SUMMARY
Drosophila Dicer-1 produces microRNAs (miRNAs) from pre-miRNA, whereas Dicer-2 generates small interfering RNAs (siRNAs) from long dsRNA. Alternative splicing of the loquacious (loqs) mRNA generates three distinct Dicer partner proteins. To understand the function of each, we constructed flies expressing Loqs-PA, Loqs-PB, or Loqs-PD. Loqs-PD promotes both endo- and exo-siRNA production by Dicer-2. Loqs-PA or Loqs-PB is required for viability, but the proteins are not fully redundant: a specific subset of miRNAs requires Loqs-PB. Surprisingly, Loqs-PB tunes where Dicer-1 cleaves pre-miR-307a, generating a longer miRNA isoform with a distinct seed sequence and target specificity. The longer form of miR-307a represses glycerol kinase and taranis mRNA expression. The mammalian Dicer-partner TRBP, a Loqs-PB homolog, similarly tunes where Dicer cleaves pre-miR-132. Thus, Dicer-binding partner proteins change the choice of cleavage site by Dicer, producing miRNAs with target specificities different from those made by Dicer alone or Dicer bound to alternative protein partners.
INTRODUCTION
In Drosophila melanogaster, distinct enzymes generate 21 nt small interfering RNAs (siRNAs) and ~21–24 nt microRNAs (miRNAs). miRNA production begins with cleavage of an RNA polymerase II primary transcript by the RNase III enzyme Drosha, aided by its dsRNA-binding partner protein, Pasha. Drosha cleavage liberates a ~60–70 nt long pre-miRNA (Lee et al., 2003; Denli et al., 2004; Gregory et al., 2004; Han et al., 2004, 2006). A second RNase III enzyme, Dicer-1 (Dcr-1), then cleaves the pre-miRNA to release a duplex comprising the mature miRNA and its miRNA*, a partially complementary small RNA derived from the opposite arm of the pre-miRNA stem. Just as Pasha collaborates with Drosha, dsRNA-binding partner proteins assist Dicers in the production of small RNAs. In Drosophila, Dcr-1 partners with Loquacious-PA (Loqs-PA) or Loquacious-PB (Förstemann et al., 2005; Saito et al., 2005; Jiang et al., 2005).
Drosophila Dicer-2 (Dcr-2) generates siRNAs (Lee et al., 2004). Dcr-2 forms a 1:1 complex with R2D2, a paralog of Loqs (Liu et al., 2003; Tomari et al., 2004). Both R2D2 and free phosphate suppress the inherent ability of Dcr-2 to process pre-miRNAs into 21 nt duplexes, restricting it to the longer dsRNA triggers associated with RNAi (Cenik et al., 2011). The Dcr-2: R2D2 heterodimer also senses the thermodynamic asymmetry of siRNA duplexes and, together with chaperone proteins, loads them into Argonaute2 (Ago2; Liu et al., 2003, 2006; Tomari et al., 2004, 2007; Pham and Sontheimer, 2005). Dcr-2 produces siRNAs from both endogenous and exogenous double-stranded RNA (dsRNA). Exogenous siRNAs (exo-siRNAs) in flies are thought to derive from long dsRNA molecules generated during viral replication (Ding, 2010) or long dsRNA introduced experimentally (Kennerdell and Carthew, 1998, 2000; Lee and Carthew, 2003). In flies and mammals, endogenous siRNAs (endo-siRNAs) derive from self-complementary hairpin transcripts (esiRNAs), convergent mRNAs (cis-NAT endo-siRNAs), or mobile elements (Yang and Kazazian, 2006; Czech et al., 2008; Ghildiyal et al., 2008; Kawamura et al., 2008; Okamura et al., 2008a, 2008b; Tam et al., 2008; Watanabe et al., 2008).
Production of esiRNAs by Dcr-2 in cultured Drosophila S2 cells requires the alternative partner protein, Loqs-PD, rather than R2D2 (Okamura et al., 2008b; Hartig et al., 2009; Zhou et al., 2009; Miyoshi et al., 2010; Hartig and Förstemann, 2011). However, it is not known whether the production of exo-siRNAs, cis-NAT-endo-siRNAs, and mobile element-derived endo-siRNAs also requires Loqs-PD. Moreover, an in vivo role for Loqs-PD in small RNA production has not been established.
The loqs gene generates four mRNA isoforms by alternative splicing: loqs-RA, loqs-RB, loqs-RC, and loqs-RD. The four mRNAs are predicted to produce four distinct proteins, Loqs-PA, Loqs-PB, Loqs-PC, and Loqs-PD (Figure 1A; Förstemann et al., 2005; Saito et al., 2005; Jiang et al., 2005; Zhou et al., 2009; Hartig et al., 2009; Miyoshi et al., 2010). The Loqs-PC protein has not yet been detected in any fly cell or tissue. In theory, an intron in the 3′ UTR of loqs-RC may target the mRNA for nonsense-mediated decay (Moore and Proudfoot, 2009).
Figure 1. Loqs-PB is Necessary and Sufficient to Maintain Female Germline Stem Cells.
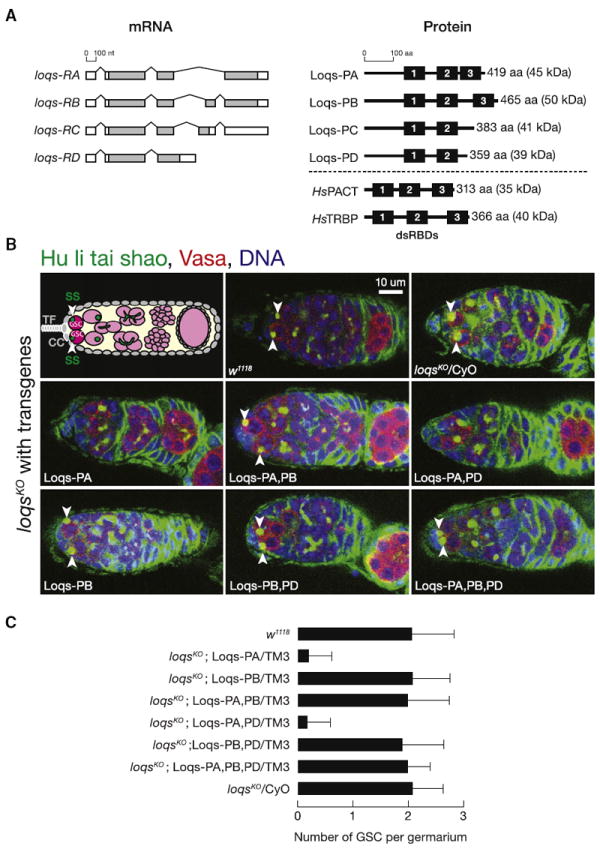
(A) Drosophila and human Dicer partners.
(B) Drosophila germarium architecture. TF, terminal filament. CC, cap cells. SS, spectrosomes. Green, fusomes. Somatic cells are gray, germline are pink. Ovaries from 10-day-old females were stained with anti-Hu li tai shao (Hts) antibody (green) to detect the spectrosome and fusome, anti-Vasa antibody (red) to identify germ cells, and DAPI (blue) to highlight DNA. White arrowheads mark GSCs.
(C) The number of GSCs per germarium (mean ± SD) was measured for each genotype in 10–15 randomly selected germaria from pooled ovaries from >30 flies. Eight z axis images, spanning 20 μm, were acquired for each germarium. See also Figure S1 and Table S1.
The largest Loqs protein isoform, Loqs-PB, comprises three double-stranded RNA-binding domains (dsRBDs); dsRBD3 is required for Loqs-PB to bind to Dcr-1 (Förstemann et al., 2005; Ye et al., 2007). Loqs-PA, which also binds to Dcr-1, lacks a part of the third dsRBD of Loqs-PB. Loqs-PD has a unique carboxy terminal sequence in place of dsRBD3 and binds to Dcr-2 rather than Dcr-1. Thus, loqs encodes at least three distinct proteins that act in either the miRNA or siRNA pathways (Förstemann et al., 2005; Jiang et al., 2005; Saito et al., 2005; Hartig et al., 2009; Zhou et al., 2009; Marques et al., 2010; Miyoshi et al., 2010; Hartig and Förstemann, 2011).
Small silencing RNA production in mammals is also catalyzed by RNase III enzymes assisted by dsRBD partner proteins. Mammalian Drosha acts with DGCR8, which contains two dsRBDs (Gregory et al., 2004). Mammalian Dicer, which produces both miRNAs and siRNAs, collaborates with TRBP and PACT, partner proteins with three dsRBDs (Figure 1A; Chendrimada et al., 2005; Gregory et al., 2004; Haase et al., 2005; Lee et al., 2006). Partnerships between dicer-like proteins and dsRBD proteins are a general theme in RNA silencing pathways. For example, plants produce four distinct Dicer enzymes, each with its own specialized dsRBD partner (Vazquez et al., 2004; Hiraguri et al., 2005; Eamens et al., 2009).
