 32547–32550 (1999). [DOI] [PubMed] [Google Scholar]
- Korlimbinis A., Aquilina J. A. & Truscott R. J. W. Protein-bound and free UV filters in cataract lenses. The concentration of UV filters is much lower than in normal lenses. Exp. Eye. Res. 85, 219–225 (2007). [DOI] [PubMed] [Google Scholar]
- Streete I. M., Jamie J. F. & Truscott R. J. W. Lenticular levels of amino acids and free UV filters differ significantly between normals and cataract patients. Invest. Ophthalmol. Vis. Sci. 45, 4091–4098 (2004). [DOI] [PubMed] [Google Scholar]
- Vazquez S., Parker N. R., Sheil M. & Truscott R. J. W. Protein-bound kynurenine decreases with the progression of age-related nuclear cataract. Invest. Ophthalmol. Vis. Sci. 45, 879–883 (2004). [DOI] [PubMed] [Google Scholar]
- Dolin P. J. Ultraviolet radiation and cataract: a review of the epidemiological evidence. Br. J. Ophthalmol. 78, 478–482 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snytnikova O. A. et al. Tryptophan and kynurenine levels in lenses of Wistar and accelerated-senescence OXYS rats. Mol. Vis. 15, 2780–2788 (2009). [PMC free article] [PubMed] [Google Scholar]
- Basak A. et al. High-resolution X-ray crystal structures of human gamma D crystallin (1.25 angstrom) and the R58H mutant (1.15 angstrom) associated with aculeiform cataract. J. Mol. Biol. 328, 1137–1147 (2003). [DOI] [PubMed] [Google Scholar]
- Kosinski-Collins M. S., Flaugh S. L. & King J. Probing folding and fluorescence quenching in human gamma D crystallin Greek key domains using triple tryptophan mutant proteins. Protein Sci. 13, 2223–2235 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das P., King J. A. & Zhou R. beta-strand interactions at the domain interface critical for the stability of human lens gamma D-crystallin. Protein Sci. 19, 131–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das P., King J. A. & Zhou R. H. Aggregation of gamma-crystallins associated with human cataracts via domain swapping at the C-terminal beta-strands. Proc. Natl. Acad. Sci. U. S. A. 108, 10514–10519 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosinski-Collins M. S. & King J. In vitro unfolding, refolding, and polymerization of human gamma D crystallin, a protein involved in cataract formation. Protein Sci. 12, 480–490 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaugh S. L., Kosinski-Collins M. S. & King J. Contributions of hydrophobic domain interface interactions to the folding and stability of human gammaD-crystallin. Protein Sci. 14, 569–581 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaugh S. L., Kosinski-Collins M. S. & King J. Interdomain side-chain interactions in human gamma D crystallin influencing folding and stability. Protein Sci. 14, 2030–2043 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaugh S. L., Mills I. A. & King J. Glutamine deamidation destabilizes human gamma D-crystallin and lowers the kinetic barrier to unfolding. J. Biol. Chem. 281, 30782–30793 (2006). [DOI] [PubMed] [Google Scholar]
- Mills-Henry I. S. Stability, unfolding, and aggregation of the gamma D and gamma S human eye lens crystallins Ph.D thesis, Massachusetts Institute of Technology, (2007). [Google Scholar]
- Zhou R. H., Huang X. H., Margulis C. J. & Berne B. J. Hydrophobic collapse in multidomain protein folding. Science 305, 1605–1609 (2004). [DOI] [PubMed] [Google Scholar]
- Young T. et al. Dewetting transitions in protein cavities. Proteins. 78, 1856–1869 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy Y. & Onuchic J. N. In Annu. Rev. Biophys. Biomol. Struct. Vol. 35 Annual Review of Biophysics 389–415 (Annual Reviews, 2006). [DOI] [PubMed] [Google Scholar]
- Berne B. J., Weeks J. D. & Zhou R. H. In Annu. Rev. Phys. Chem. Vol. 60 Annual Review of Physical Chemistry 85–103 (Annual Reviews, 2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krone M. G. et al. Role of water in mediating the assembly of Alzheimer amyloid-beta a beta 16–22 protofilaments. J. Am. Chem. Soc. 130, 11066–11072 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. X., Shi B. Y., Lu H. J., Xiu P. & Zhou R. H. Dewetting Transitions in the Self-Assembly of Two Amyloidogenic beta-Sheets and the Importance of Matching Surfaces. J. Phys. Chem. B 115, 11137–11144 (2011). [DOI] [PubMed] [Google Scholar]
- Liu P., Huang X., Zhou R. & Berne B. J. Observation of a dewetting transition in the collapse of the melittin tetramer. Nature 437, 159–162 (2005). [DOI] [PubMed] [Google Scholar]
