Abstract
Dengue viruses (DENV) are the most important arboviral pathogens in tropical and subtropical regions throughout the world, putting at risk of infection nearly a third of the global human population. Evidence from the historical record suggests a long association between these viruses and humans. The transmission of DENV includes a sylvatic, enzootic cycle between nonhuman primates and arboreal mosquitoes of the genus Aedes, and an urban, endemic/epidemic cycle between Aedes aegypti, a mosquito with larval development in peridomestic water containers, and human reservoir hosts. DENV are members of the genus Flavivirus in the Family Flaviviridae and comprise of 4 antigenically distinct serotypes (DENV-1-4). Although they are nearly identical epidemiologically, the 4 DENV serotypes are genetically quite distinct. Utilization of phylogenetic analyses based on partial and/or complete genomic sequences has elucidated the origins, epidemiology (genetic diversity, transmission dynamics and epidemic potential), and the forces that shape DENV molecular evolution (rates of evolution, selection pressures, population sizes, putative recombination and evolutionary constraints) in nature. In this review, we examine how phylogenetics have improved understanding of DENV population dynamics and sizes at various stages of infection and transmission, and how this information may influence pathogenesis and improve our ability to understand and predict DENV emergence.
1. Dengue, the preeminent human arboviral disease
Arthropod-borne viruses (arboviruses) comprise a taxonomically diverse group of viruses that are transmitted by arthropod vectors (Calisher and Karabatsos, 1988). Nearly all arboviruses have RNA genomes, probably a reflection of the genetic plasticity required to maintain transmission cycles requiring replication in disparate arthropod and vertebrate hosts. However, arboviruses are found in several RNA virus families, all of which also include members that do not rely on arthropod transmission. This suggests that the arthropod-borne transmission cycle probably arose at least several times during the course of virus evolution. All currently recognized arboviruses are found in 5 RNA virus families, including the flaviviruses (genus Flavivirus, one of 3 genera in the family Flaviviridae)(Hanley and Weaver, 2008). More than 50% of all known flaviviruses are associated with human disease, including some of the most important human pathogens, such as dengue (DENV), yellow fever, Japanese encephalitis, West Nile and tick-borne encephalitis viruses.
Among all arboviruses, the dengue viruses (DENV) are by far the most important human pathogens. An estimated 50-100 million DENV infections occur each year in tropical and subtropical regions, where more than 2.5 billion people are at risk (nearly a third of the global population)(Gubler, 1994; Gubler, 2002). Unlike many flaviviruses, DENV are highly restricted in their natural vertebrate host range, generally utilizing primates as their amplification and reservoir hosts. They are also among the most widely distributed of the flaviviruses, and all 4 DENV serotypes can be found nearly throughout the tropics where the mosquito vector Aedes aegypti is abundant, putting at risk of infection nearly a third of the global human population. DENV comprise 4 antigenically distinct serotypes (DENV-1-4), which, though epidemiologically nearly identical, are genetically quite distinct. Infection with one DENV serotype leads to lifelong protection against homologous challenge, but only brief protection against heterologous infection with a different serotype (Kurane and Ennis, 1992; Sabin, 1952). All DENV cause dengue (DEN) fever (DF), a self-limited febrile illness lasting 2-10 days. However, some patients progress to develop life threatening syndromes including dengue hemorrhagic fever (DHF) characterized by thrombocytopenia and hemorrhage, and dengue shock syndrome (DSS) due to excessive plasma leakage (Gubler, 1994; Monath, 1994). Most DHF and DSS occur following a secondary infection, and 2 principal hypotheses explain this epidemiological pattern: 1) the immune enhancement theory maintains that hemorrhage occurs in secondary infections when DENV-specific antibodies and memory T cells, resulting from primary infection with a different serotype, enhance the binding of heterologous DENV-IgG complexes to Fcγ receptors on monocytic cells; 2) the virulence hypothesis suggests that some DENV strains are intrinsically more virulent than others, and cause higher viremia leading to more severe disease. An Asian genotype of DENV-2 recently introduced into the New World may be associated with increased risk for hemorrhagic fever and shock during secondary infections (Rico-Hesse et al., 1997). Also, endemic transmission on the South Pacific islands of Tonga, involving more highly susceptible vectors than Ae. aegypti, may have resulted in less severe dengue because selection for high viremia was relaxed due to the high degree of mosquito susceptibility (Gubler et al., 1978).
2. Dengue viruses and their genomes
All DENV are members of the DEN antigenic complex in the genus Flavivirus, family Flaviviridae (Calisher et al., 1989). The inclusion of DENV in this genus is based on antigenic cross-reactivity with other flaviviruses, as well as genomic organization and sequence homology. The 4 DENV serotypes are defined based on limited cross-reactions in various serological tests. Initial genetic characterizations of DENV in all serotypes identified “topotypes” or geographic variants by T1 RNase fingerprinting (Repik et al., 1983; Trent et al., 1990). Later, nucleic acid sequencing confirmed the homology of the 4 serotypes as well as their conserved genetic organization, and allowed for the more precise and broad classification of DENV into genetically distinct groups or genotypes within each serotype (Rico-Hesse, 1990). Rico-Hesse defined DENV “genotypes” as clusters of DENV with sequence divergence not greater than 6% within the chosen genome region (in this case the E/NS1 junction), which was based on the clustering of strains for which associations could be inferred on epidemiological grounds (Rico-Hesse, 1990).
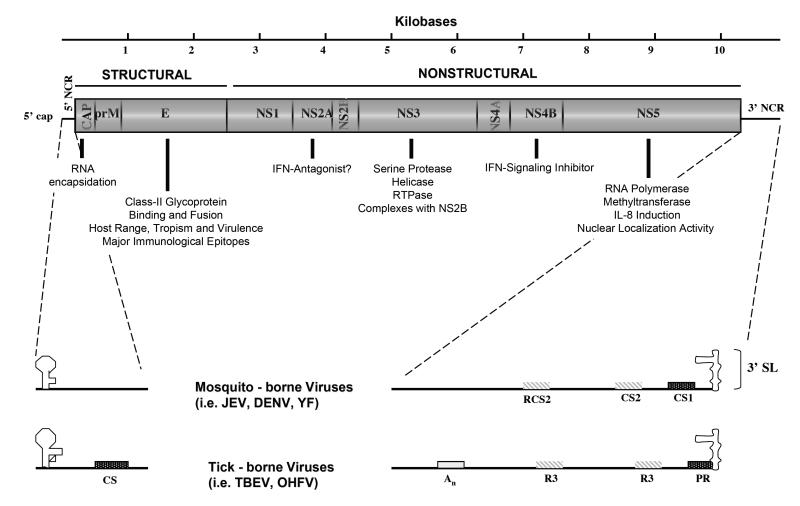
Like that of other flaviviruses, the DENV genome is a single stranded RNA molecule of positive polarity or messenger sense and about 11 kB in length that encode a single open reading frame (Lindenbach and Rice, 2003). The genomic RNA includes 5′ (ca 100 nt) and 3′ (ca. 400-800 nt) untranslated regions (UTRs)(Fig. 1). Translation of the single ORF results in a single polyprotein that is cleaved by host and virus-derived proteases to produce the structural (C-prM-E) and non-structural (NS1-NS2A-NS2B-NS3-NS4A-NS4B-NS5) proteins. The envelope (E) protein is comprised of 3 domains; domain III is believed to interact with cellular receptors for entry, and includes most important epitopes that bind to neutralizing antibodies. The 5′ and 3′ termini of the DENV genomes include untranslated sequences important for viral RNA replication and translation, and probably interact with cellular factors involved in these functions.
Fig. 1.
Cartoon depicting the DENV genome and the major functions of the gene products.
The capsid protein (C), an 11 kDa homodimer protein with an unusually high net charge (Trent, 1977), is essential for RNA genome encapsidation (Chang et al., 2001). The membrane protein (prM), a 27-31 kDa N-glycosylated protein cleaved in the trans-Golgi network (TGN) by a cell-encoded furin-like protease during the late stages of virus assembly, to release mature virions (Kuhn et al., 2002; Li et al., 2008; Yu et al., 2008). The envelope (E) protein is a 53 kDa class II N-glycosylated dimeric membrane fusion protein that mediates virus binding and fusion to host cell membrane (Johnson, Guirakhoo, and Roehrig, 1994; Modis et al., 2004; Rey et al., 1995), as well as confers protective immune responses by eliciting neutralizing, antifusion, and replication-enhancing antibodies (Chen, Maguire, and Marks, 1996; Roehrig, Bolin, and Kelly, 1998). As mentioned above, each of the monomer subunits of the E protein is composed of three distinct domains: domain I, a β-barrel structure oriented parallel to the viral membrane; domain II, a finger-like structure composed of a pair of discontinuous loops one of which is highly conserved among all flaviviruses functioning as an internal fusion peptide and is stabilized by three disulfide bridges; and the C-terminal domain III (aa 303 – 395), located in the outer lateral surface of the dimer. Domain III has been suggested to contain residues that are responsible for the determination of host range, tropism and virulence among flaviviruses (Rey et al., 1995). Mutations within domain III, such as N390D which is located within the putative glycosaminoglycan binding motif (386L-411M) responsible for the binding of DENV onto the host cell membrane via a non-Fc receptor (Chen, Maguire, and Marks, 1996), have been implicated as a potential virulence determinant in American DENV genotypes (Leitmeyer et al., 1999), and has been shown to alter virulence in mice (Sanchez and Ruiz, 1996).
Among the nonstructural proteins, NS1 is a 46 kDa dimeric N-glycosylated glycosyl-phosphatidylinositol (GPI) anchored protein that exists in both intra- and extracellular forms (Jacobs et al., 2000; Winkler et al., 1989). NS2A, is a 22 kDa protein involved in the coordination of change between RNA packaging and replication (Khromykh et al., 2001) and possibly antagonism of interferon (IFN) (Jones et al., 2005; Munoz-Jordan et al., 2003). Several recent analyses have indicated that the gene encoding for NS2A are under weak selection pressures, a process that influences virus evolution during natural transmission (Bennett et al., 2003; Vasilakis et al., 2007a; Zhang et al., 2005). NS2B, is a membrane-associated 14 kDa protein which associates with NS3 to form the viral protease complex and serves as a cofactor in the structural activation of the DENV serine protease of NS3 (Erbel et al., 2006; Leung et al., 2001). NS3, is a 70kDa multifunctional protein with trypsin-like serine protease, helicase, and RNA triphosphatase (RTPase) enzyme activities (Gorbalenya et al., 1989; Li et al., 1999) and is involved in the processing of the viral polyprotein, as well as RNA replication. NS4A and NS4B, are small hydrophobic proteins of 16 and 27 kDa, respectively, with the latter functioning as an interferon (IFN)-signaling inhibitor (Jones et al., 2005; Munoz-Jordan et al., 2003). Recent evidence demonstrated a concentration of putative positive selection in NS4B of sylvatic DENV (Vasilakis et al., 2007a), which suggests a possible role in distinguishing endemic and sylvatic DENV genotypes. Lastly, NS5 is a large multifunctional, well-conserved protein of 103 kDa with RNA cap-processing, RNA-dependent RNA polymerase (RdRp) (Ackermann and Padmanabhan, 2001; Egloff et al., 2002), interleukin-8 induction (IL-8) (Medin, Fitzgerald, and Rothman, 2005) and nuclear localization (Pryor et al., 2007; Uchil, Kumar, and Satchidanandam, 2006) activities.
3. The history of dengue disease
Although DEN reached dramatic levels of incidence that brought increased awareness in many tropical and neotropical locations following the cessation of World War II, the disease has a long history of human interaction. It is known from the historical record that a DEN-like illness with similar clinical description occurred in China as early as the 3nd Century during the Chin Dynasty [Common Era (CE) 265-420]. Similar reports were described during the 7th and 10th Century [Tang Dynasty (CE 610) and Northern Sung Dynasty (CE 992), respectively] (Gubler, 1997b). These reports described a disease called ‘water poison,’ due to its association with water-associated flying insects and whose clinical description included fever, rash, arthralgia, myalgia and hemorrhagic manifestations. After a long absence in the historical record, reports of a similar illness appeared almost seven centuries later, describing an acute illness with prolonged convalescence in the French West Indies and Panama during 1635 and 1699, respectively (Gubler, 1997b). A century later (1779-1788), several reports of DEN from Batavia (present day Jakarta), Cairo, Philadelphia, and Cadiz and Seville, Spain suggested the possibility of a DEN pandemic (Bylon, 1780; Christie, 1881; Hirsch, 1883; Pepper, 1941; Rush, 1789). These accounts described for the first time evidence for a widespread DENV geographic distribution (or at the very least of an illness very similar to DEN), reaching pandemic proportions by 1788. Interestingly, this widespread geographic distribution of DEN prevalence coincided with the increase in global commerce aided by sailing ships.