The importance of such partnerships is well established, but the individual molecular and biological functions of dsRBD partner proteins are largely unknown. Here, we define the specific functions of the three Loqs partner proteins. Using isoform-specific rescue transgenes, we reconstituted loqs knockout (loqsKO) flies (Park et al., 2007) with one, two, or all three Loqs proteins. We find that Loqs-PA and Loqs-PB, but not Loqs-PD, can rescue the lethality of loqs null mutant flies. Loqs-PB is crucial for female fertility and for producing specific subsets of miRNAs. Loqs-PD enhances the production of both endo-and exo-siRNAs. In the absence of Loqs-PB, Dcr-1 produces aberrant products from pre-miR-307a, pre-miR-87, and pre-miR-316. miR-307a and miR-87 reside in the 3′ arm of their pre-miRNA, so Dcr-1 cleavage defines their 5′ end. In fly ovaries, Loqs-PB is required for Dcr-1 to generate miR-307a bearing the correct seed sequence to repress its targets glycerol kinase (Gk) and taranis (tara). Finally, in vitro dicing assays and the sequences of miRNAs from Trbp−/− mouse embryonic fibroblasts (MEFs) suggest that the mammalian Loqs ortholog TRBP similarly acts to ensure the correct choice of pre-miRNA cleavage sites by Dicer.
RESULTS
loqs Isoform-Specific Flies
To define the in vivo function of each loqs isoform, we constructed flies harboring a single-copy transgene encompassing the cDNA of each loqs isoform under the control of the endogenous loqs promoter (Figure S1A). Double and triple transgene strains (Loqs-PA,PB; Loqs-PA,PD; Loqs-PB,PD; and Loqs-PA,PB,PD) were created by meiotic recombination (Figure S1B). The transgenic, FLAG-tagged Loqs-PA, Loqs-PB, and Loqs-PD proteins were expressed at levels equal to or greater than the level of the corresponding endogenous protein in genetically matched control (w1118) or loqsKO/CyO heterozygous flies (Figure S1C). loqsKO mutant flies are embryonic lethal (Park et al., 2007). Loqs-PA or Loqs-PB, but not Loqs-PD, restored embryonic viability, suggesting that a defect in miRNA biogenesis underlies the lethality observed in loqsKO embryos.
Loqs-PB Is Necessary and Sufficient for Germline Stem Cell Maintenance
Male loqsKO adult flies rescued with either Loqs-PA or Loqs-PB were as fertile as control flies (w1118 or loqsKO/CyO; Table S1). In contrast, female loqsKO flies rescued with either Loqs-PA (4.3 eggs per female per day, none of which hatched) or Loqs-PA together with Loqs-PD (6.0 eggs per female per day, 3% hatched) laid far fewer eggs than w1118 or loqsKO/CyO (~36–45 eggs per female per day, of which 48%–54% hatched; Table S1). Few of the eggs laid by loqsKO;Loqs-PA/TM3 (6%) or loqsKO; Loqs-PA,PD/TM3 (14%) females had normal dorsal appendages; 88%–91% of the eggs laid by w1118 or loqsKO/CyO females had normal dorsal appendages. In contrast, Loqs-PB alone was sufficient to rescue the number of eggs laid per female per day (38), the rate of hatching (53%), and patterning of the dorsal appendages (91% normal; Table S1).
loqsf00791/loqsKO (loqsf00791 is a hypomorphic allele) ovaries fail to maintain their germline stem cells (GSCs). A transgene expressing Loqs-PB but not a transgene expressing Loqs-PA rescues the GSC loss (Park et al., 2007). However, these experiments preceded the discovery of Loqs-PD and its role in endo-siRNA biogenesis, leaving open the possibility that Loqs-PA in combination with Loqs-PD might rescue GSC maintenance. To test this possibility, we counted the number of germline stem cells (GSCs) per germarium. Ovaries were immuostained to detect Hu Li Tai Shao (Hts), a component of the GSC-specific spectrosome (Lin et al., 1994) and Vasa, which marks germ cells (Figure 1B). GSCs were identified by their anterior-positioned, round spectrosome and by their apposition to somatic cap cells or terminal filament cells, which lack Vasa. Control (w1118) flies had 2.1 ± 0.8 GSCs per germarium (mean ± SD; Figure 1C), whereas loqsKO; Loqs-PA/TM3 and loqsKO;Loqs-PA,PD/TM3 flies had fewer (0.2 ± 0.4 GSCs per germaria for each). Ovaries from all genotypes expressing Loqs-PB had a normal complement of GSCs: loqsKO;Loqs-PB/TM3 (2.1 ± 0.7), loqsKO;Loqs-PA,PB/TM3 (2.0 ± 0.7), loqsKO;Loqs-PB,PD/TM3 (1.9 ± 0.7), loqsKO;Loqs-PA,PB,PD/TM3 (2.0 ± 0.4), and loqsKO/CyO (2.1 ± 0.5).
Our data support earlier conclusions that among the Loqs isoforms, Loqs-PB alone provides the necessary and sufficient Loqs function to maintain GSCs (Park et al., 2007). Consistent with the idea that Loqs-PA and Loqs-PB have distinct functions in the production of miRNAs, the relative abundance of their mRNAs, loqs-RA and loqs-RB, varies widely among tissues and developmental stages (Figure S1D). For example, loqs-RA is lower at earlier stages of development and increases with developmental time. In adult flies, testes predominantly express loqs-RA, whereas ovaries express more loqs-RB than loqs-RA (Figure S1D).
Normal Exo-siRNA Accumulation Requires Loqs-PD In Vivo
In cultured Drosophila S2 cells, Loqs-PD is required for efficient esiRNA production (Hartig et al., 2009; Zhou et al., 2009; Miyoshi et al., 2010). Does Loqs-PD play a role in RNAi in adult flies?
The GMR-wIR transgene produces during eye development an inverted repeat hairpin RNA corresponding to white exon 3. Dcr-2 processes the wIR hairpin into siRNAs (Lee and Carthew, 2003; Vagin et al., 2006), which in turn silence white expression, causing the eye to be white or orange instead of red. Because white encodes an ABC transporter required for the entry of red pigment precursor into cells, the reduction in the absorbance at 480 nm of pigment extracted from the eye provides a quantitative measure of white silencing. In wild-type flies (Canton S) and flies heterozygous for the RNAi pathway genes dcr-2, r2d2, or ago2, the wIR transgene produced white eyes (A480 = 0.02–0.03; Figure 2A, females; Figure S2A, males). In contrast, the eyes from wIR transgenic female flies homozygous for dcr-2, r2d2, or ago2 mutations (A480 = 0.9–1.0) were similar to those of wild-type (mean A480 = 1.2).
Figure 2. Loqs-PB Helps Dcr-1 Produce a Subset of miRNAs, Whereas Loqs-PD Helps Dcr-2 Produce Endo-siRNAs and Exo-siRNAs.
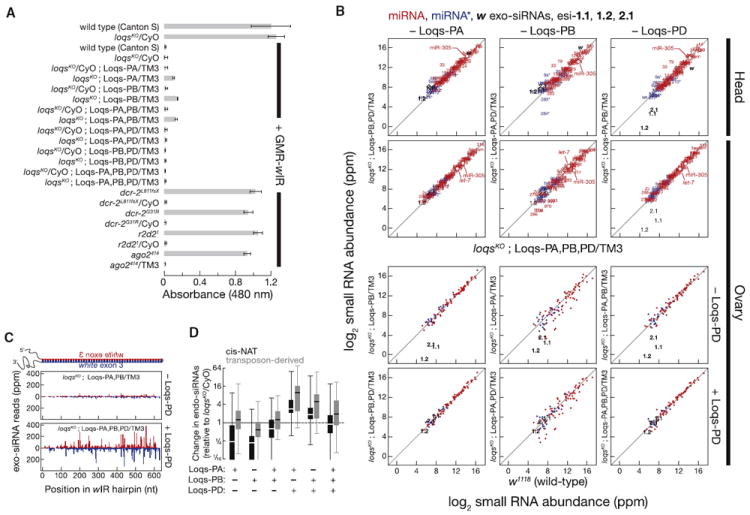
(A) Silencing of white mRNA by a long hairpin RNA trigger corresponding to white exon 3 (GMR-wIR) was measured by using eye pigment from 3- to 4-day-old female fly eyes. Data are mean ± SD for five biological replicates. Figure S2A presents data from male heads.
(B–D) Sequencing of small RNAs from female heads expressing the wIR RNAi trigger and from ovaries.
(B) Normalized number of reads (ppm) for esi-1.1, esi-1.2, and esi-2.1, white siRNAs, miRNA (red) and miRNA* (blue) from fly heads and ovaries. Only miRNAs and miRNA* strands whose abundance was > 100 ppm in at least one library were analyzed. Figure S2C presents additional head data.