- Wu Y., Vadrevu R., Kathuria S., Yang X. & Matthews C. R. A tightly packed hydrophobic cluster directs the formation of an off-pathway sub-millisecond folding intermediate in the alpha subunit of tryptophan synthase, a TIM barrel protein. J. Mol. Biol. 366, 1624–1638 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins M. D., Quillin M. L., Hummer G., Matthews B. W. & Gruner S. M. Structural rigidity of a large cavity-containing protein revealed by high-pressure crystallography. J. Mol. Biol. 367, 752–763 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasaiah J. C., Garde S. & Hummer G. In Annu. Rev. Phys. Chem. Vol. 59 Annual Review of Physical Chemistry 713–740 (Annual Reviews, 2008). [DOI] [PubMed] [Google Scholar]
- Collins M. D., Hummer G., Quillin M. L., Matthews B. W. & Gruner S. M. Cooperative water filling of a nonpolar protein cavity observed by high-pressure crystallography and simulation. Proc. Natl. Acad. Sci. U. S. A. 102, 16668–16671 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day R. & Garcia A. E. Water penetration in the low and high pressure native states of ubiquitin. Proteins. 70, 1175–1184 (2008). [DOI] [PubMed] [Google Scholar]
- Hua L., Huang X. H., Liu P., Zhou R. H. & Berne B. J. Nanoscale dewetting transition in protein complex folding. J. Phys. Chem. B 111, 9069–9077 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche J. et al. Cavities determine the pressure unfolding of proteins. Proc. Natl. Acad. Sci. U. S. A. 109, 6945–6950 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha S. K. & Udgaonkar J. B. Direct evidence for a dry molten globule intermediate during the unfolding of a small protein. Proc. Natl. Acad. Sci. U. S. A. 106, 12289–12294 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow C. D., Nguyen N., Pande V. S. & Gruebele M. Absolute comparison of simulated and experimental protein-folding dynamics. Nature 420, 102–106 (2002). [DOI] [PubMed] [Google Scholar]
- Beauchamp K. A., Ensign D. L., Das R. & Pande V. S. Quantitative comparison of villin headpiece subdomain simulations and triplet-triplet energy transfer experiments. Proc. Natl. Acad. Sci. U. S. A. 108, 12734–12739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A. A. & Garcia A. E. Mechanism of enhanced mechanical stability of a minimal RNA kissing complex elucidated by nonequilibrium molecular dynamics simulations. Proc. Natl. Acad. Sci. U. S. A. 109, E1530–E1539 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L. Q., Chen M. G. & Yang W. Random walk in orthogonal space to achieve efficient free-energy simulation of complex systems. Proc. Natl. Acad. Sci. U. S. A. 105, 20227–20232 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z. X., Reddy G., O'Brien E. P. & Thirumalai D. Collapse kinetics and chevron plots from simulations of denaturant-dependent folding of globular proteins. Proc. Natl. Acad. Sci. U. S. A. 108, 7787–7792 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossfield A., Feller S. E. & Pitman M. C. A role for direct interactions in the modulation of rhodopsin by omega-3 polyunsaturated lipids. Proc. Natl. Acad. Sci. U. S. A. 103, 4888–4893 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du D. G., Zhu Y. J., Huang C. Y. & Gai F. Understanding the key factors that control the rate of beta-hairpin folding. Proc. Natl. Acad. Sci. U. S. A. 101, 15915–15920 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R. H., Eleftheriou M., Royyuru A. K. & Berne B. J. Destruction of long-range interactions by a single mutation in lysozyme. Proc. Natl. Acad. Sci. U. S. A. 104, 5824–5829 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips J. C. et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks B. R. et al. CHARMM: the biomolecular simulation program. J. Comput. Chem. 30, 1545–1614 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neria E. & Karplus M. A position dependent friction model for solution reactions in the high friction regime: Proton transfer in triosephosphate isomerase (TIM). J. Chem. Phys. 105, 10812–10818 (1996). [Google Scholar]
- Jorgensen W. L., Chandrasekhar J., Madura J., Impey R. W. & Klein M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983). [Google Scholar]
- Darden T. A., York D. M. & Pedersen L. G. Particle mesh Ewald: An NlogN method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 (1993). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information