The historical record also indicates that a second series of DEN or DEN-like pandemics cris-crossed the globe, from Africa to India to Oceania to the Americas, from 1823 to 1916 (Brown, 1977; Christie, 1881; Hirsch, 1883; More, 1904), each lasting for 3-7 years. Although it is impossible to know which serotype was involved, these outbreaks were probably caused by the same DENV serotype and were transported among geographic regions by the slave trade and commerce (Brown, 1977; Christie, 1881; Gubler, 1997b). Further credence for commerce-fueled spread comes from Leichtenstern, who first recognized DEN as a disease of seaports and coastal regions that could also spread inland along rivers, like the Ganges and the Indus in India, or the Mississippi in the United States (Leichtenstern, 1896). As described below (see DENV transmission Cycles and Control of Disease), the increased prevalence of DEN was also facilitated by the invasion of the tropics by the African Ae. aegypti mosquito vector, most likely due to the movement of people (i.e. slave trade, commerce, migration) and their water storage containers by sailing ships (Daniels, 1908; Smith, 1956; Stanton, 1920; Steadman, 1828; White, 1934). Subsequently, these factors (commerce and new vector invasion of the tropics) altered dramatically DENV behavior in Southeast Asia, the Indian subcontinent and the Philippines from the sudden onset of urban epidemics to endemicity.
A new relationship between DENV and humans dawned with the onset of World War II, which brought immense ecologic, demographic, and epidemiologic changes leading to the establishment of scientific commissions to study the disease, its etiologic agent and development of diagnostic tests (Hota, 1952; Sabin, 1952; Sabin and Schlesinger, 1945). However, the cessation of World War II led to uncontrolled urbanization, where inadequate housing, water distribution systems, as well as sewer and waste management, allowed for the vector (Ae. aegypti) to reach high densities and facilitated dispersal of the DENV serotypes among diverse geographic regions. Although rare occurrences of severe and fatal hemorrhagic disease associated with DEN were reported as early as the 1780 Philadelphia (Rush, 1789) and 1927-29 Greek epidemics (Copanaris, 1928), the ecologic and demographic changes of the 20th Century created ideal conditions for the emergence of DHF in Southeast Asia (Hammon, Rudnick, and Sather, 1960; Hammon et al., 1960).
The initiation of an Ae. aegypti eradication program to combat urban epidemics of yellow fever under the auspices of the Pan American Health Organization (PAHO), led to a quiescence of DEN in the Americas for the next 20 years. However, abandonment of this program during the early 1970s allowed for the gradual reinfestation by Ae. aegypti, a process that continued well into the 1990s. An important characteristic of DEN epidemics in the Americas during the 1960s and 1970s was the circulation of a single serotype at any given time within a given region (hypoendemicity). However, during the 1960’s, a dramatic increase in DENV activity was observed in many tropical locations throughout the rest of the world (Carey et al., 1971; Ehrenkranz et al., 1971; Russell et al., 1966; Saugrain et al., 1970), with a series of epidemics associated with an increased incidence of disease severity (Balaya et al., 1969; Basaca-Sevilla and Halstead, 1966; Halstead et al., 1965; Halstead and Yamarat, 1965). By the end of the 1960’s, it was evident all 4 DENV serotypes shared overlapped in a spatiotemporal manner (hyper-endemicity) throughout Southeast Asia and the Indian subcontinent. Subsequent, prospective field studies by Halstead and colleagues in Thailand suggested an inherent association between secondary infections and the severity of DEN disease (Halstead et al., 1967; Russell, Udomsakdi, and Halstead, 1967), which formed the basis for the development of the antibody-dependent enhancement theory (ADE) of DEN pathogenesis (Halstead, Chow, and Marchette, 1973).
Although the historical record indicated the presence of DEN in Africa as early as the 19th Century (Christie, 1881; Hirsch, 1883), relatively little was known about DEN prevalence (Edington, 1927; Kokernot, Smithburn, and Weinbren, 1956) until a surveillance program established by the Rockefeller Foundation at the University of Ibadan, Nigeria in 1964, documented endemic transmission of DENV-1 and DENV-2 in humans (Anonymous, 1969; Carey et al., 1971). Even today, a major limitation in our understanding of DEN prevalence and disease burden in Africa is the absence of effective surveillance, although there is evidence for increased DEN incidence fueled by an increase in circulation of all 4 serotypes (Botros et al., 1989; Fagbami and Fabiyi, 1976; Fagbami, Monath, and Fabiyi, 1977; Gonzalez et al., 1985; Gubler et al., 1986; Hyams et al., 1986; Johnson et al., 1982; Nogueira et al., 1991; Rodier et al., 1996; Saleh et al., 1985; Saluzzo et al., 1986). Curiously, this increased incidence has not been associated with any increase in disease severity, except on rare occasions (Gubler et al., 1986).
However, the introduction of DENV genotypes from Southeast Asia into the Americas in 1981 (Kouri et al., 1983; Rico-Hesse, 1990) was associated with a significant increase in DEN severity during subsequent epidemics (Alvarez et al., 2006; Cabrera-Batista et al., 2005; Kouri et al., 1989; Nogueira, de Araujo, and Schatzmayr, 2007; Rigau-Perez, Vorndam, and Clark, 2001; Uzcategui et al., 2001). Retrospective observations from the Cuban epidemic of 1981 were instrumental in inferring the putative role of host genetics (Bravo, Guzman, and Kouri, 1987; Kouri, Guzman, and Bravo, 1987), gender and age (Guzman et al., 1984) in influencing the severity of disease. Later studies also implicated nutritional status (Kalayanarooj and Nimmannitya, 2005) and immune enhancement (Halstead, 2003).
A new era in human-DENV relationships began by the 1990s and the beginning of the 21st Century, when the global distribution of all DENV serotypes had been nearly completed. This process was facilitated by the gradual convergence of several factors, including expanding urban populations, increased vector density due to unsustainable control programs, and the increase in commercial air travel facilitating the rapid movement of viremic humans. These factors facilitated the rapid and dramatic re-emergence of DEN associated with increased disease severity throughout the tropics. Although the intensity of several epidemics peaked globally in 1998 (Aziz et al., 2002; Corwin et al., 2001; Cunha et al., 1999; Ha et al., 2000; Harris et al., 2000; Thomas, Strickman, and Vaughn, 2003), the incidence rates of severe DEN have increased significantly throughout all hyperendemic regions (Cordeiro et al., 2007; Kusriastuti and Sutomo, 2005; Ooi, Goh, and Gubler, 2006; PAHO, 2007; WHO, 2006).
Although the information presented above suggests a long relationship between humans and DENV due to infections from strains circulating in urban settings throughout the tropics, little is known about the interaction of sylvatic DENV and humans. The pioneering studies of Smith (Smith, 1956) presented for the first time serological evidence for the existence of an ecologically distinct transmission cycle. Smith proposed a connecting link (Ae. albopictus) to the prevalence of mostly subclinical or mild DEN among the rural population of peninsular Malaysia. Subsequent studies by Rudnick and colleagues (Rudnick, 1986; Rudnick and Lim, 1986) confirmed the existence of an ecologically distinct DENV transmission cycle with the isolation of DENV-1, -2, and -4 from sylvatic habitats.
Retrospective serologic studies of isolated, forest-inhabiting human populations first suggested an interaction between sylvatic DENV and humans (Rudnick, 1986; Rudnick and Lim, 1986). Currently, our understanding of human illness after infection with sylvatic DENV comes from 5 documented case histories of human DENV infections by an ecologically and genetically distinct sylvatic genotype of DENV-2 (Monlun et al., 1992; Robin et al., 1980; Saluzzo et al., 1986). Conducting similar DEN seroprevalence studies among nonhuman primates and human communities within rainforests, Fagbami suggested similar inferences regarding zoonotic DENV to those put forward by Rudnick and Smith in Malaysia (Fagbami, Monath, and Fabiyi, 1977). Moreover, recent phylogenetic evidence (Vasilakis, Tesh, and Weaver, 2008) confirmed that human contact with sylvatic DENV also took place in Ibadan, Nigeria during 4 years of DEN activity in the 1960’s (Carey et al., 1971). Entomologic studies suggest that the linking vector between sylvatic DENV and human communities located within or near the forest may be Ae. furcifer, a gallery forest-dwelling mosquito that disperses into rural peridomestic habitats (Diallo et al., 2003; Diallo et al., 2005).
Our understanding of the DENV clinical presentation (DF or DHF/DSS) in humans has been greatly enhanced from the early 3rd century, when it was first associated with water-associated flying insects, to the 20th Century where their ecology and transmission cycle were elucidated. As described above, DENV have had a close relationship with humans for about 2,000 years, which only during the past few decades has intensified due to altered ecologic conditions, as well as human behavior (global commerce, urbanization, large population movements) and unstainable vector control programs. Currently, all DENV serotypes have reached nearly global hyperendemicity and will likely continue to cause epidemics of various intensities and pathogenic severity. Today, about a third of the global human population is at risk for DENV infection mostly in urban and periurban areas throughout the tropics. By current estimates, approximately 100 million DENV infections occur annually, leading to 500,000 cases of DHF and 20,000 deaths. Thus DENV have become the preeminent arboviral pathogens of humans.
4. The origins of dengue viruses
The origins of DENV have been the subject of speculation for decades. Phylogenetic relationships to other flaviviruses provide little insight because the closest relatives to DENV occur in several continents (Kuno et al., 1998). Gubler (Gubler, 1997a) hypothesized that endemic DENV evolved from sylvatic strains in Africa or Asia that utilize nonhuman primate hosts and gallery forest-dwelling Aedes vectors (not the endemic/epidemic vectors Ae. aegypti or Ae. albopictus). The sylvatic cycle is presumed to be ancestral because efficient interhuman transmission is thought to require a minimum human population size of 10,000-1 million, which did not exist until about 4,000 years ago when urban civilizations arose (Gubler, 1997a).
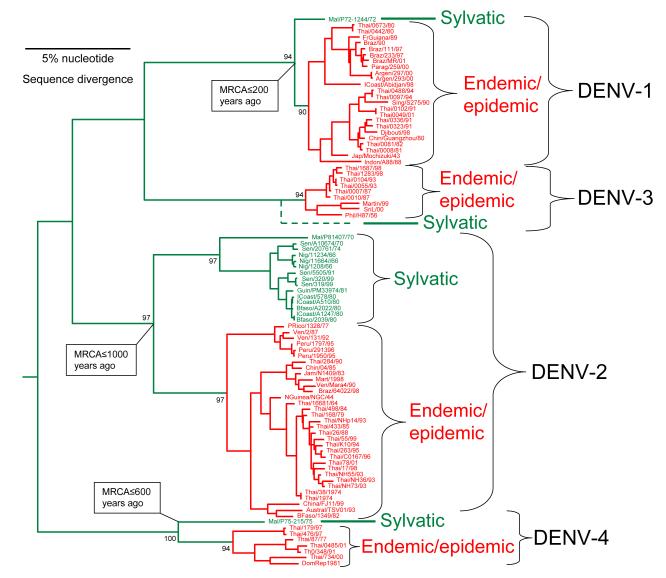
The first support for the hypothesis that DENV was originally zoonotic came from phylogenetic studies of DENV-2, which demonstrated that strains from sylvatic habitats in West Africa were distinct from all others (Rico-Hesse, 1990). Wang et al. (Wang et al., 2000) provided more comprehensive phylogenetic support for the hypothesis of a sylvatic DENV origin, when they sequenced the complete E genes of DENV-1, -2 and -4 isolated in tropical forest habitats in Malaysia by Rudnick et al. during the 1960’s and 1970’s (Rudnick, 1965; Rudnick, Marchette, and Garcia, 1967), as well as DENV-2 from sylvatic cycles in West Africa (Robin et al., 1980). As in the previous studies (Rico-Hesse, 1990), the African sylvatic DENV-2 were distinct from all endemic/epidemic (henceforth referred to as endemic) isolates, differing by 19% in their nucleotide sequences (Fig. 2). The Malaysian sylvatic isolates of DENV-1, -2 and -4 were also distinct from both endemic isolates collected from humans and urban vectors, and from the African sylvatic strains; the Malaysian DENV-2 isolates differ from the epidemic strains by 17%, while the DENV-1 and -4 Malaysian sylvatic strains differ from their epidemic counterparts by about 7% and 14%, respectively. Although sylvatic DENV-3 was not isolated in Malaysia by Rudnick and colleagues, the presence of DENV-3 antibodies in non-human, canopy dwelling primates indicated that a sylvatic DENV-3 cycle probably exists there (Rudnick, 1978).
Fig. 2.
Phylogenetic tree of DENV strains from all 4 serotypes derived from complete open reading frames available in the GenBank library. The phylogeny was inferred using Bayesian analysis (one million reiterations) and all horizontal branches are scaled according to the number of substitutions per site. Bayesian probability values are shown for key nodes. Virus strains are coded by abbreviated country of collection/strain name/year of collection.