(C) Abundance and 5′ position of wIR-derived siRNAs in heads. Antisense siRNAs are shown in red, sense in blue.
(D) Change in cis-NAT and transposon-derived endo-siRNA abundance in heads.
Hypomorphic loqsf007911 homozygous mutant flies show a quantitative defect in white silencing (Förstemann et al., 2005). loqsKO homozygous mutant female flies bearing the wIR and rescued with Loqs-PA or Loqs-PB or both showed similar quantitative defects in white silencing (Figure 2A), as evidenced by their significantly higher 480 nm absorbance (mean ± SD ranged from 0.11 ± 0.00 to 0.16 ± 0.01 with p values from 5.9 × 10−11 to 6.3 × 10−5). The Loqs-PD transgene (loqsKO; Loqs-PA,PD/TM3, loqsKO;Loqs-PB,PD/TM3, or loqsKO;Loqs-PA,PB,PD/TM3) quantitatively rescued silencing (A480 = 0.01 ± 0.01 for all three genotypes; Figure 2A). Similar results were obtained by using male flies (Figure S2A). Thus, Loqs-PD significantly (p value = 1.6 × 10−2, loqsKO;Loqs-PA,PB/TM3 compared to loqsKO;Loqs-PA,PB,PD/TM3) enhanced RNAi triggered by long dsRNA in vivo.
Next, we sequenced 18–29 nt RNA (Tables S2A and S2B) from the heads of female flies bearing the wIR transgene. The steady-state level of white exo-siRNAs produced from the wIR decreased in loqsKO;Loqs-PA/TM3, loqsKO;Loqs-PB/TM3, and loqsKO;Loqs-PA,PB/TM3 rescue flies, compared with wild-type or heterozygous controls (Figures 2B and S2C). In contrast, loqsKO;Loqs-PA,PD/TM3, loqsKO;Loqs-PB,PD/TM3, and loqsKO; Loqs-PA,PB,PD/TM3 flies had normal or elevated amounts of white siRNAs. Loqs-PD appeared to play no role in miRNA accumulation: we detected no significant change (p value ≫ 0.05; Wilcoxon test) in the global abundance of miRNAs and miRNA* strands in the absence of Loqs-PD (loqsKO;Loqs-PA,PB/TM3 versus loqsKO/CyO). Northern hybridization of RNA from the heads of adult females expressing the wIR confirmed the sequencing results (Figure S2B). Loss of Loqs-PD decreased the abundance of white siRNA reads across the entire wIR hairpin sequence for both sense and antisense siRNAs, suggesting a sequence-independent role for Loqs-PD in facilitating exo-siRNA production by Dcr-2 (Figure 2C).
Endo-siRNA Accumulation Requires Loqs-PD
esi-1.1, esi-1.2, and esi-2.1 are the major endo-siRNAs derived from structured loci, genes that produce long, repetitive transcripts with extensive intramolecular base pairing. Like exo-siRNAs, the steady-state levels of these three esiRNAs decreased in loqsKO mutant flies rescued with Loqs-PA or Loqs-PB or both, compared with loqsKO/CyO heterozygotes (Figures 2B, S2B, and S2C). In fact, loqsKO;Loqs-PA,PB/TM3 flies accumulated less esi-2.1 than ago2414 homozygous null mutants, suggesting a critical role for Loqs-PD in esiRNA production in vivo (Figure S2B). The presence of the Loqs-PD transgene rescued the abundance of these three esiRNAs in heads (Figures 2B, S2B, and S2C). esiRNA accumulation in ovaries similarly required Loqs-PD (Figures 2B, and S3 and Tables S3A and S3B).
Cis-NAT endo-siRNA accumulation also required Loqs-PD in heads and ovaries (Figures 2D and S2D). Our data were more equivocal for transposon-derived endo-siRNAs: these decreased when Loqs-PB was the only isoform present, but were unaltered compared to loqsKO/CyO heterozygotes when Loqs-PA was expressed (Figure 2D). In contrast, overexpression of Loqs-PD increased both cis-NAT and transposon-derived endo-siRNAs, suggesting that Loqs-PD acts in the production of transposon-derived endo-siRNAs as it does for exo-siRNA, esiRNA, and cis-NAT endo-siRNA biogenesis.
The absence of Loqs-PD altered the efficiency of dicing, but did not alter the choice of cleavage site by Dcr-2. Without Loqs-PD, fewer siRNAs accumulated, but those that remained were predominantly 21 nt long for white exo-siRNAs and 22 nt for esi-2.1. In heads, 84% of white siRNAs were 21 nt in loqsKO/CyO heterozygotes, whereas 78% were 21 nt in loqsKO;Loqs-PA,PB/TM3 flies. For esi-2.1, 81% were 22 nt in loqsKO/CyO heterozygotes; 75% were 22 nt in loqsKO Loqs-PA,PB/TM3 flies.
Loqs-PD Decreases the KM of Dcr-2 for Pre-esiRNA
To understand how Loqs-PD enhances steady-state accumulation of endo-siRNAs in vivo, we analyzed the processing of an esiRNA precursor (pre-esi-2.1) by purified, recombinant Dcr-2 supplemented with purified, recombinant Loqs-PD (Table S4 and Figure S4A). Adding Loqs-PD decreased the KM of Dcr-2 from 135 ± 32 nM to 31 ± 5 nM (p value = 0.028). In contrast, the kcat of Dcr-2 alone (0.073 ± 0.003 min−1) was not significantly changed (p value > 0.05) by the addition of Loqs-PD (0.085 ± 0.007 min−1). Thus, Loqs-PD decreased the substrate concentration at which Dcr-2 was half-maximally active, but had no detectable effect on the rate of enzyme turnover. Overall, Loqs-PD increased enzyme efficiency (kcat/KM) ~5-fold. Loqs-PD similarly enhances KM (decreased ~14-fold) but not kcat for Dcr-2 processing long dsRNA (Cenik et al., 2011). We conclude that Loqs-PD functions in both endo-siRNA and exo-siRNA production by decreasing the concentration of substrate required for Dcr-2 to efficiently produce siRNA.
Loqs-PB Contributes Differentially to the Production of Individual miRNAs
loqs was discovered as a gene required for efficient miRNA production in vivo. Either Loqs-PA or Loqs-PB suffices to rescue the embryonic lethality observed in loqsKO homozygotes, and in ovaries some miRNAs such as let-7, miR-1, miR-184, miR-263a, miR-312, or miR-996, accumulated to wild-type levels when either Loqs-PA or Loqs-PB was the only Loqs protein present (Figures 2B and S3A–S3C). In contrast, the abundance of miRNAs such as miR-79, miR-283, miR-305, miR-311, and miR-318 decreased in ovaries of loqsKO mutant flies rescued with Loqs-PA but achieved wild-type levels when rescued with Loqs-PB. If Loqs-PB enhances the processing of these pre-miR-NAs by Dcr-1 in vivo, then their pre-miRNAs might accumulate in the absence of Loqs-PB. Although pre-miRNA accumulation is diagnostic for a defect in dicing, a failure to accumulate pre-miRNA could suggest that Loqs-PB acts at a step after dicing but might simply indicate that the pre-miRNA is unstable. Neither pre-miR-283 nor pre-miR-305 accumulated in loqsKO;Loqs-PA/TM3 flies, relative to loqsKO;Loqs-PB/TM3 or in w1118 (Figure S3A). In contrast, miR-79, miR-311, and miR-318 decreased and their corresponding pre-miRNAs increased in loqsKO;Loqs-PA/TM3, compared with loqsKO;Loqs-PB/TM3 and w1118 (Figure S3A). For these miRNAs, our data suggest that Loqs-PB enhances the Dcr-1-catalyzed conversion of pre-miRNAs to miRNA/miRNA* duplexes. Similarly, in heads from loqs mutant flies rescued with Loqs-PA rather than Loqs-PB, the abundance of miR-277 and miR-305 declined and the corresponding pre-miRNAs accumulated (Figures 2B and S2B).
Loqs-PB Decreases the KM and Increases the kcat of Dcr-1 for Pre-miRNA
To understand how Loqs-PB contributes to the processing of miRNAs like miR-305, we measured the KM and kcat of purified, recombinant Dcr-1 and of Dcr-1 supplemented with purified, recombinant Loqs-PA or Loqs-PB, using pre-miR-305 and pre-let-7 as substrates (Table S4 and Figures S4B and S4C). For pre-miR-305, Loqs-PB decreased the KM (Dcr-1 alone, KM = 2.2 ± 0.1 nM versus Dcr-1 + Loqs-PB, KM = 1.4 ± 0.1 nM; p value = 5.5 × 10−4) and increased turnover (Dcr-1 alone, kcat = 0.11 ± 0.00 min−1 versus Dcr-1 + Loqs-PB, kcat = 0.15 ± 0.00 min−1, p value = 2.4 × 10−4), more than doubling dicing efficiency (i.e., kcat/KM). In contrast, Loqs-PA decreased KM (Dcr-1 + Loqs-PA, KM = 1.5 ± 0.3 nM, p value = 0.030), but did not alter kcat (Dcr-1 + Loqs-PA, kcat = 0.11 ± 0.00 min−1, p value > 0.05).