Using various methods and estimates of nucleotide substitution rates, various investigators have estimated that the endemic DENV-2 genotypes diverged from the sylvatic forms about 40-600 years ago (Twiddy, Holmes, and Rambaut, 2003; Vasilakis et al., 2007a; Wang et al., 2000). The African and Malaysian sylvatic lineages diverged slightly later, as did the sylvatic and epidemic DENV-1 and -4 lineages (Fig. 2). The greater diversity of sylvatic DENV in Malaysia compared to Africa suggests that an ancestor of all DENV arose in the former region (although the exact location cannot be determined due to limited sampling) and diverged in the relatively distant past into the 4 serotypes recognized today.
The 4 independent epidemic DENV emergence events, presumably involving host range changes from arboreal Aedes mosquitoes to Ae. albopictus and later to Ae. aegypti (Gubler, 1997a), suggest that adaptation to new vectors and vertebrate hosts may have occurred repeatedly (Fig. 3). Because Ae. aegypti did not occur in Asia and Oceania at that time prior to the establishment of shipping trade, Ae. albopictus was probably the original human DENV vector. Acquisition of Ae. aegypti as a vector may have occurred only during the past few centuries as commerce via sailing ships distributed this mosquito throughout the tropics (Gubler, 1997a).
Fig. 3.
Cartoon depicting the hypothetical evolutionary history of endemic/epidemic DENV emergence from zoonotic transmission cycles.
The emergence of distinct DENV serotypes, prior to emergence of the extant epidemic lineages, was most likely facilitated by the allopatric and perhaps ecological partitioning of ancestral sylvatic DENV strains in different non-human primate populations. This scenario is supported by the limited cross-protection against heterologous challenge exhibited by current strains of DENV. Maintenance of the current levels of antigenic and genetic diversity among the 4 DENV serotypes may be selected by immune enhancement, if each virus gains replication efficiency in the human and/or nonhuman primate host due to limited cross-reactive immunity (Ferguson, Anderson, and Gupta, 1999). Spatial-temporal travelling waves of DHF incidence, which have a three-year period in Thailand, appear to move radially at a speed of 148 km per month (Cummings et al., 2004). The resulting oscillations in DEN disease may be explained by immune enhancement without any external influences (Billings et al., 2007).
5. DENV transmission cycles and control of disease
Two distinct DENV transmission cycles occur: (1) Endemic DENV circulate among humans, which serve as both reservoir and amplification hosts, and peridomestic Ae. Aegypti and Ae. albopictus, with other Aedes spp. serving as secondary vectors. The efficiency of the endemic cycle, which is now completely independent both evolutionarily and ecologically from the ancestral, sylvatic cycles (see section on genetic relationships below), is greatly enhanced by the ecology and behavior of Ae. aegypti. This species lays its eggs in water storage containers and refuse in close proximity to human dwellings, readily enters human habitations, and takes multiple bloodmeals during each gonotrophic cycle for both egg production and general nutrition. All of these behaviors increase vector competence and transmission to multiple human hosts (Harrington, Edman, and Scott, 2001; Weaver et al., 2004). Movement of people and their water storage containers during the 17th-19th centuries probably spread Ae. aegypti from its origin in Africa throughout the tropics. After World War II, this species increased in prevalence and distribution in Asia and the Pacific Islands. Ae. aegypti was partially eradicated from tropical America in the 1940s and 50s, but has now reinfested most of the neotropics (Gubler, 1997a). Although Ae. aegypti plays a greater role in urban transmission, Ae. albopictus and some other secondary Aedes spp. vectors are sometimes more susceptible to experimental infection (Gubler, 1988) and in some situations are more important vectors (Ali et al., 2003). Ae. albopictus, another treehole species of sylvatic origin albeit in Asia, has spread widely in the world since the 1970s including to the U.S., Latin America, tropical Africa, the Pacific Islands and Europe (Deubel and Murgue, 2001). Although less anthropophagic than Ae. aegypti, it is considered an important secondary vector of DENV and possibly of greater importance in the early historical stages of endemic dengue emergence. Considerable variation in susceptibility and transmission efficiency has also been demonstrated among geographic populations of both Ae. aegypti and Ae. albopictus (Gubler et al., 1979; Gubler and Rosen, 1976).
(2) Sylvatic DENV circulate among non-human primate reservoir hosts and several different Aedes mosquitoes in forested habitats of West Africa and Malaysia (Gubler, 1988; Gubler, 1994). While sylvatic DENV transmission cycles have received little study, some important features have been determined. In Malaysia, where all 4 serotypes of DENV are probably maintained in canopy-dwelling Ae. niveus mosquitoes and most likely non-human primates (Rudnick, 1965; Rudnick, 1978; Rudnick, 1984b; Rudnick, Marchette, and Garcia, 1967), DENV-1, -2 and -4 have been isolated from sentinel monkeys, and seroconversion has been demonstrated against DENV-1, -2 and -3. In West Africa, sylvatic DENV-2 has been isolated from the arboreal species Ae. africanus, Ae. leuteocephalus, Ae. opok, Ae. taylori and Ae. furcifer (Cordellier et al., 1983; Roche et al., 1983), as well as from 5 humans in eastern Senegal (Monlun et al., 1992; Robin et al., 1980; Saluzzo et al., 1986). All sylvatic isolates are genetically distinct from all endemic isolates, and are isolated evolutionarily (Gubler, 1997a; Rico-Hesse, 1990; Wang et al., 2000)(see discussion of genetics below). Intriguingly, although an African origin for Ae. aegypti is suggested by (i) the abundance in Africa of closely related Aedes (Stegomyia) spp., (ii) the lack of related Stegomyia subgenus Aedes mosquitoes in the Americas, and (iii) the existence only in Africa of sylvatic Ae. aegypti (Ae. aegypti formosus)(Carter, 1930; Christophers, 1960; Dyar, 1928; Powell, Tabachnick, and Arnold, 1980; Tabachnick and Powell, 1979), the sylvatic, African forms of DENV probably do not utilize Ae. aegypti formosus as an important vector. Indeed, this subspecies is relatively refractory to infection (Black et al., 2002; Diallo et al., 2008; Diallo et al., 2005).
Control of DENV spread relies primarily on reduction of vector populations by elimination of peridomestic water sources and insecticide applications. This strategy succeeded in most neotropical locations during the 1950s-1970s during campaigns to eliminate yellow fever, but was not sustained. Ae. aegypti has recently reinfested most neotropical and semi-neotropical locations, and DEN has returned with a vengeance (Gubler, 2002; Monath, 1994). Worldwide, changing lifestyles have enhanced Ae. aegypti densities by providing larval breeding sites for this anthropophilic and peridomestic species. Increases in air travel have resulted in the movement of virus strains among transmission foci, providing opportunities for sequential infection of populations by 2 or more serotypes, resulting in an increased incidence of DHF and DSS in many areas. No effective antiviral drugs are available to treat dengue infections. Several promising DEN vaccine candidates have been developed, but are not yet available for general use. A particular challenge with DEN vaccination is avoiding immune enhancement, which could occur if a vaccine is not protective against all 4 serotypes (Guy and Almond, 2008).
6. Contributions of phylogenetics to the understanding of dengue epidemiology
Early genetic comparisons of DENV strains relied on T1-RNase-resistant oligonucleotide fingerprinting, also called RNA fingerprinting. Initial studies of the 4 DENV serotypes revealed few shared T1-resistent oligonucleotides (Vezza et al., 1980). Repik et al. (Repik et al., 1983) first delineated topotypes of DENV-1 by showing that isolates from the same geographic region were very similar, but differed from those from other areas. Trent et al. also identified topotypes of DENV-2, and showed that different topotypes are not selectively the cause of severe DEN disease in Thailand (Trent et al., 1989). Evolution of the DENV-2 virus genome from 1962-1986 was gradual, with a slow accumulation of changes in the pattern of oligonucleotides through time, suggesting a balance of genetic drift and natural selection. Similar results followed soon thereafter for DENV-4 (Henchal et al., 1986).
During the late 1980’s, direct sequencing of viral RNA largely supplanted RNA fingerprinting for DENV molecular epidemiologic studies due to the greater efficiency and ability to characterize more precisely individual mutations. Rico-Hesse used RNA sequencing of DENV-1 and DENV-2 isolates to demonstrate additional evolutionary relationships between etiologic isolates of disease outbreaks, as well as the distinct nature of DENV-2 isolates from sylvatic cycles in West Africa (Rico-Hesse, 1990). Similar studies of NS1 gene sequences from DENV-2 strains revealed no correlation between disease severity, serological response, or year of isolation and NS1 nucleotide or amino acid sequences (Blok et al., 1991).
Phylogenetic studies of many mosquito-borne flaviviruses have revealed relatively long time periods between lineage divergence, suggesting a “boom and bust” pattern of intense diversification followed by a “pruning” of the tree via extinction of many lineages (Zanotto et al., 1996b). The best examples of this pattern are the four serotypes of DENV, which appear to be undergoing a period of rapid radiation, consistent with their rapid emergence since World War II (Holmes and Twiddy, 2003).
As the efficiency of viral RNA genome sequencing advanced with the advent of reverse transcription-polymerase chain reaction (RT-PCR) amplification and automated sequencing, more extensive analyses using longer sequences generated more robust phylogenies. Early studies of all 4 DENV serotypes demonstrated multiple lineages, often geographically-based, sometimes with implications for disease severity, as follows:
a. DENV-1
Early studies of DENV-1 evolution utilized direct RNA sequencing of 240 nucleotides across the E/NS1 gene junction. Five distinct genotypes were recognized, and these were later confirmed with complete E gene sequences showing a maximum nucleotide divergence of 6% among them (Goncalvez et al., 2002; Rico-Hesse, 1990).
Current phylogenies based on complete DENV-1 E gene sequences (Fig. 4) confirm the previously identified lineages as follows: (i) genotype I, representing strains from Southeast Asia, China and East Africa; (ii) genotype II, representing strains from Thailand collected in the 1950s and 1960s; (iii) genotype III, representing the sylvatic strain collected in Malaysia; (iv) genotype IV, representing strains from the West Pacific islands and Australia; and (v) genotype V, representing all strains collected in the Americas, strains from West Africa, and a limited number of strains collected from Asia.
Fig. 4.
Phylogenetic relationships of DENV-1 strains from the GenBank library. The phylogeny was inferred based on complete E gene nucleotide sequences and Bayesian analysis (one million reiterations) using MrBayes 3.1 (Ronquist and Huelsenbeck, 2003). Bayesian probability values are shown for key nodes. Virus strains are coded by abbreviated country of collection/strain name/year of collection.
b. DENV-2
DENV-2 evolution using homologous 240 nt sequences. As with DENV-1, 5 distinct genotypes were recognized, including one comprising the West African sylvatic strains (Rico-Hesse, 1990). Similar analyses using complete E gene sequences produced more robust results with similar genotypic groupings (Lewis et al., 1993). Extensive analyses of DENV-2 strains from Thailand revealed the simultaneous circulation of many DENV-2 variants simultaneously, with both Thai genotypes isolated from both DF and DHF cases (Rico-Hesse et al., 1998). Although the 240 nt E/NS1 gene sequences produced trees with many low bootstrap values, probably reflecting few informative nucleotides in the relatively short sequences, the conclusion was drawn that only 4 robust DENV-2 genotypes could be identified. Later, an analysis of E gene sequences from 147 DENV-2 isolates extended the range of the newly defined “Cosmopolitan” genotype to nearly global distribution, and identified 2 new genotypes that appeared to be restricted to Asia (Twiddy et al., 2002).
Current phylogenies of DENV based on complete E gene sequences (Fig. 5) confirm 5 major genotypes: (i) the Asian genotype, or Asian genotype 1, representing strains from Malaysia and Thailand, and Asian genotype 2 representing strains from Vietnam, China, Taiwan, Sri Lanka and the Philippines; (ii) the cosmopolitan genotype, representing strains of wide geographic distribution including Australia, East and West Africa, the Pacific and Indian ocean islands, the Indian subcontinent and the Middle East; (iii) the American genotype, representing strains from Latin America and older strains collected from the Caribbean, the Indian subcontinent and Pacific Islands in the 1950s and 1960s; (iv) the Southeast Asian/American genotype, representing strains from Thailand and Vietnam and strains collected in the Americas over the last 20 years; and (v) the sylvatic genotype, representing strains collected from humans, forest mosquitoes or sentinel monkeys in West Africa and Southeast Asia (Lewis et al., 1993; Rico-Hesse et al., 1997; Twiddy et al., 2002; Vasilakis, Tesh, and Weaver, 2008; Wang et al., 2000) (Fig. 5).
Fig. 5.
Phylogenetic relationships of DENV-2 strains from the GenBank library. The phylogeny was inferred based on complete E gene nucleotide sequences and Bayesian analysis (one million reiterations) using MrBayes 3.1 (Ronquist and Huelsenbeck, 2003). Bayesian probability values are shown for key nodes. Virus strains are coded by abbreviated country of collection/strain name/year of collection.
c. DENV-3
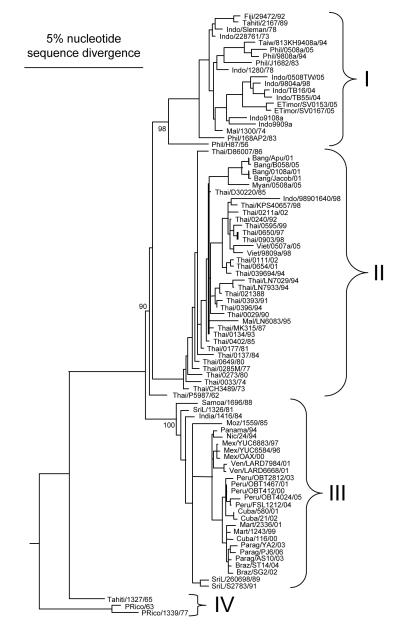
Focusing on the E gene, Lanciotti et al. (Lanciotti et al., 1994)identified geographically independent evolution of DENV-3 in 4 regions: subtype I was identified in Indonesia, Malaysia, the Philippines and the South Pacific islands; subtype II included strain from Thailand; subtype III consisted of viruses from Sri Lanka, India, Africa and Samoa; and subtype IV included of strains from Puerto Rico and Tahiti. Similar results were also obtained using NS3 gene sequences (Chow, Seah, and Chan, 1994).
Current DENV-3 phylogenies complete E gene sequences (Fig. 6) confirm the above described 4 genotypes plus the sylvatic genotype, as follows: (i) genotype I, representing strains from Indonesia, Malaysia, the Philippines and recent isolates from the South Pacific islands; (ii) genotype II, representing strains from Thailand, Vietnam and Bangladesh; (iii) genotype III, representing strains from Sri Lanka, India, Africa and Samoa. However, the complete genome phylogenetic analysis clearly includes the 1962 strain from Thailand within this genotype (Chao et al., 2005); and (iv) genotype IV, representing strains from Puerto Rico, Latin and central America and the 1965 Tahiti strain. Sylvatic strains of DENV-3 have not been isolated to date, but they are believed to exist in Malaysia, based on the seroconversion of sentinel monkeys (Rudnick, 1984a).
Fig. 6.
Phylogenetic relationships of DENV-3 strains from the GenBank library. The phylogeny was inferred based on complete E gene nucleotide sequences and Bayesian analysis (one million reiterations) using MrBayes 3.1 (Ronquist and Huelsenbeck, 2003). Bayesian probability values are shown for key nodes. Virus strains are coded by abbreviated country of collection/strain name/year of collection.
d. DENV-4
Initial analyses revealed that DENV-4 strains exhibited greater sequence conservation than the other DENV serotypes (92%) and 96-100% conservation in E protein amino acids. DEN-4 viruses were separated into two subtypes from: 1) the Philippines, Thailand and Sri Lanka, and; 2) Indonesia, Tahiti, the Caribbean, Central and South America (Lanciotti, Gubler, and Trent, 1997).
Current DENV-4 phylogenies complete E gene sequences (Fig. 7) delineate 4 major genotypes: (i) genotype I, representing strains from Thailand, the Philippines, Sri Lanka, and Japan (imported into Japan from Southeast Asia); (ii) genotype II, representing strains from Indonesia, Malaysia, Tahiti, the Caribbean and the Americas. Subsequent analysis with additional strains revealed putative evidence of intra-serotypic recombination amongst DENV-4 from independent ancestral lineages (most likely Indonesia 1976 and Malaysia 1969) may have contributed to the emergence of a distinct genotype, representing all Malaysian strains (AbuBakar, Wong, and Chan, 2002). Genotype II has become well established in the Caribbean since its introduction in the area in the early 1980s from Southeast Asia (Bennett et al., 2003; Foster et al., 2003); (iii) genotype III, representing recent Thai strains that are distinct from other Thai isolates (Klungthong et al., 2004); and (iv) genotype IV, representing the sylvatic strains from Malaysia.
Fig. 7.
Phylogenetic relationships of DENV-4 strains from the GenBank library. The phylogeny was inferred based on complete E gene nucleotide sequences and Bayesian analysis (one million reiterations) using MrBayes 3.1 (Ronquist and Huelsenbeck, 2003). Bayesian probability values are shown for key nodes. Virus strains are coded by abbreviated country of collection/strain name/year of collection.
e. Epidemic initiations and lineage replacements
The initiation of DEN epidemics has also been attributed to DENV strain introductions, as revealed by phylogenetics. Partial sequences of DENV-2 strains from the 1981 Cuban epidemic, the first in the Western hemisphere to include documented DHF cases, showed a close relationship to older DENV-2 strains from Asia, and more distant relationships to contemporary isolates from Jamaica and Vietnam. Partial genomic sequences derived from paraffin-embedded autopsy tissues of severe Cuban cases confirmed this DHF etiology (Sariol et al., 1999). DENV-4 was also first reported in the Americas in 1981. Bayesian coalescent studies revealed that, for both DENV-2 and DENV-4, the initial introductions were characterized by an exponential increase in the number of viral lineages, followed by constant genetic diversity (Carrington et al., 2005). DENV-4 apparently underwent a more rapid rate of population growth than DENV-2, allowing for a more efficient spread. Phylogenetic studies showed that DENV-2 outbreaks in Puerto Rico were associated with subtype IIIb, which was originally introduced from Asia. This lineage continued to circulate in Puerto Rico until 1994, when another was introduced from somewhere in the Western Hemisphere (Bennett et al., 2006). Positive selection has apparently acted on a several of amino acid sites in the E protein, some of which may have altered virulence based on their association with epidemics.
A similar history of DENV introduction and spread was reported from Brazil. Partial E/NS1 nucleotide sequences of DENV-1 and -2 isolates suggested that, since their initial introduction from 1981-1988, these lineages have continued to evolve without the introduction of new genotypes (Pires Neto et al., 2005).
A 1993-1994 Malaysian DEN epidemic was traced to the introduction of subtype II DENV-3 from Thailand, where it was isolated from 1962-1987. In contrast, earlier DENV-3 Malaysian isolates belonged to subtype I (Kobayashi et al., 1999). Similar studies in India suggest that genotype V of DENV-2 was replaced by genotype IV between 1967 and 1996 (Singh et al., 1999). DENV-3 strains isolated from Bangladesh in 2000 and 2001 were most closely related to recently emerged DENV-3 from neighboring Thailand and Myanmar, but distinct from those isolated in India and Sri Lanka (Podder et al., 2006). DENV-3, subtype III, which apparently originated in the Indian subcontinent, appears to have spread into Africa during the 1980s and from Africa into Latin America during the 1990s. Furthermore, DENV-3, subtype III isolates from mild versus severe disease outbreaks are phylogenetically distinct, suggesting differences in virulence among subtype III clades (Messer et al., 2003). Recently, Indian DEN epidemics have been determined phylogenetically to be linked to DENV-3 subtype III, which apparently replaced the earlier circulating DENV-2 (subtype IV) (Dash et al., 2006).
A detailed investigation of a DEN epidemic that occurred in French Polynesia from August 1996 to April 1997 revealed the introduction of DENV-2 after 7 years of DENV-3 circulation. Index cases were detected close to the international airport of Tahiti, and accompanying seroepidemiologic studies suggested transmission primarily within houses (Deparis et al., 1998). A 1996-1998 Guatemalan DEN epidemic was caused by DENV-3 strains that were apparently imported from Asia, Africa, or from a Pacific island (Usuku et al., 2001). Several 2002 DENV-4 outbreaks in north Queensland, Australia, were traced phylogenetically to Thailand or Indonesia as their likely origins (Hanna et al., 2003). Two Australian outbreaks of DEN occurred in 2003-2004, one that spread from Cairns to Townsville, the other from the Torres Strait to Cairns. Phylogenetic analyses indicated that both epidemics were initiated by viremic travelers from Papua New Guinea (Hanna et al., 2006).
Studies of E gene sequences from 2000-2001 Venezuelan DENV-3 isolates revealed that they are closely related to genotype III isolates previously detected Panama and Nicaragua, which have since spread through Central America and Mexico. This genotype apparently replaced the genotype V strain that circulated in Venezuela in the late 1960s and 1970s (Uzcategui et al., 2003). Similar studies indicate that DENV-3 genotype III was introduced into Brazil from the Caribbean Islands at least twice, and then into Paraguay from Brazil on at least three occasions (Aquino et al., 2008; Aquino et al., 2006). Ecuadorian DENV strains have also been linked phylogenetically to isolates of Caribbean origin (Regato et al., 2008).
Nearly simultaneous 2001 outbreaks of DENV-1 in Myanmar and several distant sites in the Pacific were shown phylogenetically to be caused by 3 different genotypes (I, II, and III). These results suggested multiple introductions from a variety of locations in Asia (Nuegoonpipat et al., 2004). Phylogenetic studies of isolates from the Myanmar outbreak, involving over 15,000 cases of DHF/DSS and 192 deaths, incriminated 2 lineages of DENV-1 that had diverged from an earlier, now extinct, lineage sometime before 1998 (Thu et al., 2004). Further phylogenetic analyses indicated that the two new lineages did not arise from the previously circulating DENV-1 viruses nor did they imply any differences in the fitness of the pre- and post-2001 viral populations, suggesting that a stochastic interepidemic event led to the replacement (Myat Thu et al., 2005). More recently, DENV-1 outbreaks occurred in Hawaii and Tahiti in 2001-2002. Phylogenetic analyses indicated that most Hawaiian isolates were imported from Tahiti and belonged to subtype IV (Imrie et al., 2006). One Hawaiian isolate from a Hawaii resident with a travel history to Samoa was linked to a DENV-1 strain previously isolated from another visitor to Samoa, indicating that 2 distinct strains of DENV-1 were introduced into Hawaii in 2001. A 2004 Chinese DEN epidemic in was linked to DENV-1 importation via a traveler returning from Thailand (Xu et al., 2007).
Phylogenetic studies have also revealed the apparent extinction of DENV genotypes and replacements at several locations. Extensive phylogenetic studies of DENV from Puerto Rico revealed marked evolutionary shifts in DENV-4 populations. Although viral lineages tend to be temporally clustered there, the most common genotype at a given time and place apparently arises from a previously rare lineage (Bennett et al., 2003). Recent replacements may, in part, have resulted from positive selection of three amino acid replacements in the NS2A gene. DENV-4 was introduced into the Caribbean on a single occasion in the early 1980s, and has readily dispersed in the region since, resulting in virus stains related more by time of isolation than by location (Foster et al., 2003).
The most extensive studies of DENV lineage replacements have been conducted in Thailand. Strains of DENV-2 circulating there prior to 1992 were apparently replaced by two new lineages that have not been identified elsewhere, suggesting their local evolution, and similar DENV-3 extinctions may have occurred there previously (Wittke et al., 2002). No evidence of positive selection in the DENV E gene was detected in either case. The authors speculated that, because these replacements occurred during interepidemic periods, genetic bottlenecks and genetic drift were responsible. DENV-1 also underwent a lineage turnover in Thailand, where a major clade replacement in genotype I occurred during the mid-1990s. The preceding increase in DENV-1 clade diversity was associated with a concomitant decrease in DENV-4 prevalence, while the subsequent clade replacement was associated with a decline in DENV-1 and an increase in DENV-4 (Zhang et al., 2005). The authors hypothesized that intraserotypic DENV diversification predominates at times of relative serotype abundance, and that clade replacements result from differential susceptibility to cross-reactive immunity from other serotypes. Modeling studies indicate that moderately cross-protective immunity gives rise to persistent out-of-phase oscillations similar to those observed between DENV-1 and -4, but that weak cross-protection (e.g. among DENV-1, -2, and -3) or cross-enhancement only produces in-phase patterns. Comprehensive analyses of DENV-2 revealed that 3 genotypes have circulated in Thailand, with Asian genotype predominating since 1991.
Recently, these comprehensive studies of DENV in Thailand have been extended to a smaller spatial scale by monitoring prospectively school children in Kamphaeng Phet during 2001 (Jarman et al., 2008). Phylogenetic studies of E gene sequences of DENV-1, -2, and -3 isolates revealed genetic variation in both time and space, with multiple DENV lineages circulating within individual schools, suggesting the frequent viral dispersal. Frequent introductions of DENV-2 sequences into the area occurred. The sequences tended to cluster within individual schools, but gene flow was also observed among nearby schools. Little sequence evolution was observed in DENV lineages within individual schools over this short timescale.
f. Genetic DENV correlates of DEN severity
Many investigators have compared the sequences of DENV isolates obtained from patients who experienced severe disease (DHF and/or DSS) with those isolated from cases of DEN fever. Most of these studies have identified genetic variation among strains, but not consistent sequences differences that correlated with disease severity (Blok et al., 1991; dos Santos et al., 2002; Mangada and Igarashi, 1998; Pandey et al., 2000; Raekiansyah et al., 2005; Sistayanarain et al., 1996; Uzcategui et al., 2001). One study of DENV-2 infections in northeastern Thailand reported that subtype III strains may result in milder disease irrespective of the immune status (primary or secondary DENV infection) compared with subtypes I or II, but this conclusion was based on a relatively small number of isolates (Pandey and Igarashi, 2000). Genetic studies of DENV-3 strains from a 2004 Indonesia epidemic indicated that they group with strains isolated in 1998 during a major epidemic in Sumatra, yet no newly-acquired amino acid changes were detected (Ong et al., 2008). These results suggested that a particular genotype with inherent epidemic potential reemerged in 2004.