For pre-let-7, Loqs-PB decreased the KM of Dcr-1 (Dcr-1, KM = 3.0 ± 0.2 nM versus Dcr-1 + Loqs-PB, KM = 1.8 ± 0.2 nM, p value = 2.0 × 10−3) without altering turnover (Dcr-1, kcat = 0.52 ± 0.01 min−1 versus Dcr-1 + Loqs-PB, kcat = 0.49 ± 0.02 min−1, p value > 0.05). For pre-let-7, Loqs-PA did not detectably alter either KM (Dcr-1 + Loqs-PA, KM = 2.7 ± 0.7 nM, p value > 0.05) or enzyme turnover (Dcr-1 + Loqs-PA, kcat = 0.51 ± 0.02 min−1, p value > 0.05). Our data suggest a model in which Loqs-PB makes a greater contribution than Loqs-PA to the dicing of “difficult” pre-miRNA substrates such as pre-miR-305, perhaps because for these pre-miRNAs, Loqs-PB enhances Dcr-1 substrate binding while favoring product release.
Loqs-PB Tunes Where Dcr-1 Cleaves Some Pre-miRNAs
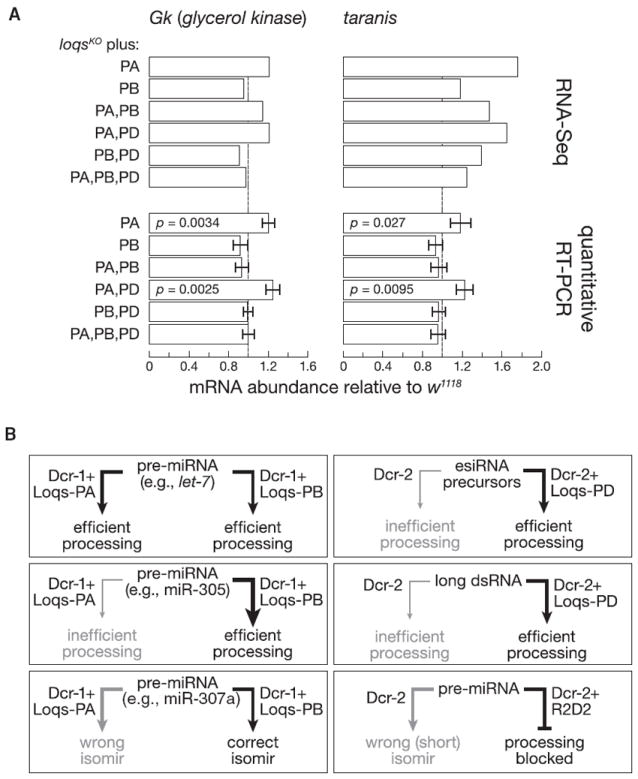
Dicing by Dcr-1 determines miRNA length. In addition to showing changes in miRNA abundance, sequencing of loqsKO mutants rescued with Loqs protein isoforms revealed changes in miRNA isoform (isomir) distributions for several miRNAs. In particular, the mean length of three miRNAs in ovaries differed between loqsKO mutants rescued with Loqs-PA versus Loqs-PB: in the absence of Loqs-PB, miR-307a was shorter and miR-316 and miR-9b were longer (Figure 3A–3D). miR-307a was also shorter and miR-316 was longer in loqsf00791 hypomorphic mutants.
Figure 3. Loqs-PB Changes the Size Distribution of Small RNAs Cleaved from Pre-miR-307a In Vivo.
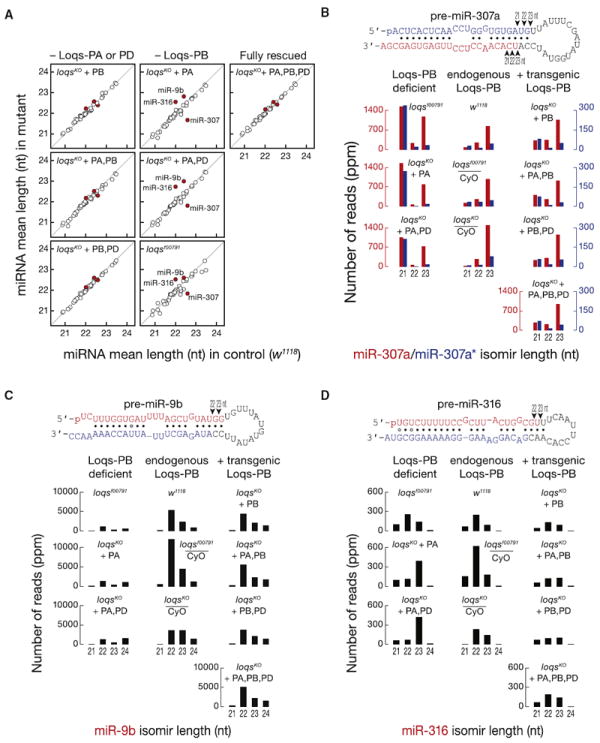
The sequence, length, and abundance of ovary small RNAs from control and mutant females bearing Loqs-expressing transgenes was measured by deep sequencing.
(A) Mean length of all miRNA isomirs in the transgenic, mutant flies compared to control (w1118). Only miRNAs whose total isomir abundance was more than 100 ppm in w1118 were analyzed.
(B) Normalized abundance of each isomir of miR-307a* (blue) and miR-307a (red) for each genotype. Here and below, we considered only isomirs whose sequence variation reflected a change solely in the site Dicer cleaved.
(C) Abundance of miR-9b isomirs.
(D) Abundance of miR-316 isomirs.
See also Figure S5.
miR-307a resides in the 3′ arm of its pre-miRNA, so Dcr-1 defines its 5′ end. miR-307a isomirs differ mainly at their 5′ ends, a difference that alters their seed sequences, changing their repertoire of mRNA targets. In both ovaries (Figure 3B) and heads (Figure S5A), flies lacking Loqs-PB accumulate a 21 nt long miR-307a isomir (miR-307a21-mer) whose seed sequence begins at position 4 of the canonical 23 nt long miR-307a isomir (miR-307a23-mer). loqsKO flies rescued with Loqs-PB, w1118 flies, and loqsKO/CyO heterozygotes produce mainly miR-307a23-mer (Figures 3B, S3B, and S5A). The absence of Loqs-PB also alters the 5′ end of the less abundant (i.e., <100 ppm) miR-87, decreasing the canonical 24 nt miR-8724-mer and increasing the 23 nt miR-8723-mer in both heads and ovaries (Figures S5C and S5D). miR-87 resides in the 3′ arm of pre-miR-87, so miR-8724-mer and miR-8723-mer have different seed sequences.
miR-316 and miR-9b reside in the 5′ arm of their pre-miRNAs, so Dcr-1 cleavage defines their 3′ ends. loqsKO mutants rescued with Loqs-PA produce miR-316 and miR-9b isomirs that differ from the canonical miRNA at their 3′ termini (Figures 3C, 3D, and S5B), so their seed sequences are unchanged.
The different lengths of these miRNAs in loqsKO;Loqs-PA/TM3 and loqsKO;Loqs-PB/TM3 flies likely reflects a direct influence of Loqs-PB on the choice of cleavage site by Dcr-1. We incubated pre-miR-307a with Dcr-1 alone or Dcr-1 supplemented with Loqs-PA or Loqs-PB. (Figures 4 and S6A–S6C). Dcr-1 alone and Dcr-1 + Loqs-PA produced mostly 21 nt miR-307a; more 22 and 23 nt isomirs were made when Dcr-1 was supplemented with Loqs-PB (Figures 4 and S6A and S6B). Similarly, miR-307a*, which derives from the 5′ arm of pre-miR-307a, was longer when Loqs-PB, but not Loqs-PA, was included (Figures 4A–4C and S6A–S6C). The addition of Loqs-PA or Loqs-PB had no effect on the length of let-7 or let-7* produced by Dcr-1 from pre-let-7 (Figures 4A and 4B). Our data suggest that Loqs-PB repositions pre-miR-307a on Dcr-1 so as to favor production of miR-307a23-mer over miR-307a21-mer.
Figure 4. Purified Loqs-PB, but Not Loqs-PA, Tunes the Length of Small RNAs from Pre-miR-307a.