The first strong evidence for genetic determinants of DEN severity came from studies of DENV-2, which indicated that the Asian genotype is associated with severe disease whereas sympatric American genotype isolates are not (Rico-Hesse et al., 1997). Detailed genomic sequence analyses of 11 strains from these 2 genotypes revealed 6 deduced amino acid charge differences between them, in the prM, E, NS4B, and NS5 genes. In addition, consistent sequence differences observed within the 5′ and 3′ untranslated regions (UTRs) were predicted to change RNA secondary structures (Leitmeyer et al., 1999). The authors hypothesized that the primary determinants of severe DEN reside in (i) amino acid 390 of the E protein, which may alter interactions with cellular receptors; (ii) in nt 68-80 of the 5′ UTR, which may be involved in translation initiation; and (iii) in the upstream 300 nucleotides of the 3′ UTR, which may regulate viral replication via the formation of replicative intermediates.
The abovementioned phylogenetic evidence of selection in the Americas for the more virulent Southeast Asian DENV-2 lineage has been corroborated by experimental data. The southeast Asian DENV-2 lineage strains associated with DHF/SSS generally replicate more efficiently in human dendritic cells, which are probably important human targets for DENV replication, than the American lineage strains (Cologna, Armstrong, and Rico-Hesse, 2005). Furthermore, 15 DENV-2 strains of both lineages of were compared their ability to infect and disseminate in several populations of Ae. aegypti (Armstrong and Rico-Hesse, 2003). Overall disseminated infection rates were higher for the SE Asian genotype strains, suggesting that both the human hosts and the endemic vector have selected for the more virulent SE Asian genotype strains.
5. Molecular evolution of dengue viruses: insights from phylogenetic and experimental studies
a. Selection pressures
In addition to elucidating patterns of DENV transmission and beginning to unravel determinants of severe disease, phylogenetic studies have also provided insights into their process of molecular evolution during natural transmission. As with nearly all arboviruses and other kinds of viruses as well, the identification of predominantly synonymous mutations during the natural evolution [see references above as well as (King et al., 2008)] of DENV implied that purifying selection (selection against most mutations, especially those that encode amino acid changes) dominated their evolution. Presumably, the DENV proteins have evolved to a high level of fitness for their alternate infection of mosquitoes and primates, which results in most amino acid changes being deleterious and purified quickly from the population by selection.
The first comprehensive attempt to identify selection pressures on DENV involved 147 DENV-2 strains (Twiddy et al., 2002). Comparisons of synonymous and nonsynonymous substitution rates in the E gene suggested that different DENV-2 genotypes have experienced different selection pressures. Maximum likelihood methods that evaluate differences in dN/dS ratios among codons suggested positive selection in the Cosmopolitan genotype and one of the two Asian genotypes. One mutation that appeared to be under positive selection, E-390, may affect transmissibility. However, no evidence of positive selection was detected during the emergence of the endemic DENV-2 lineage from the ancestral, sylvatic strains (Twiddy et al., 2002). Further studies with all 4 DENV serotypes identified several E gene codons in DENV-2, -3 and -4 but not -1 that apparently are subject to relatively weak positive selection (Twiddy, Woelk, and Holmes, 2002). Most of these sites were located in, or near to, potential T- or B-cell epitopes, suggesting positive immune selection. Although most other genes exhibited strong purifying selection, several positively selected amino acid substitutions were also identified in the NS2B and NS5 genes of DENV-2.
The effect of humoral immune selection on DENV evolution has received relatively little support. DENV-1 and -3 are reported to have lost cytotoxic lymphocyte epitopes during their evolution, based on computer epitope predictions and maximum parsimony analyses (Hughes, 2001).
b. Rates of DENV evolution
Early attempts to infer rates of DENV evolution employed simple genetic distance measurements and regression analyses (Lanciotti, Gubler, and Trent, 1997; Wang et al., 2000; Zanotto et al., 1996a). These methods generally yielded rate estimates of 6-8 × 10−4 subs/site/year. Newer, more sophisticated maximum likelihood methods applied to E gene sequences have produced similar estimates for DENV evolutionary rates and suggest that evolution generally approximates a molecular clock, although some minor rate differences occur (Twiddy, Holmes, and Rambaut, 2003). Rates estimated for the 4 DENV serotypes range from 4.55 × 10−4 for DENV-1 to 11.58 × 10−4 for genotype 3 of DENV-3. Statistical comparisons suggest that DENV-3 is evolving significantly faster than both DENV-1 and DENV-2. Comprehensive, genomic analyses of DENV-2 suggest that both the evolutionary rates and patterns of selection are similar for endemic versus sylvatic lineages, with the only exception being a uniquely high frequency of positive selection in the NS4B genes of sylvatic strains (Vasilakis et al., 2007a). These estimates are within a range determined for a variety of zoonotic arboviruses, and the alternating replication in arthropods and vertebrates, as well as the modulation of viral genome replication during persistent vector infections, may contribute to the slow rates of arboviruses (Hanley and Weaver, 2008).
c. DENV population sizes
The effects of population size on DENV has also been addressed by many phylogenetic studies. By comparing multiple DENV-2 isolates from humans infected both in 1980 and 1987, Sittisombut et al. (Sittisombut et al., 1997) first suggested that a genetic bottleneck and subsequent expansion of one or a limited number of subtype IIIa strains may have occurred during the intervening 6 years.
In addition to the epidemiological data indicating a dramatic increase in DENV circulation in many areas of the tropics, phylogenetic analyses have also documented increased genetic diversity consistent with viral population expansion. Combined with the growing evidence that DENV strains differ in virulence, this trend predicts the evolution of an expanded range of pathogenic properties (Holmes and Burch, 2000).
d. Recombination
The role of recombination in DENV evolution remains controversial. Phylogenetic methods such as bootscanning have been used to identify recombination in DENV-1 (Aaskov et al., 2007; AbuBakar, Wong, and Chan, 2002; Chen et al., 2008; Tolou et al., 2001; Uzcategui et al., 2001). The strongest evidence comes from a New Caledonian who was infected with both genotypes I and II, as well as a recombinant (Aaskov et al., 2007). Because recombination of flaviviruses has not been achieved experimentally despite concerted efforts (K.A. Hanley, personal communication), caution should be exercised when inferring conclusions about these putative recombination events. Several conditions should be met in order for natural recombination leading to the transmission of a recombinant strain to be conclusively confirmed. First, the recombinant crossover should be demonstrated in a single PCR amplicon following cloning to ensure that it occurs in a single cDNA molecule. Second, the recombination should be demonstrated repeatedly in clonal populations of viable virus (e.g. a plaque harvest or limited endpoint dilution isolate). And third, the recombinant should be maintained during post-recombination evolution (Tolou et al., 2001).
e. DENV quasispecies
Like other RNA viruses, DENV is believed to undergo low fidelity replication, resulting in virus populations described as quasispecies, or mutant swarms surrounding a master sequence (Domingo et al., 1985; Holland and Domingo, 1998). The presence of DENV quasispecies has been confirmed by sequencing multiple clones derived from RT-PCR products, including from unpassaged, natural isolates (Aviles et al., 2003; Wang et al., 2002). These quasispecies have been found to include significant proportions of genomes with stop codons in their E genes, as well as mixed genotypes and recombinants thereof (Craig et al., 2003). Using RT-PCR, concurrent mixed serotype (DENV-2 and DENV-3) human infections were detected in 2/21 patients studied in southern Taiwan in 2000 (Wang et al., 2003). A 2001 genetic DENV-1 variant containing a stop codon in the E gene, detected in both humans and mosquitoes from Myanmar, is reported to have spread to all individuals sampled (Aaskov et al., 2006). The maintenance of long-term transmission of these defective RNA genomes would require complementation via coinfection of host cells with DENV supplying a functional E gene. Therefore, high multiplicities of infection would need to be maintained at all stages of transmission and dissemination through both human hosts and mosquito vectors, which seems unlikely based on the bottlenecks at the stage of mosquito infection inferred from studies of other mosquito-borne flaviviruses (Scholle et al., 2004) or alphaviruses (Smith et al., 2008).
f. Constraints on DENV evolution imposed by mosquito-borne transmission
DENV are maintained by horizontal arthropod-borne (mosquito-borne or tick-borne) transmission among vertebrate hosts. As a consequence, DENV must replicate alternately in the very different environments created by invertebrate and vertebrate hosts. Flaviviruses, as well as other arthropod-borne viruses, exhibit a lower rate of sequence evolution than many other RNA viruses, such as poliovirus or HIV, that are maintained in single host transmission cycles (Jenkins et al., 2002; Parvin et al., 1986; Wolfs et al., 1990). The genetic stability of arboviruses presents a paradox, since it is known that the RNA-dependent RNA polymerase (RdRp) of RNA viruses does not possess proofreading activity (Holland et al., 1982; Steinhauer, Domingo, and Holland, 1992), and the absence of a repair mechanism leads to approximately one mutational event per genome replication. Thus, one could argue that the observed arbovirus stability could be attributed to their unique requirement for replication in divergent hosts, which could impose additional selective constraints on arboviruses compared to single-host viruses. Arboviruses may have adapted to replicate efficiently in either host through selection for a compromise genome, where optimal replication in one host involves a fitness tradeoff for the alternate host (Coffey et al., 2008; Cooper and Scott, 2001; Greene et al., 2005; Weaver et al., 1999; Weaver, Rico-Hesse, and Scott, 1992).
However, some experimental evidence does not support the notion that alternating host replication produces negative fitness trade-offs (Novella et al., 1999). Alternatively, the observed arbovirus stability could also be attributed to the establishment of persistent infections, a phenomenon most common in tick-borne arboviruses, where replication is probably modulated by RNA interference (Sanchez-Vargas et al., 2004). To date, many phylogenetic analyses suggest the presence of strong purifying selection on arboviruses in nature, supporting strong selective constraints on flavivirus evolution (Holmes, 2003; Jerzak et al., 2005; Klungthong et al., 2004; Woelk and Holmes, 2002). Consequently, arboviruses evolve at a slower rate than other RNA viruses transmitted by a single host (Jenkins et al., 2002).
As described above, DENV have been shown to circulate within hosts in nature as a population of closely related genomes around a dominant master genome sequence, indicating that DENV are maintained as a quasispecies. Therefore the existence of extensive genetic diversity within DENV populations suggests that genome variants within these populations may play a role in viral fitness, replication and their ability to successfully adapt to new and dynamic environments.
A study by Chen (Chen, Wu, and Chiou, 2003), examined the genetic diversity of DENV-2 variants after they were subjected to serial passage in mammalian (Vero), mosquito (C6/36) or alternating passage between them. Unlike serial passage in Vero cells or alternating passage between Vero and C6/36 cells, serial passage (20 or 30 times) in C6/36 cells did not result in any nucleotide change, suggesting that mosquito cells have a minimal effect on DENV evolution. However, since the conclusions of this study were based upon consensus sequences of a limited fraction of the DENV genome (2,541 nucleotides of the E/NS1 region), rather that characterization of the mutant spectrum of examined region, no meaningful inferences could be derived about the role of mosquitoes in DENV evolution.
A more recent study (Vasilakis and Weaver, unpublished), examined whether the human-to-mosquito transmission cycle may constrain DENV evolution by imposing constraints on optimal replication in either host, if fitness tradeoffs occur. Passaged but uncloned, as well as clonal DENV were serially passaged either in one cell type to eliminate host alternation, or in an alternating passage series between vertebrate liver (Huh-7) and mosquito (C6/36 Ae. albopictus) cells. DENV that were allowed to specialize in single host cells (10 passages) exhibited fitness gains but fitness losses in the bypassed cells, and most viruses passaged in 10 alternating cycles exhibited detectable fitness gains in both cells. Detection of several amino acid changes that were common in both passage series suggested convergent evolution via positive selection. These data were consistent with the hypothesis that releasing arboviruses from the alternating host cell transmission and replication cycle will result in the acquisition of higher fitness for the retained host cell (vertebrate or invertebrate), than will occur in an alternating cell cycle. However, the data provided limited support to the hypothesis that DENV and other arboviruses must adopt a fitness compromise, where optimal replication in one host cell or involves a fitness tradeoff for the alternate host cell. Support for this hypothesis was mainly provided by the alternate passage of the uncloned viruses, whereas in the cloned viruses significant fitness increases were observed in both cell types. Two possible explanations could account for this observation: (i) acquisition of host cell-specific and/or amphi-cell-specific adaptations could account for the ability of the alternating host cell passaged cloned viruses to acquire fitness gains almost similar to those observed from exclusive passage in the single host cell line and (ii) fitness increases in the alternate passage of the cloned viruses may be attributed to a low starting fitness due to the genetic bottleneck effects of the biological cloning, a phenomenon that has been observed previously (Chao, 1990; Duarte et al., 1992; Duarte et al., 1993; Escarmis et al., 1996; Weaver et al., 1999).