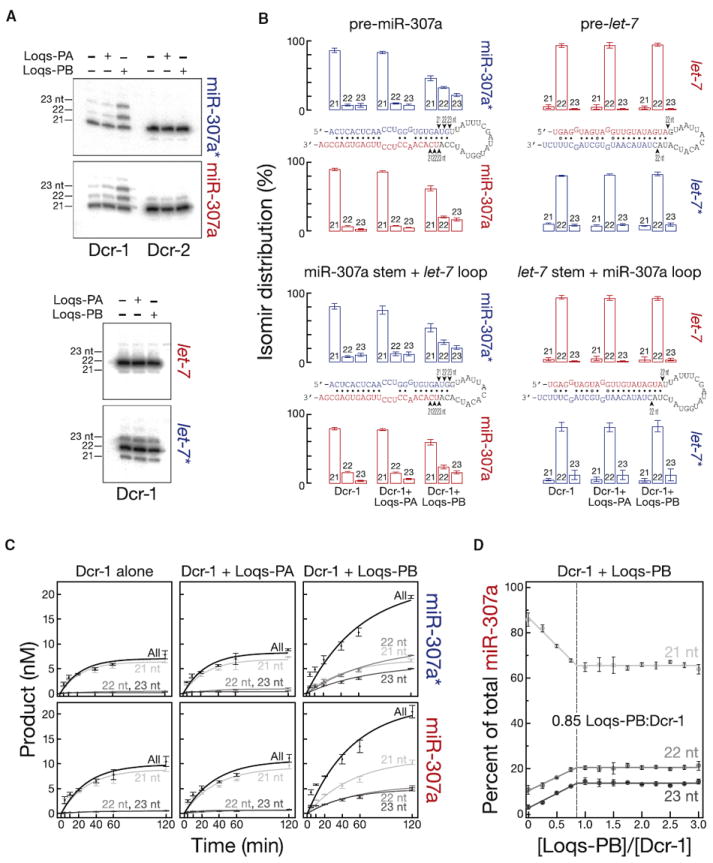
(A) Pre-miR-307a and pre-let-7 (100 nM) were incubated with recombinant Dcr-1 or Dcr-2 (10 nM) supplemented with Loqs-PA or Loqs-PB (10 nM) for 2 hr. The products were resolved by denaturing polyacrylamide electrophoresis and detected by northern hybridization.
(B) Quantification of three independent replicates of the experiment in (A) and Figures S6H and S6I. Chimeric pre-miRNA comprised the pre-miR-307a stem and the pre-let-7 loop or the pre-let-7 stem and the pre-miR-307a loop.
(C) Pre-miR-307 (30 nM) was incubated with purified, recombinant Dcr-1 alone (8 nM) or supplemented with Loqs-PA or Loqs-PB (8 nM). For sites of the 32P-radiolabel see Figure S6A.
(D) The stoichiometry of Loqs-PB in complex with Dcr-1 was determined by titration. Products were detected by quantitative northern hybridization. The inflection point in the curves, 0.85 molecules of Loqs-PB per molecule of Dcr-1, suggests the two proteins form a 1:1 complex.
Data are reported as mean ± SD for three independent trials. See also Figures S6 and S7.
Inclusion of Loqs-PB similarly favored production of miR-8724-mer over miR-8723-mer from pre-miR-87 (Figures S6D and S6E). In contrast, Loqs-PB favored production from pre-miR-316 of a shorter, 22 nt isomir of miR-316 (Figure S7); miR-8724-mer and the shorter isomir of miR-316 predominate in vivo in wild-type flies (Figures 3D, S5B, S5C, and S5D).
The effect of Loqs-PB on cleavage site choice within the pre-miRNA stem was specific for Dcr-1. Neither Loqs-PA nor Loqs-PB affected the cleavage site chosen by Dcr-2 (Figure 4A).
Loqs-PB Forms a 1:1 Complex with Dcr-1
The precise stoichiometry between Dicer partners and Dicer has been established only for the 1:1 R2D2:Dcr-2 heterodimer (Tomari et al., 2004, Supporting Online Material, page 14). We used titration to estimate the stoichiometry of Loqs-PB bound to Dcr-1 during pre-miRNA processing. We incubated 5 nM Dcr-1 with increasing concentrations of Loqs-PB (0–15 nM) and 100 nM pre-miR-307a and measured the isomirs produced (Figures 4D, S6B, and S6C). After 2 hr, 86% ± 3% of the miR-307a made by Dcr-1 alone was the 21 nt isoform (Figure 4D). The percentage of 22 and 23 nt miR-307a isomirs increased with increasing Loqs-PB, then plateaued once Dcr-1 and Loqs-PB were essentially equimolar. The results were similar when we monitored the production of miR-307a* isomirs (Figures S6B and S6C). We conclude that Dcr-1 and Loqs-PB form a 1:1 complex.
Tuning of Dcr-1 Cleavage by Loqs-PB Requires the Pre-miR-307a Stem but Not the Loop
To begin to determine the features required for Loqs-PB to alter where Dcr-1 cleaves pre-miR-307a, we created two chimeric pre-miRNAs: one combined the pre-miR-307a stem and the pre-let-7 loop, the other combined the pre-let-7 stem and the pre-miR-307a loop (Figure 4B, S6H, and S6I). Loqs-PB changed the site at which Dcr-1 cleaved the chimera containing the pre-miR-307a stem but not the chimera with the pre-let-7 stem. Thus, the pre-miR-307a stem, not the loop, is necessary and sufficient for Loqs-PB to redirect where Dcr-1 cleaves.
Loqs-PB Does Not Require a 5′ Terminal Phosphate to Influence Dcr-1 Cleavage Site Choice
Both human Dicer and Drosophila Dcr-1 use the 5′ terminal phosphate to align a pre-miRNA in their active sites (Park et al., 2011); without a 5′ phosphate, cleavage site choice is less precise. To ask whether Loqs-PB requires a 5′ terminal phosphate to influence the choice of cleavage site by Dcr-1, we compared the products from 5′ phosphorylated (Figures 4A, 4B, S6A, and S6B) and hydroxy pre-miR-307a (Figures S6J and S6K). Loqs-PB did not require a 5′ terminal phosphate to influence where Dcr-1 cuts pre-miR-307a: for Dcr-1 alone, 66% ± 1% of 5′ hydroxy miR-307a* was 21 nt and 25% ± 1% was 23 nt, but when Dcr-1 was supplemented with Loqs-PB, 24% ± 2% of 5′ hydroxy miR-307a* was 21 nt and 65% ± 4% was 23 nt (Figures S6J and S6K).
miR-307a21-mer and miR-307a23-mer Have Distinct Target Specificities
miR-307a21-mer and miR-307a23-mer have overlapping but distinct seed sequences. To test whether both miR-307a isoforms are functional and whether they have different target specificities, we measured their ability to silence a Renilla reniformis (Rr) luciferase reporter in cultured Drosophila S2 cells.
Endogenous miR-307a in S2 cells is below the limit of detection of northern hybridization (data not shown), and only 28 pre-miR-307a-mapping sequence reads per million (ppm) were detected in S2 cells (Han et al., 2011). For comparison, the tenth most abundant miRNA in S2 cells, miR-305, was present at 11,523 ppm in the same library, and 46 miRNAs were more abundant than miR-307a. The low level of miR-307a allowed us to introduce the 21-mer and 23-mer isoforms as miR-307a/miR-307a* duplexes in S2 cells by transfection.
We constructed Rr luciferase reporters containing in their 3′ UTRs six tandem, bulged sites (i.e., fully complementary sites containing mismatches at positions 9–11; Figure 5A). Each reporter plasmid was cotransfected with an increasing amount of 21-mer or 23-mer miR-307a/miR-307a* duplex. miR-307a21-mer and miR-307a23-mer each suppressed the miR-307a21-mer and the miR-307a23-mer complementary reporters (Figure 5B). This was expected, because the two isomirs are both complementary to the two reporters.
Figure 5. miR-307a21-mer and miR-307a23-mer Have Distinct Targets.
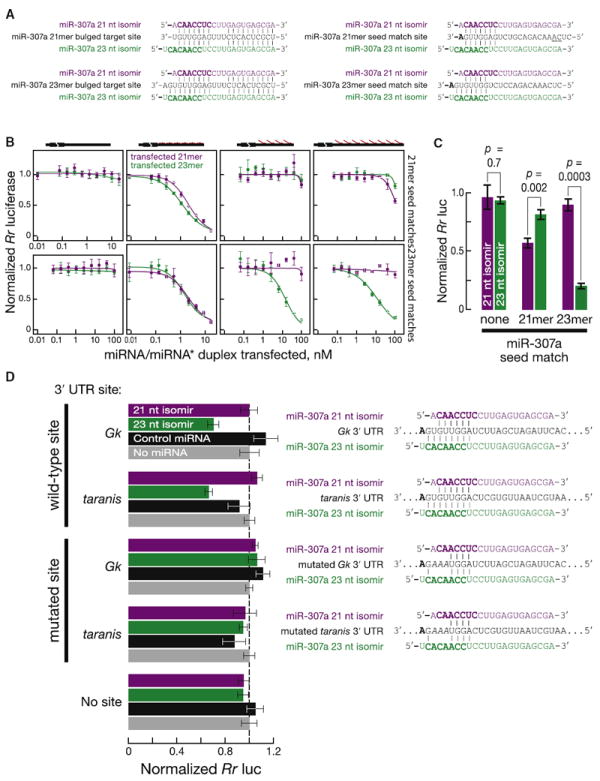
(A) Pairing of miR-307a21-mer (purple) and miR-307a23-mer (green) to luciferase reporter target sites (black).