Interestingly, exclusive DENV-2 passage in the vertebrate host cell or alternating host cells led to the emergence of a qualitatively and quantitatively different mutation spectrum than exclusive passage in the invertebrate host cell line, whereas in the former the mutant spectrum was distributed throughout the genome, in the latter mutations located exclusively within the non-structural genes and 3′ - NCR. These observations support the notion that selection occurs mainly within the vertebrate host (Ciota et al., 2008). However, an inherent limitation of utilizing consensus sequences to generate inferences with regard to the fitness of viral populations is that consensus sequences only reflect the majority nucleotide at any given position of the viral genome, and do not represent the true range of the quasispecies spectrum of a master fit sequence that dominates the ensemble. Consequently, it is impossible to account for mutations present at a low frequency in the quasispecies ensemble that remain undetected, since low frequency viral sequences within the mutant spectrum may have dominant phenotypes. In fact, a recent West Nile virus (WNV) study demonstrated the importance of the mutant spectrum’s size in the establishment of the adaptive phenotype (Ciota et al., 2007).
6. Research trends and needs
Genetic studies of DENV isolates from mosquitoes and patients have made strong progress toward understanding patterns of spread and disease. Considerable progress has also been made toward understanding the patterns of genotype abundance and replacements that sometimes lead to new epidemics and that may regulate transmission dynamics. The continued advances in the efficiency, speed and cost of DNA sequencing are now opening new opportunities to revisit many aspects of DENV epidemiology and evolution by examining DENV strains from natural hosts at the population level. For example, new “deep sequencing” technology can efficiently sequence a large number of RNA viral genomes from a single host to provide more complete information on the quasispecies rather than just determining consensus sequences from RT-PCR amplicons. This will enable a more thorough understanding of the population dynamics and sizes of DENV at various stages of infection and transmission, and could also lead to the identification of sequence variants, present as minority populations, that are associated with DHF/DSS.
Recent progress has also been made by retrospective genetic studies of DENV from the sylvatic cycles. This work, combined with experimental studies of replication in models for human infection and in mosquitoes, suggests that the sylvatic genotypes have the capability to cause human infection and diseases, and can readily reemerge in the future unless effective vaccination is developed and sustained (Vasilakis et al., 2008; Vasilakis et al., 2007a; Vasilakis et al., 2007b; Vasilakis, Tesh, and Weaver, 2008). However, understanding the ecology and epidemiology of sylvatic DENV is essential to understanding and predicting emergence, yet has been largely ignored. Except for the pioneering studies of Rudnick and colleagues in Malaysia during the 1970s (Rudnick, 1984b), nothing is known of the distribution and ecology of sylvatic DENV in Asia, and human infections have never been identified or characterized. Longitudinal mosquito surveillance in Senegal has provided information on the temporal patterns of sylvatic DENV-2 amplification there (Diallo et al., 2003), and human infections including small epidemics caused by sylvatic strains have been characterized in West Africa (Vasilakis, Tesh, and Weaver, 2008). However, the ecological contact between sylvatic cycles and human populations is poorly studied, and the range of human disease and, most importantly viremia, caused by sylvatic DENV strains is unknown. Therefore, advances in understanding DENV emergence and predicting locations and times with the greatest potential will require comprehensive, prospective epidemiological and ecological studies in enzootic locations of West Africa and Asia. Such studies might even lead to the identification of additional serotypes of DENV that have not yet made their way into the urban transmission cycle.
Finally, the prospect of effective tetravalent DENV vaccination (Guy and Almond, 2008), which could control or even eradicate the urban cycles that rely only on human reservoir hosts, heightens the need for understanding the ease with which the sylvatic strains can reemerge to reinitiate urban transmission cycles.
Acknowledgements
We thank Shannan Rossi for expert graphic design of Figure 1. SCW’s dengue research is supported by NIH grant AI069145.
References
- Aaskov J, Buzacott K, Field E, Lowry K, Berlioz-Arthaud A, Holmes EC. Multiple recombinant dengue type 1 viruses in an isolate from a dengue patient. J Gen Virol. 2007;88(Pt 12):3334–40. doi: 10.1099/vir.0.83122-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aaskov J, Buzacott K, Thu HM, Lowry K, Holmes EC. Long-term transmission of defective RNA viruses in humans and Aedes mosquitoes. Science. 2006;311(5758):236–8. doi: 10.1126/science.1115030. [DOI] [PubMed] [Google Scholar]
- AbuBakar S, Wong PF, Chan YF. Emergence of dengue virus type 4 genotype IIA in Malaysia. J Gen Virol. 2002;83(Pt 10):2437–42. doi: 10.1099/0022-1317-83-10-2437. [DOI] [PubMed] [Google Scholar]
- Ackermann M, Padmanabhan R. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J Biol Chem. 2001;276(43):39926–37. doi: 10.1074/jbc.M104248200. [DOI] [PubMed] [Google Scholar]
- Ali M, Wagatsuma Y, Emch M, Breiman RF. Use of a geographic information system for defining spatial risk for dengue transmission in Bangladesh: role for Aedes albopictus in an urban outbreak. Am. J. Trop. Med. Hyg. 2003;69(6):634–40. [PubMed] [Google Scholar]
- Alvarez M, Rodriguez-Roche R, Bernardo L, Vazquez S, Morier L, Gonzalez D, Castro O, Kouri G, Halstead SB, Guzman MG. Dengue hemorrhagic Fever caused by sequential dengue 1-3 virus infections over a long time interval: Havana epidemic, 2001-2002. Am J Trop Med Hyg. 2006;75(6):1113–7. [PubMed] [Google Scholar]
- University of Ibadan Arbovirus Research Project Group B arboviruses. 1969. Anonymous.
- Aquino JD, Tang WF, Ishii R, Ono T, Eshita Y, Aono H, Makino Y. Molecular epidemiology of dengue virus serotypes 2 and 3 in Paraguay during 2001-2006: The association of viral clade introductions with shifting serotype dominance. Virus Res. 2008 doi: 10.1016/j.virusres.2008.07.011. [DOI] [PubMed] [Google Scholar]
- Aquino VH, Anatriello E, Goncalves PF, EV DAS, Vasconcelos PF, Vieira DS, Batista WC, Bobadilla ML, Vazquez C, Moran M, Figueiredo LT. Molecular epidemiology of dengue type 3 virus in Brazil and Paraguay, 2002-2004. Am J Trop Med Hyg. 2006;75(4):710–5. [PubMed] [Google Scholar]
- Armstrong PM, Rico-Hesse R. Efficiency of dengue serotype 2 virus strains to infect and disseminate in Aedes aegypti. Am J Trop Med Hyg. 2003;68(5):539–44. doi: 10.4269/ajtmh.2003.68.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviles G, Meissner J, Mantovani R, St Jeor S. Complete coding sequences of dengue-1 viruses from Paraguay and Argentina. Virus Res. 2003;98(1):75–82. doi: 10.1016/j.virusres.2003.08.018. [DOI] [PubMed] [Google Scholar]
- Aziz MM, Hasan KN, Hasanat MA, Siddiqui MA, Salimullah M, Chowdhury AK, Ahmed M, Alam MN, Hassan MS. Predominance of the DEN-3 genotype during the recent dengue outbreak in Bangladesh. Southeast Asian J Trop Med Public Health. 2002;33(1):42–8. [PubMed] [Google Scholar]
- Balaya S, Paul SD, D’Lima LV, Pavri KM. Investigations on an outbreak of dengue in Delhi in 1967. Indian J Med Res. 1969;57(4):767–74. [PubMed] [Google Scholar]
- Basaca-Sevilla V, Halstead SB. Recent virological studies on haemorrhagic fever and other arthropod-borne virus infections in the Philippines. J Trop Med Hyg. 1966;69(9):203–8. [PubMed] [Google Scholar]
- Bennett SN, Holmes EC, Chirivella M, Rodriguez DM, Beltran M, Vorndam V, Gubler DJ, McMillan WO. Selection-driven evolution of emergent dengue virus. Mol Biol Evol. 2003;20(10):1650–8. doi: 10.1093/molbev/msg182. [DOI] [PubMed] [Google Scholar]
- Bennett SN, Holmes EC, Chirivella M, Rodriguez DM, Beltran M, Vorndam V, Gubler DJ, McMillan WO. Molecular evolution of dengue 2 virus in Puerto Rico: positive selection in the viral envelope accompanies clade reintroduction. J Gen Virol. 2006;87(Pt 4):885–93. doi: 10.1099/vir.0.81309-0. [DOI] [PubMed] [Google Scholar]
- Billings L, Schwartz IB, Shaw LB, McCrary M, Burke DS, Cummings DA. Instabilities in multiserotype disease models with antibody-dependent enhancement. J Theor Biol. 2007;246(1):18–27. doi: 10.1016/j.jtbi.2006.12.023. [DOI] [PubMed] [Google Scholar]
- Black W. C. t., Bennett KE, Gorrochotegui-Escalante N, Barillas-Mury CV, Fernandez-Salas I, de Lourdes Munoz M, Farfan-Ale JA, Olson KE, Beaty BJ. Flavivirus susceptibility in Aedes aegypti. Arch. Med. Res. 2002;33(4):379–88. doi: 10.1016/s0188-4409(02)00373-9. [DOI] [PubMed] [Google Scholar]
- Blok J, Gibbs AJ, McWilliam SM, Vitarana UT. NS 1 gene sequences from eight dengue-2 viruses and their evolutionary relationships with other dengue-2 viruses. Arch Virol. 1991;118(3-4):209–23. doi: 10.1007/BF01314031. [DOI] [PubMed] [Google Scholar]
- Botros BA, Watts DM, Soliman AK, Salib AW, Moussa MI, Mursal H, Douglas C, Farah M. Serological evidence of dengue fever among refugees, Hargeysa, Somalia. J Med Virol. 1989;29(2):79–81. doi: 10.1002/jmv.1890290202. [DOI] [PubMed] [Google Scholar]
- Bravo JR, Guzman MG, Kouri GP. Why dengue haemorrhagic fever in Cuba? 1. Individual risk factors for dengue haemorrhagic fever/dengue shock syndrome (DHF/DSS) Trans R Soc Trop Med Hyg. 1987;81(5):816–20. doi: 10.1016/0035-9203(87)90041-1. [DOI] [PubMed] [Google Scholar]
- Brown AWA. Yellow Fever, Dengue and Dengue Haemorrhagic Fever. In: Howe GM, editor. A World Geography of Human Diseases. Academic Press; New York: 1977. pp. 271–317. [Google Scholar]
- Bylon D. Verhandelungen van het bataviaasch Genootschop der Konsten in Wetenschappen. Batavia: 1780. Korte Aantekening, wegens eene Algemeene Ziekte, Doorgaans Genaamd de Knokkel-Koorts; pp. 17–30. [Google Scholar]
- Cabrera-Batista B, Skewes-Ramm R, Fermin CD, Garry RF. Dengue in the Dominican Republic: epidemiology for 2004. Microsc Res Tech. 2005;68(3-4):250–4. doi: 10.1002/jemt.20225. [DOI] [PubMed] [Google Scholar]
- Calisher CH, Karabatsos N. Arbovirus serogroups: Definition and geographic distribution. In: Monath TP, editor. The Arboviruses: Epidemiology and Ecology, Vol. I. CRC Press; Boca Raton, Florida: 1988. pp. 19–57. [Google Scholar]
- Calisher CH, Karabatsos N, Dalrymple JM, Shope RE, Porterfield JS, Westaway EG, Brandt WE. Antigenic relationships between flaviviruses as determined by cross-neutralization tests with polyclonal antisera. J Gen Virol. 1989;70(Pt 1):37–43. doi: 10.1099/0022-1317-70-1-37. [DOI] [PubMed] [Google Scholar]
- Carey DE, Causey OR, Reddy S, Cooke AR. Dengue viruses from febrile patients in Nigeria, 1964-68. Lancet. 1971;1(7690):105–6. doi: 10.1016/s0140-6736(71)90840-3. [DOI] [PubMed] [Google Scholar]
- Carrington CV, Foster JE, Pybus OG, Bennett SN, Holmes EC. Invasion and maintenance of dengue virus type 2 and type 4 in the Americas. J Virol. 2005;79(23):14680–7. doi: 10.1128/JVI.79.23.14680-14687.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter HR, Carter LA, Frost WH, editors. Yellow fever :an epidemiological and historical study of its place of origin. Williams and Wilkins Co.; Baltimore, Maryland: 1930. [Google Scholar]
- Chang CJ, Luh HW, Wang SH, Lin HJ, Lee SC, Hu ST. The heterogeneous nuclear ribonucleoprotein K (hnRNP K) interacts with dengue virus core protein. DNA Cell Biol. 2001;20(9):569–77. doi: 10.1089/104454901317094981. [DOI] [PubMed] [Google Scholar]
- Chao DY, King CC, Wang WK, Chen WJ, Wu HL, Chang GJ. Strategically examining the full-genome of dengue virus type 3 in clinical isolates reveals its mutation spectra. Virol J. 2005;2:72. doi: 10.1186/1743-422X-2-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao L. Fitness of RNA virus decreased by Muller’s ratchet. Nature. 1990;348(6300):454–5. doi: 10.1038/348454a0. [DOI] [PubMed] [Google Scholar]
- Chen SP, Yu M, Jiang T, Deng YQ, Qin CF, Han JF, Qin ED. Identification of a recombinant dengue virus type 1 with 3 recombination regions in natural populations in Guangdong province, China. Arch Virol. 2008;153(6):1175–9. doi: 10.1007/s00705-008-0090-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WJ, Wu HR, Chiou SS. E/NS1 modifications of dengue 2 virus after serial passages in mammalian and/or mosquito cells. Intervirology. 2003;46(5):289–95. doi: 10.1159/000073208. [DOI] [PubMed] [Google Scholar]
- Chen Y, Maguire T, Marks RM. Demonstration of binding of dengue virus envelope protein to target cells. J Virol. 1996;70(12):8765–72. doi: 10.1128/jvi.70.12.8765-8772.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow VT, Seah CL, Chan YC. Comparative analysis of NS3 sequences of temporally separated dengue 3 virus strains isolated from southeast Asia. Intervirology. 1994;37(5):252–8. doi: 10.1159/000150386. [DOI] [PubMed] [Google Scholar]
- Christie J. On epidemics of dengue fever: their diffusion and etiology. Glasgow Medical Journal. 1881;16:161–176. [PMC free article] [PubMed] [Google Scholar]
- Christophers SR. Aedes aegypti (L.) The yellow fever mosquito; its life history, bionomics and structure. Cambridge University Press; London: 1960. [Google Scholar]
- Ciota AT, Lovelace AO, Jia Y, Davis LJ, Young DS, Kramer LD. Characterization of mosquito-adapted West Nile virus. J Gen Virol. 2008;89(Pt 7):1633–42. doi: 10.1099/vir.0.2008/000893-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciota AT, Ngo KA, Lovelace AO, Payne AF, Zhou Y, Shi P-Y, Kramer LD. Role of the mutant spectrum in adaptation and replication of West Nile virus. J. Gen. Virol. 2007;88:865–874. doi: 10.1099/vir.0.82606-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey LL, Vasilakis N, Brault AC, Powers AM, Tripet F, Weaver SC. Arbovirus evolution in vivo is constrained by host alternation. Proc Natl Acad Sci U S A. 2008;105(19):6970–5. doi: 10.1073/pnas.0712130105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cologna R, Armstrong PM, Rico-Hesse R. Selection for virulent dengue viruses occurs in humans and mosquitoes. J Virol. 2005;79(2):853–9. doi: 10.1128/JVI.79.2.853-859.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper LA, Scott TW. Differential evolution of eastern equine encephalitis virus populations in response to host cell type. Genetics. 2001;157(4):1403–12. doi: 10.1093/genetics/157.4.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copanaris P. L’epidemie de dengue en Grece au cours de l’ete 1928. Office International d’Hygiene Publique Bulletin. 1928;20:1590–1601. [Google Scholar]
- Cordeiro MT, Schatzmayr HG, Nogueira RM, Oliveira VF, Melo WT, Carvalho EF. Dengue and dengue hemorrhagic fever in the State of Pernambuco, 1995-2006. Rev Soc Bras Med Trop. 2007;40(6):605–611. doi: 10.1590/s0037-86822007000600001. [DOI] [PubMed] [Google Scholar]
- Cordellier R, Bouchite B, Roche JC, Monteny N, Diaco B, Akoliba P. Circulation silvatique du virus Dengue 2 en 1980, dans les savannes sub-soudaniennes du Cote d’Ivoire. Cah. ORSTOM Ser. Entomo. Med. Parasitol. 1983;21:165. [Google Scholar]
- Corwin AL, Larasati RP, Bangs MJ, Wuryadi S, Arjoso S, Sukri N, Listyaningsih E, Hartati S, Namursa R, Anwar Z, Chandra S, Loho B, Ahmad H, Campbell JR, Porter KR. Epidemic dengue transmission in southern Sumatra, Indonesia. Trans R Soc Trop Med Hyg. 2001;95(3):257–65. doi: 10.1016/s0035-9203(01)90229-9. [DOI] [PubMed] [Google Scholar]
- Craig S, Thu HM, Lowry K, Wang XF, Holmes EC, Aaskov J. Diverse dengue type 2 virus populations contain recombinant and both parental viruses in a single mosquito host. J Virol. 2003;77(7):4463–7. doi: 10.1128/JVI.77.7.4463-4467.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings DA, Irizarry RA, Huang NE, Endy TP, Nisalak A, Ungchusak K, Burke DS. Travelling waves in the occurrence of dengue haemorrhagic fever in Thailand. Nature. 2004;427(6972):344–7. doi: 10.1038/nature02225. [DOI] [PubMed] [Google Scholar]
- Cunha RV, Schatzmayr HG, Miagostovich MP, Barbosa AM, Paiva FG, Miranda RM, Ramos CC, Coelho JC, dos Santos FB, Nogueira RM. Dengue epidemic in the State of Rio Grande do Norte, Brazil, in 1997. Trans R Soc Trop Med Hyg. 1999;93(3):247–9. doi: 10.1016/s0035-9203(99)90008-1. [DOI] [PubMed] [Google Scholar]
- Daniels CW. Notes on the mosquitoes on the river and coast districts on the eastern side of the Peninsula. Stud. Inst. Med. Res. Malaya. 1908;3:262–266. [Google Scholar]
- Dash PK, Parida MM, Saxena P, Abhyankar A, Singh CP, Tewari KN, Jana AM, Sekhar K, Rao PV. Reemergence of dengue virus type-3 (subtype-III) in India: implications for increased incidence of DHF & DSS. Virol J. 2006;3:55. doi: 10.1186/1743-422X-3-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deparis X, Roche C, Murgue B, Chungue E. Possible dengue sequential infection: dengue spread in a neighbourhood during the 1996/97 dengue-2 epidemic in French Polynesia. Trop Med Int Health. 1998;3(11):866–71. doi: 10.1046/j.1365-3156.1998.00330.x. [DOI] [PubMed] [Google Scholar]
- Deubel V, Murgue B. Dengue. In: Service MW, editor. The Encyclopedia of Arthropod-transmitted Infections. CAB International; Wallingford, UK: 2001. pp. 133–143. [Google Scholar]
- Diallo M, Ba Y, Faye O, Soumare ML, Dia I, Sall AA. Vector competence of Aedes aegypti populations from Senegal for sylvatic and epidemic dengue 2 virus isolated in West Africa. Trans R Soc Trop Med Hyg. 2008;102(5):493–8. doi: 10.1016/j.trstmh.2008.02.010. [DOI] [PubMed] [Google Scholar]
- Diallo M, Ba Y, Sall AA, Diop OM, Ndione JA, Mondo M, Girault L, Mathiot C. Amplification of the sylvatic cycle of dengue virus type 2, Senegal, 1999-2000: entomologic findings and epidemiologic considerations. Emerg Infect Dis. 2003;9(3):362–7. doi: 10.3201/eid0903.020219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diallo M, Sall AA, Moncayo AC, Ba Y, Fernandez Z, Ortiz D, Coffey LL, Mathiot C, Tesh RB, Weaver SC. Potential role of sylvatic and domestic african mosquito species in dengue emergence. Am J Trop Med Hyg. 2005;73(2):445–9. [PubMed] [Google Scholar]
- Domingo E, Martinez-Salas E, Sobrino F, de la Torre JC, Portela A, Ortin J, Lopéz-Galindez C, Péres-Brena P, Villanueva N, Nájera R, VandePol S, Steinhauer D, DePolo N, Holland JJ. The quasispecies (extremely heterogeneous) nature of viral RNA genome populations: biological relevance -- a review. Gene. 1985;40:1–8. doi: 10.1016/0378-1119(85)90017-4. [DOI] [PubMed] [Google Scholar]
- dos Santos CN, Rocha CF, Cordeiro M, Fragoso SP, Rey F, Deubel V, Despres P. Genome analysis of dengue type-1 virus isolated between 1990 and 2001 in Brazil reveals a remarkable conservation of the structural proteins but amino acid differences in the non-structural proteins. Virus Res. 2002;90(1-2):197–205. doi: 10.1016/s0168-1702(02)00180-6. [DOI] [PubMed] [Google Scholar]
- Duarte E, Clarke D, Moya A, Domingo E, Holland J. Rapid fitness losses in mammalian RNA virus clones due to Muller’s ratchet. Proc Natl Acad Sci U S A. 1992;89(13):6015–9. doi: 10.1073/pnas.89.13.6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte EA, Clarke DK, Moya A, Elena SF, Domingo E, Holland J. Many-trillionfold amplification of single RNA virus particles fails to overcome the Muller’s ratchet effect. J Virol. 1993;67(6):3620–3. doi: 10.1128/jvi.67.6.3620-3623.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyar HG. The mosquitoes of the Americas. Carnegie Institute of Washington; Washington: 1928. [Google Scholar]
- Edington AD. Dengue”, as seen in the recent epidemic in Durban. The Journal of the Medical Association of South Africa. 1927;1:446–448. [Google Scholar]
- Egloff MP, Benarroch D, Selisko B, Romette JL, Canard B. An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. Embo J. 2002;21(11):2757–68. doi: 10.1093/emboj/21.11.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenkranz NJ, Ventura AK, Cuadrado RR, Pond WL, Porter JE. Pandemic dengue in Caribbean countries and the southern United States--past, present and potential problems. N Engl J Med. 1971;285(26):1460–9. doi: 10.1056/NEJM197112232852606. [DOI] [PubMed] [Google Scholar]
- Erbel P, Schiering N, D’Arcy A, Renatus M, Kroemer M, Lim SP, Yin Z, Keller TH, Vasudevan SG, Hommel U. Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus. Nat Struct Mol Biol. 2006;13(4):372–3. doi: 10.1038/nsmb1073. [DOI] [PubMed] [Google Scholar]
- Escarmis C, Davila M, Charpentier N, Bracho A, Moya A, Domingo E. Genetic lesions associated with Muller’s ratchet in an RNA virus. J Mol Biol. 1996;264(2):255–67. doi: 10.1006/jmbi.1996.0639. [DOI] [PubMed] [Google Scholar]
- Fagbami AH, Fabiyi A. Epidemiology of dengue infections in Nigeria: virus isolations and clinical observations, 1972-1975. J Trop Med Hyg. 1976;79(10):226–8. [PubMed] [Google Scholar]
- Fagbami AH, Monath TP, Fabiyi A. Dengue virus infections in Nigeria: a survey for antibodies in monkeys and humans. Trans R Soc Trop Med Hyg. 1977;71(1):60–5. doi: 10.1016/0035-9203(77)90210-3. [DOI] [PubMed] [Google Scholar]
- Foster JE, Bennett SN, Vaughan H, Vorndam V, McMillan WO, Carrington CV. Molecular evolution and phylogeny of dengue type 4 virus in the Caribbean. Virology. 2003;306(1):126–34. doi: 10.1016/s0042-6822(02)00033-8. [DOI] [PubMed] [Google Scholar]
- Goncalvez AP, Escalante AA, Pujol FH, Ludert JE, Tovar D, Salas RA, Liprandi F. Diversity and evolution of the envelope gene of dengue virus type 1. Virology. 2002;303(1):110–9. doi: 10.1006/viro.2002.1686. [DOI] [PubMed] [Google Scholar]
- Gonzalez JP, Du Saussay C, Gautun JC, McCormick JB, Mouchet J. Dengue in Burkina Faso (ex-Upper Volta): seasonal epidemics in the urban area of Ouagadougou] Bull Soc Pathol Exot Filiales. 1985;78(1):7–14. [PubMed] [Google Scholar]