(B) Silencing of Renilla luciferase reporters bearing 3′ UTR target sites for miR-307a21-mer or miR-307a23-mer. Reporters had six sites with mismatches at positions 9–11, or four or eight sites matching the seed of miR-307a21-mer or miR-307a23-mer. Renilla luciferase expression relative to a firefly luciferase internal control was determined at different concentrations of transfected miRNA/miRNA*. Data were normalized to a transfection control without miR-307a/miR-307*.
(C) The reporter data for Renilla mRNA bearing eight or no target sites matching the seeds of miR-307a21-mer or miR-307a23-mer was analyzed to determine the extent of repression at the highest concentration of transfected miR-307/miR-307* duplex.
(D) Data for Renilla mRNA bearing the 3′ UTR of Gk or tara mRNA. miRNAs were transfected at 20 nM.
Data are mean ± SD for three independent trials.
To test whether miR-307a21-mer and miR-307a23-mer have distinct targeting specificities, we constructed reporter constructs with four or eight t1A, 7 nt seed-matching sites. Each seed match was complementary to the seed sequence of one of the two miR-307 isoforms (Figure 5A). The miR-307a21-mer seed-match reporter was more efficiently repressed by the 21-mer than by the 23-mer (Figures 5B and 5C). Conversely, the miR-307a23-mer seed-match reporter was more efficiently repressed by the 23-mer than by the 21-mer. Control reporters were not repressed by either isoform. Thus, miR-307a21-mer and miR-307a23-mer have distinct target specificities.
miR-307a23-mer, but Not miR-307a21-mer, Represses Gk and tara
The mRNAs for glycerol kinase (Gk) and taranis (tara), a trithorax group protein, are the only 8-mer (t1A plus 7 seed-matching nucleotides) targets predicted by TargetScan (Lewis et al., 2005) for miR-307a23-mer. To test whether Gk and tara mRNAs are direct targets of miR-307a23-mer, we constructed luciferase reporters with the 3′ UTR of Gk or tara. Both reporter constructs were silenced by transfection of miR-307a23-mer but not by miR-307a21-mer or a control miRNA (Figure 5D; for miR-307a23-mer, p value = 0.004 for Gk and 2 × 10−4 for tara; for miR-307a21-mer, p value > 0.05 for both). Mutating the seed-matching sites in the Gk or tara 3′ UTR eliminated silencing by miR-307a23-mer (all p values > 0.05).
To further test the idea that miR-307a23-mer but not miR-307a21-mer represses Gk and tara, we used RNA sequencing to measure the consequences of changing the miR-307a isoform distribution in vivo. We sequenced poly(A)+ RNA from ovaries of control and loqsKO mutant flies rescued with Loqs isoform-specific transgenes (Figure 6A). Both Gk and tara mRNAs were derepressed in the mutant flies lacking Loqs-PB, compared with control and rescue flies expressing Loqs-PB; quantitative RT-PCR confirmed these results (Figure 6A). We conclude that Loqs-PB is required to produce the correct miR-307a isomir to regulate specific mRNAs in vivo (Figure 6B).
Figure 6. miR-307a23-mer, but Not miR-307a21-mer, Represses Gk and tara.
(A) Relative abundance of Gk and tara mRNA in ovaries of control and mutant flies with Loqs-expressing transgenes, determined by quantitative RT-PCR (mean ± SD for three biological replicates) and mRNA-Seq.
(B) Proposed functions of Dicer partner proteins in flies.
TRBP Changes Where Dicer Cleaves Pre-miR-132
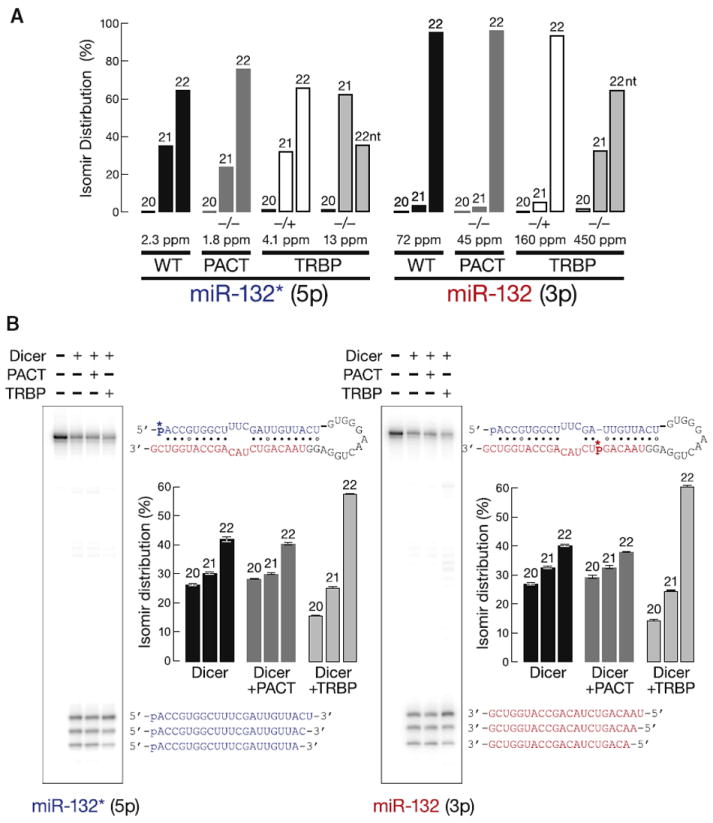
Mammals produce two partner proteins, PACT and TRBP (Chendrimada et al., 2005; Haase et al., 2005; Lee et al., 2006) for their single Dicer, whose main role in somatic cells is the production of miRNAs (Hutvágner et al., 2001). To assess the role of PACT and TRBP in establishing where mammalian Dicer cleaves pre-miRNAs, we sequenced small RNAs from wild-type, Pact−/−, Trbp−/+, and Trbp−/− MEFs (Tables S5A and S5B). Of the 809 miRNAs detected in wild-type MEFs, the 21 nt isomirs of miR-132 and miR-132* were more abundant and the 22 nt isomirs were less abundant in Trbp−/− MEFs compared to Trbp−/+, Pact−/−, or wild-type MEFs. The 22 nt miR-132 isoform, which is the annotated form in miRBase, predominated in all MEF genotypes, but was lowest in the cells lacking TRBP (Figure 7A). miR-132 derives from the 3′ arm of its pre-miRNA. Thus, changing where Dicer cleaves changes its seed sequence and alters the repertoire of targets it regulates.
Figure 7. Mammalian TRBP Tunes Where Dicer Cleaves Pre-miR-132.
(A) Length distribution of miR-132 and miR-132* isomirs in MEFs measured by sequencing.
(B) Pre-miR-132 (100 nM), bearing a single 32P-radiolabel (asterisk), was incubated for 2 hr with purified, recombinant human Dicer (10 nM) or Dicer supplemented with PACT or TRBP (10 nM). Data are mean ± SD for four independent trials.
See also Table S5.
To test whether partner proteins directly influence where mammalian Dicer cleaves, we monitored the production of small RNAs from human pre-miR-132. (Human pre-miR-132 differs from mouse pre-miR-132 at only a single loop nucleotide.) In the presence of TRBP, purified, recombinant human Dicer produced more 22 nt and less 20 nt miRNA and miRNA* products from pre-miR-132, compared with Dicer alone or Dicer plus PACT (Figure 7B). We conclude that, like Loqs-PB in flies, TRBP in mammals can directly influence where Dicer cleaves a pre-miRNA.
DISCUSSION
The simplest interpretation of our data that Loqs-PA and Loqs-PB both decrease the KM of Dcr-1 for pre-miRNA substrates is that Loqs partner proteins increase the affinity of Dcr-1 for some pre-miRNA substrates. For other pre-miRNAs, Loqs-PB can also increase Dcr-1 enzyme turnover. Figure 6B summarizes the functions of Dicer partner proteins in small RNA biogenesis in flies: preventing Dcr-2 from processing pre-miRNAs (R2D2), promoting the processing of “foreign” dsRNA and of esiRNAs and other endo-siRNA precursors (Loqs-PD), enhancing the efficiency of pre-miRNA processing by Dcr-1 (Loqs-PA and PB), and tuning cleavage site choice by Dcr-1 for a small number of pre-miRNAs (Loqs-PB).
The last two functions are surprising because the number of pre-miRNAs that require Loqs-PB and not Loqs-PA for the efficient production of their miRNAs is small and even fewer require Loqs-PB to produce the correct isomir. Nonetheless, our results suggest that the loss of GSCs in flies lacking Loqs-PB reflects the reduced abundance of specific miRNAs such as miR-79, miR-283, miR-305, miR-311, and miR-318 or perhaps the production of the wrong isomirs for miR-307a, miR-87, or miR-316. Among these, miR-318 is the most abundant miRNA in ovaries (140,000 ppm in w1118 ovaries but only 8 ppm in heads). Future experiments using Loqs-independent versions of pre-miR-318 and other miRNAs should help test this idea.
The amino acid sequence of Loqs-PB suggests it is slightly more related to PACT (28% identity; 34% similarity) than to TRBP (24% identity; 33% similarity). Nonetheless, our data suggest that the functional homolog of Loqs-PB in mammals is TRBP. Like Loqs-PA and Loqs-PB, TRBP may act generally to enhance the binding of Dicer to pre-miRNA. Such a role for TRBP is consistent with earlier observations that the two cleavage reactions catalyzed by mammalian Dicer are less well coordinated in the absence of TRBP (Koscianska et al., 2011). But like Loqs-PB, TRBP also helps Dicer produce specific isoforms of a one or more miRNAs. This novel function of Dicer partner proteins may be widely conserved, enabling plants and animals to effectively and accurately dice difficult but important pre-miRNA substrates. Perhaps the Drosha-binding partner Pasha in flies or DGCR8 in mammals similarly tunes pri-miRNA cleavage site choice by Drosha.
The stem, but not the loop, of pre-miR-307a enables Loqs-PB to influence where Dcr-1 cleaves. One possible explanation for how Loqs-PB and TRBP change where Dcr-1 and Dicer cleave is that the mismatches and internal loop causes the stems of pre-miR-307a, pre-miR-87, and mammalian pre-miR-132 to be longer than the corresponding A-form helix; binding of Loqs-PB or TRBP might then “shrink” the stems. In contrast, Loqs-PB binding may extend of the stem of pre-miR-316. Because Loqs-PB does not change where Dcr-2 cleaves pre-miR-307a, Loqs-PB likely acts only when bound to Dcr-1.
Our data suggest that the effect of Loqs-PB is biologically relevant. The long miR-307a isomir predominates in wild-type flies, and miR-307a23-mer but not miR-307a21-mer can repress the Gk and tara mRNAs in vivo. Why has evolution failed to select for easier-to-dice variants of pre-miR-307a and pre-miR-87 in flies and pre-miR-132 in mice, so that a specific partner is no longer needed to ensure production of the “right” isomir? The persistence of the miR-307a21-mer in wild-type flies, for example, suggests that most, if not all, abundant isomirs have distinct biological functions and that the optimal function of miRNAs like miR-307a, miR-87, and miR-132 requires a defined ratio of isomirs.
Although we failed to detect altered regulation of predicted miR-307a21-mer targets in ovaries from flies lacking Loqs-PB, miR-307a21-mer may have functions in other tissues or times in development. In flies, mice, and humans, the relative abundance of miRNA isomirs generated by different Dicer cleavage sites, including 5′ isomirs with distinct seed sequences, varies among tissues and developmental stages (Azuma-Mukai et al., 2008; Fernandez-Valverde et al., 2010; Lee et al., 2010). Perhaps the abundance of Loqs-PB versus Loqs-PA or TRBP versus PACT is regulated across development and differentiation to ensure the correct relative abundance of isomirs from various pre-miRNAs, much as the ratios of alternatively spliced mRNAs are regulated in different tissues and cell types. Our loqsKO flies rescued with transgenes producing individual Loqs isoforms should facilitate the testing of this idea in vivo.
EXPERIMENTAL PROCEDURES
Transgenic Flies
P{Loqs-PA}, P{Loqs-PB}, and P{Loqs-PD} rescuing transgenes containing the loqs promoter were generated by appending the ~2.5 kbp upstream and ~0.8 kbp downstream genomic sequences to the Loqs-PA, Loqs-PB, or Loqs-PD cDNA in pattB (Bischof et al., 2007). A 3x FLAG tag was added to the amino-terminus of each protein. Transgenes were integrated into a single site in the genome (Bischof et al., 2007), and the mini-white gene (w+mC) removed by using Cre-Lox. Double and triple rescue flies were generated by meiotic recombination.
Immunohistochemistry and Microscopy
Egg chamber fixation and whole-mount antibody labeling were performed as described (Theurkauf, 1994; Li et al., 2009). Samples were analyzed with a Leica TCS-SP inverted, laser-scanning microscope. Images were acquired in 2.89 μm increments along the z axis in order to determine the number of GSC within the germarium. All images were acquired at the same settings and are presented in the figure at the same scale.
Recombinant Protein Expression and Purification
His6-Dicer-1 containing the HRV3C protease recognition site after the aminoterminal His6-tag was expressed in Sf9 insect cells by using the Bac-to-Bac system (Invitrogen). cDNA encoding Loqs-PA, Loqs-PB, PACT, or TRBP was inserted between the SalI and NotI restriction sites of a modified pCold I vector (Takara, Otsu, Shiga, Japan) containing the HRV3C protease recognition site. Amino-terminally His6-tagged Loqs-PA, Loqs-PB, PACT, and TRBP proteins were expressed in Escherichia coli BL21 codon Plus RIL (Agilent Technologies). Proteins were purified by using Ni-Sepharose (GE Healthcare, Pittsburgh, PA, USA), followed by HRV3C protease cleavage to remove the His6-tag, and then further purified by chromatography by using HiTrap SP and HiTrap Heparin (GE Healthcare). Recombinant protein concentrations were determined by quantitative amino acid analysis of protein isolated from an SDS-polyacrylamide gel (Keck Biotechnology Resource Laboratory, New Haven, CT, USA).
Dicing
Dicing reactions were performed as described (Cenik et al., 2011) at 25°C for fly and 37°C for human Dicer. Dcr-1, Dcr-2, or human Dicer was used with or without an equimolar amount of purified recombinant partner protein.
To determine the inflection point n in the stoichiometric titration (Figures 4D and S6C), the data were fit to:
When r ≥ n, f(r) = m + b; when r < n, f(r) = m * r/n + b; where b = y intercept and m + b = yplateau, the y intercept of the plateau.
Sequencing
miRNA libraries were as described (Li et al., 2009; Ghildiyal et al., 2010; Ameres et al., 2010; Han et al., 2011; Zhang et al., 2011). Strand-specific mRNA libraries were prepared (Wang et al., 2011) by using adapters and primers in Table S6. Data are available from SRA (accession number SRP014526).
Supplementary Material
Acknowledgments
We thank Haruhiko and Mikiko Siomi for strains and antibodies; Richard Carthew, Qinghua Liu, and the Bloomington Stock Center for strains; Ganes Sen and Robert Braun for MEF cells; Ian MacRae for human Dicer plasmid; Lin Wang and Lauren Dedow for RNA-Seq library protocols; Chengjian Li for help with small RNA libraries; Wei Wang for bioinformatics help; Alicia Boucher, Cindy Tipping, and Shengmei Ma for fly husbandry; and Traci Hall, Gang Lu, and members of the Zamore laboratory for discussion, advice, and comments on the manuscript. This work was supported in part by the National Institutes of Health (GM62862 and GM65236) and a JSPS Research Fellowship for Research Abroad and a Charles A. King Trust Postdoctoral Fellowship to R.F. P.D.Z. is a member of the scientific advisory board of Regulus Therapeutics.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, seven figures, and six tables and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2012.09.027.
ACCESSION NUMBERS
The GEO accession number for the small RNA libraries reported in this paper is GSE37443, and the SRA accession number for the RNA-Seq libraries reported in this paper is SRP014526.
References
- Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng Z, Zamore PD. Target RNA-directed trimming and tailing of small silencing RNAs. Science. 2010;328:1534–1539. doi: 10.1126/science.1187058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma-Mukai A, Oguri H, Mituyama T, Qian ZR, Asai K, Siomi H, Siomi MC. Characterization of endogenous human Argonautes and their miRNA partners in RNA silencing. Proc Natl Acad Sci USA. 2008;105:7964–7969. doi: 10.1073/pnas.0800334105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci USA. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broderick JA, Salomon WE, Ryder SP, Aronin N, Zamore PD. Argonaute protein identity and pairing geometry determine cooperativity in mammalian RNA silencing. RNA. 2011;17:1858–1869. doi: 10.1261/rna.2778911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenik ES, Fukunaga R, Lu G, Dutcher R, Wang Y, Tanaka Hall TM, Zamore PD. Phosphate and R2D2 restrict the substrate specificity of Dicer-2, an ATP-driven ribonuclease. Mol Cell. 2011;42:172–184. doi: 10.1016/j.molcel.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R, et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- Ding S-W. RNA-based antiviral immunity. Nat Rev Immunol. 2010;10:632–644. doi: 10.1038/nri2824. [DOI] [PubMed] [Google Scholar]
- Eamens AL, Smith NA, Curtin SJ, Wang MB, Waterhouse PM. The Arabidopsis thaliana double-stranded RNA binding protein DRB1 directs guide strand selection from microRNA duplexes. RNA. 2009;15:2219–2235. doi: 10.1261/rna.1646909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Valverde SL, Taft RJ, Mattick JS. Dynamic isomiR regulation in Drosophila development. RNA. 2010;16:1881–1888. doi: 10.1261/rna.2379610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förstemann K, Tomari Y, Du T, Vagin VV, Denli AM, Bratu DP, Klattenhoff C, Theurkauf WE, Zamore PD. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler EL, Zapp ML, Weng Z, Zamore PD. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008;320:1077–1081. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Xu J, Seitz H, Weng Z, Zamore PD. Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway. RNA. 2010;16:43–56. doi: 10.1261/rna.1972910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Haase AD, Jaskiewicz L, Zhang H, Lainé S, Sack R, Gatignol A, Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- Han BW, Hung JH, Weng Z, Zamore PD, Ameres SL. The 3′-to-5′ exoribonuclease Nibbler shapes the 3′ ends of microRNAs bound to Drosophila Argonaute1. Curr Biol. 2011;21:1878–1887. doi: 10.1016/j.cub.2011.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartig JV, Förstemann K. Loqs-PD and R2D2 define independent pathways for RISC generation in Drosophila. Nucleic Acids Res. 2011;39:3836–3851. doi: 10.1093/nar/gkq1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartig JV, Esslinger S, Böttcher R, Saito K, Förstemann K. Endo-siRNAs depend on a new isoform of loquacious and target artificially introduced, high-copy sequences. EMBO J. 2009;28:2932–2944. doi: 10.1038/emboj.2009.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraguri A, Itoh R, Kondo N, Nomura Y, Aizawa D, Murai Y, Koiwa H, Seki M, Shinozaki K, Fukuhara T. Specific interactions between Dicer-like proteins and HYL1/DRB-family dsRNA-binding proteins in Arabidopsis thaliana. Plant Mol Biol. 2005;57:173–188. doi: 10.1007/s11103-004-6853-5. [DOI] [PubMed] [Google Scholar]
- Hutvágner G, McLachlan J, Pasquinelli AE, Bálint É, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Jiang F, Ye X, Liu X, Fincher L, McKearin D, Liu Q. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes Dev. 2005;19:1674–1679. doi: 10.1101/gad.1334005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada TN, Siomi MC, Siomi H. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature. 2008;453:793–797. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- Kennerdell JR, Carthew RW. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell. 1998;95:1017–1026. doi: 10.1016/s0092-8674(00)81725-0. [DOI] [PubMed] [Google Scholar]
- Kennerdell JR, Carthew RW. Heritable gene silencing in Drosophila using double-stranded RNA. Nat Biotechnol. 2000;18:896–898. doi: 10.1038/78531. [DOI] [PubMed] [Google Scholar]
- Koscianska E, Starega-Roslan J, Krzyzosiak WJ. The role of Dicer protein partners in the processing of microRNA precursors. PLoS ONE. 2011;6:e28548. doi: 10.1371/journal.pone.0028548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Carthew RW. Making a better RNAi vector for Drosophila: use of intron spacers. Methods. 2003;30:322–329. doi: 10.1016/s1046-2023(03)00051-3. [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN. The role of PACT in the RNA silencing pathway. EMBO J. 2006;25:522–532. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee LW, Zhang S, Etheridge A, Ma L, Martin D, Galas D, Wang K. Complexity of the microRNA repertoire revealed by next-generation sequencing. RNA. 2010;16:2170–2180. doi: 10.1261/rna.2225110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, Seitz H, Horwich MD, Syrzycka M, Honda BM, et al. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell. 2009;137:509–521. doi: 10.1016/j.cell.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Yue L, Spradling AC. The Drosophila fusome, a germline-specific organelle, contains membrane skeletal proteins and functions in cyst formation. Development. 1994;120:947–956. doi: 10.1242/dev.120.4.947. [DOI] [PubMed] [Google Scholar]
- Liu Q, Rand TA, Kalidas S, Du F, Kim HE, Smith DP, Wang X. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921–1925. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- Liu X, Jiang F, Kalidas S, Smith D, Liu Q. Dicer-2 and R2D2 coordinately bind siRNA to promote assembly of the siRISC complexes. RNA. 2006;12:1514–1520. doi: 10.1261/rna.101606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques JT, Kim K, Wu PH, Alleyne TM, Jafari N, Carthew RW. Loqs and R2D2 act sequentially in the siRNA pathway in Drosophila. Nat Struct Mol Biol. 2010;17:24–30. doi: 10.1038/nsmb.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi K, Miyoshi T, Hartig JV, Siomi H, Siomi MC. Molecular mechanisms that funnel RNA precursors into endogenous small-interfering RNA and microRNA biogenesis pathways in Drosophila. RNA. 2010;16:506–515. doi: 10.1261/rna.1952110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Okamura K, Balla S, Martin R, Liu N, Lai EC. Two distinct mechanisms generate endogenous siRNAs from bidirectional transcription in Drosophila melanogaster. Nat Struct Mol Biol. 2008a;15:581–590. doi: 10.1038/nsmb.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Chung WJ, Ruby JG, Guo H, Bartel DP, Lai EC. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature. 2008b;453:803–806. doi: 10.1038/nature07015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JK, Liu X, Strauss TJ, McKearin DM, Liu Q. The miRNA pathway intrinsically controls self-renewal of Drosophila germline stem cells. Curr Biol. 2007;17:533–538. doi: 10.1016/j.cub.2007.01.060. [DOI] [PubMed] [Google Scholar]
- Park JE, Heo I, Tian Y, Simanshu DK, Chang H, Jee D, Patel DJ, Kim VN. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature. 2011;475:201–205. doi: 10.1038/nature10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham JW, Sontheimer EJ. Molecular requirements for RNA-induced silencing complex assembly in the Drosophila RNA interference pathway. J Biol Chem. 2005;280:39278–39283. doi: 10.1074/jbc.M509202200. [DOI] [PubMed] [Google Scholar]
- Rowe TM, Rizzi M, Hirose K, Peters GA, Sen GC. A role of the double-stranded RNA-binding protein PACT in mouse ear development and hearing. Proc Natl Acad Sci USA. 2006;103:5823–5828. doi: 10.1073/pnas.0601287103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Ishizuka A, Siomi H, Siomi MC. Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biol. 2005;3:e235. doi: 10.1371/journal.pbio.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, Hannon GJ. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theurkauf WE. Immunofluorescence analysis of the cytoskeleton during oogenesis and early embryogenesis. Methods Cell Biol. 1994;44:489–505. doi: 10.1016/s0091-679x(08)60928-0. [DOI] [PubMed] [Google Scholar]
- Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD. A protein sensor for siRNA asymmetry. Science. 2004;306:1377–1380. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- Tomari Y, Du T, Zamore PD. Sorting of Drosophila small silencing RNAs. Cell. 2007;130:299–308. doi: 10.1016/j.cell.2007.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- Vazquez F, Gasciolli V, Crété P, Vaucheret H. The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr Biol. 2004;14:346–351. doi: 10.1016/j.cub.2004.01.035. [DOI] [PubMed] [Google Scholar]
- Wang L, Si Y, Dedow LK, Shao Y, Liu P, Brutnell TP. A low-cost library construction protocol and data analysis pipeline for Illumina-based strand-specific multiplex RNA-seq. PLoS ONE. 2011;6:e26426. doi: 10.1371/journal.pone.0026426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- Yang N, Kazazian HHJ., Jr L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat Struct Mol Biol. 2006;13:763–771. doi: 10.1038/nsmb1141. [DOI] [PubMed] [Google Scholar]
- Ye X, Paroo Z, Liu Q. Functional anatomy of the Drosophila microRNA-generating enzyme. J Biol Chem. 2007;282:28373–28378. doi: 10.1074/jbc.M705208200. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Xu J, Koppetsch BS, Wang J, Tipping C, Ma S, Weng Z, Theurkauf WE, Zamore PD. Heterotypic piRNA Ping-Pong requires qin, a protein with both E3 ligase and Tudor domains. Mol Cell. 2011;44:572–584. doi: 10.1016/j.molcel.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Fernald RD. Comprehensive algorithm for quantitative real-time polymerase chain reaction. J Comput Biol. 2005;12:1047–1064. doi: 10.1089/cmb.2005.12.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J, Peters AH, Lee K, Braun RE. A double-stranded RNA binding protein required for activation of repressed messages in mammalian germ cells. Nat Genet. 1999;22:171–174. doi: 10.1038/9684. [DOI] [PubMed] [Google Scholar]
- Zhou R, Czech B, Brennecke J, Sachidanandam R, Wohlschlegel JA, Perrimon N, Hannon GJ. Processing of Drosophila endo-siRNAs depends on a specific Loquacious isoform. RNA. 2009;15:1886–1895. doi: 10.1261/rna.1611309. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