- Gorbalenya AE, Donchenko AP, Koonin EV, Blinov VM. N-terminal domains of putative helicases of flavi- and pestiviruses may be serine proteases. Nucleic Acids Res. 1989;17(10):3889–97. doi: 10.1093/nar/17.10.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene IP, Wang E, Deardorff ER, Milleron R, Domingo E, Weaver SC. Effect of alternating passage on adaptation of sindbis virus to vertebrate and invertebrate cells. J Virol. 2005;79(22):14253–60. doi: 10.1128/JVI.79.22.14253-14260.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler DJ. Dengue. In: Monath TP, editor. The Arboviruses: Epidemiology and Ecology, Vol. II. 5 vols. CRC Press; Boca Raton, Florida: 1988. pp. 223–260. [Google Scholar]
- Gubler DJ. Perspectives on the prevention and control of dengue hemorrhagic fever. Gaoxiong YiXue Ke Xue Za Zhi. 1994;10(Suppl):S15–8. [PubMed] [Google Scholar]
- Gubler DJ. Dengue and dengue hemorrhagic fever: its history and resurgence as a global public health problem. In: Gubler DJ, Kuno G, editors. Dengue and Dengue Hemorrhagic Fever. CAB International; New York: 1997a. pp. 1–22. [Google Scholar]
- Gubler DJ. Dengue and dengue hemorrhagic fever:its history and resurgence as a global public health problem. In: Gubler DJ, Kuno G, editors. Dengue and Dengue Hemorrhagic Fever. CABI Publishing; Oxon: 1997b. pp. 1–22. [Google Scholar]
- Gubler DJ. Epidemic dengue/dengue hemorrhagic fever as a public health, social and economic problem in the 21st century. Trends Microbiol. 2002;10(2):100–3. doi: 10.1016/s0966-842x(01)02288-0. [DOI] [PubMed] [Google Scholar]
- Gubler DJ, Nalim S, Tan R, Saipan H, Sulianti Saroso J. Variation in susceptibility to oral infection with dengue viruses among geographic strains of Aedes aegypti. Am. J. Trop. Med. Hyg. 1979;28(6):1045–52. doi: 10.4269/ajtmh.1979.28.1045. [DOI] [PubMed] [Google Scholar]
- Gubler DJ, Reed D, Rosen L, Hitchcock JR., Jr. Epidemiologic, clinical, and virologic observations on dengue in the Kingdom of Tonga. Am. J. Trop. Med. Hyg. 1978;27(3):581–9. doi: 10.4269/ajtmh.1978.27.581. [DOI] [PubMed] [Google Scholar]
- Gubler DJ, Rosen L. Variation among geographic strains of Aedes albopictus in susceptibility to infection with dengue viruses. Am. J. Trop. Med. Hyg. 1976;25(2):318–25. doi: 10.4269/ajtmh.1976.25.318. [DOI] [PubMed] [Google Scholar]
- Gubler DJ, Sather GE, Kuno G, Cabral JR. Dengue 3 virus transmission in Africa. Am J Trop Med Hyg. 1986;35(6):1280–4. doi: 10.4269/ajtmh.1986.35.1280. [DOI] [PubMed] [Google Scholar]
- Guy B, Almond JW. Towards a dengue vaccine: progress to date and remaining challenges. Comp Immunol Microbiol Infect Dis. 2008;31(2-3):239–52. doi: 10.1016/j.cimid.2007.07.011. [DOI] [PubMed] [Google Scholar]
- Guzman MG, Kouri G, Morier L, Soler M, Fernandez A. A study of fatal hemorrhagic dengue cases in Cuba, 1981. Bull Pan Am Health Organ. 1984;18(3):213–20. [PubMed] [Google Scholar]
- Ha DQ, Tien NT, Huong VT, Loan HT, Thang CM. Dengue epidemic in southern Vietnam, 1998. Emerg Infect Dis. 2000;6(4):422–5. doi: 10.3201/eid0604.000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halstead SB. Neutralization and antibody-dependent enhancement of dengue viruses. Adv Virus Res. 2003;60:421–67. doi: 10.1016/s0065-3527(03)60011-4. [DOI] [PubMed] [Google Scholar]
- Halstead SB, Voulgaropoulos EM, Tien NH, Udomsakdi S. Dengue hemorrhagic fever in South Vietnam: report of the 1963 outbreak. Am J Trop Med Hyg. 1965;14(5):819–30. doi: 10.4269/ajtmh.1965.14.819. [DOI] [PubMed] [Google Scholar]
- Halstead SB, Yamarat C. Recent Epidemics Of Hemorrhagic Fever In Thailand. Observations Related To Pathogenesis Of A “New” Dengue Disease. Am J Public Health Nations Health. 1965;55:1386–95. doi: 10.2105/ajph.55.9.1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammon WM, Rudnick A, Sather G. Viruses associated with epidemic hemorrhagic fevers of the Philippines and Thailand. Science. 1960;131:1102–1103. doi: 10.1126/science.131.3407.1102. [DOI] [PubMed] [Google Scholar]
- Hammon WM, Rudnick A, Sather G, Rogers KD, Morse LJ. New hemorrhagic fevers of childern in the Philippines and Thailand. Trans. Assoc. Am. Phys. 1960;73:140–155. [PubMed] [Google Scholar]
- Hanley KA, Weaver SC. Arbovirus Evolution. In: Domingo E, Parrish CR, Holland JJ, editors. Origin and Evolution of Viruses. Elsevier; Oxford: 2008. pp. 351–392. [Google Scholar]
- Hanna JN, Ritchie SA, Hills SL, Pyke AT, Montgomery BL, Richards AR, Piispanen JP. Dengue in north Queensland, 2002. Commun Dis Intell. 2003;27(3):384–9. doi: 10.33321/cdi.2003.27.66. [DOI] [PubMed] [Google Scholar]
- Hanna JN, Ritchie SA, Richards AR, Taylor CT, Pyke AT, Montgomery BL, Piispanen JP, Morgan AK, Humphreys JL. Multiple outbreaks of dengue serotype 2 in north Queensland, 2003/04. Aust N Z J Public Health. 2006;30(3):220–5. doi: 10.1111/j.1467-842x.2006.tb00861.x. [DOI] [PubMed] [Google Scholar]
- Harrington LC, Edman JD, Scott TW. Why do female Aedes aegypti (Diptera: Culicidae) feed preferentially and frequently on human blood? J Med Entomol. 2001;38(3):411–22. doi: 10.1603/0022-2585-38.3.411. [DOI] [PubMed] [Google Scholar]
- Harris E, Videa E, Perez L, Sandoval E, Tellez Y, Perez ML, Cuadra R, Rocha J, Idiaquez W, Alonso RE, Delgado MA, Campo LA, Acevedo F, Gonzalez A, Amador JJ, Balmaseda A. Clinical, epidemiologic, and virologic features of dengue in the 1998 epidemic in Nicaragua. Am J Trop Med Hyg. 2000;63(1-2):5–11. doi: 10.4269/ajtmh.2000.63.5. [DOI] [PubMed] [Google Scholar]
- Henchal EA, Repik PM, McCown JM, Brandt WE. Identification of an antigenic and genetic variant of dengue-4 virus from the Caribbean. Am J Trop Med Hyg. 1986;35(2):393–400. doi: 10.4269/ajtmh.1986.35.393. [DOI] [PubMed] [Google Scholar]
- Hirsch A. Dengue, a comparatively new desease: its symptoms. In: Creighton C, editor. Handbook of Geographical and Historical Pathology. Vol. I. III vols. Syndenham Society; 1883. pp. 55–81. [Google Scholar]
- Holland J, Domingo E. Origin and evolution of viruses. Virus Genes. 1998;16(1):13–21. doi: 10.1023/a:1007989407305. [DOI] [PubMed] [Google Scholar]
- Holland J, Spindler K, Horodyski F, Grabau E, Nichol S, VandePol S. Rapid evolution of RNA genomes. Science. 1982;215(4540):1577–85. doi: 10.1126/science.7041255. [DOI] [PubMed] [Google Scholar]
- Holmes EC. Patterns of intra- and interhost nonsynonymous variation reveal strong purifying selection in dengue virus. J Virol. 2003;77(20):11296–8. doi: 10.1128/JVI.77.20.11296-11298.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes EC, Burch SS. The causes and consequences of genetic variation in dengue virus. Trends Microbiol. 2000;8(2):74–7. doi: 10.1016/s0966-842x(99)01669-8. [DOI] [PubMed] [Google Scholar]
- Holmes EC, Twiddy SS. The origin, emergence and evolutionary genetics of dengue virus. Infect Genet Evol. 2003;3(1):19–28. doi: 10.1016/s1567-1348(03)00004-2. [DOI] [PubMed] [Google Scholar]
- Hota S. Experimental studies on dengue. I. Isolation, identification and modification of the virus. J. Infect. Dis. 1952;90:1–9. doi: 10.1093/infdis/90.1.1. [DOI] [PubMed] [Google Scholar]
- Hughes AL. Evolutionary change of predicted cytotoxic T cell epitopes of dengue virus. Infect Genet Evol. 2001;1(2):123–30. doi: 10.1016/s1567-1348(01)00013-2. [DOI] [PubMed] [Google Scholar]
- Hyams KC, Oldfield EC, Scott RM, Bourgeois AL, Gardiner H, Pazzaglia G, Moussa M, Saleh AS, Dawi OE, Daniell FD. Evaluation of febrile patients in Port Sudan, Sudan: isolation of dengue virus. Am J Trop Med Hyg. 1986;35(4):860–5. doi: 10.4269/ajtmh.1986.35.860. [DOI] [PubMed] [Google Scholar]
- Imrie A, Zhao Z, Bennett SN, Kitsutani P, Laille M, Effler P. Molecular epidemiology of dengue in the Pacific: introduction of two distinct strains of dengue virus type-1 [corrected] into Hawaii. Ann Trop Med Parasitol. 2006;100(4):327–36. doi: 10.1179/136485906X105589. [DOI] [PubMed] [Google Scholar]
- Jacobs MG, Robinson PJ, Bletchly C, Mackenzie JM, Young PR. Dengue virus nonstructural protein 1 is expressed in a glycosyl-phosphatidylinositol-linked form that is capable of signal transduction. Faseb J. 2000;14(11):1603–10. doi: 10.1096/fj.14.11.1603. [DOI] [PubMed] [Google Scholar]
- Jarman RG, Holmes EC, Rodpradit P, Klungthong C, Gibbons RV, Nisalak A, Rothman AL, Libraty DH, Ennis FA, Mammen MP, Jr., Endy TP. Microevolution of Dengue viruses circulating among primary school children in Kamphaeng Phet, Thailand. J Virol. 2008;82(11):5494–500. doi: 10.1128/JVI.02728-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins GM, Rambaut A, Pybus OG, Holmes EC. Rates of molecular evolution in RNA viruses: a quantitative phylogenetic analysis. J Mol Evol. 2002;54(2):156–65. doi: 10.1007/s00239-001-0064-3. [DOI] [PubMed] [Google Scholar]
- Jerzak G, Bernard KA, Kramer LD, Ebel GD. Genetic variation in West Nile virus from naturally infected mosquitoes and birds suggests quasispecies structure and strong purifying selection. J Gen Virol. 2005;86(Pt 8):2175–83. doi: 10.1099/vir.0.81015-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AJ, Guirakhoo F, Roehrig JT. The envelope glycoproteins of dengue 1 and dengue 2 viruses grown in mosquito cells differ in their utilization of potential glycosylation sites. Virology. 1994;203(2):241–9. doi: 10.1006/viro.1994.1481. [DOI] [PubMed] [Google Scholar]
- Johnson BK, Ocheng D, Gichogo A, Okiro M, Libondo D, Kinyanjui P, Tukei PM. Epidemic dengue fever caused by dengue type 2 virus in Kenya: preliminary results of human virological and serological studies. East Afr Med J. 1982;59(12):781–4. [PubMed] [Google Scholar]
- Jones M, Davidson A, Hibbert L, Gruenwald P, Schlaak J, Ball S, Foster GR, Jacobs M. Dengue virus inhibits alpha interferon signaling by reducing STAT2 expression. J Virol. 2005;79(9):5414–20. doi: 10.1128/JVI.79.9.5414-5420.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalayanarooj S, Nimmannitya S. Is dengue severity related to nutritional status? Southeast Asian J Trop Med Public Health. 2005;36(2):378–84. [PubMed] [Google Scholar]
- Khromykh AA, Varnavski AN, Sedlak PL, Westaway EG. Coupling between replication and packaging of flavivirus RNA: evidence derived from the use of DNA-based full-length cDNA clones of Kunjin virus. J Virol. 2001;75(10):4633–40. doi: 10.1128/JVI.75.10.4633-4640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King CC, Chao DY, Chien LJ, Chang GJ, Lin TH, Wu YC, Huang JH. Comparative analysis of full genomic sequences among different genotypes of dengue virus type 3. Virol J. 2008;5:63. doi: 10.1186/1743-422X-5-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klungthong C, Zhang C, Mammen MP, Jr., Ubol S, Holmes EC. The molecular epidemiology of dengue virus serotype 4 in Bangkok, Thailand. Virology. 2004;329(1):168–79. doi: 10.1016/j.virol.2004.08.003. [DOI] [PubMed] [Google Scholar]
- Kobayashi N, Thayan R, Sugimoto C, Oda K, Saat Z, Vijayamalar B, Sinniah M, Igarashi A. Type-3 dengue viruses responsible for the dengue epidemic in Malaysia during 1993-1994. Am J Trop Med Hyg. 1999;60(6):904–9. doi: 10.4269/ajtmh.1999.60.904. [DOI] [PubMed] [Google Scholar]
- Kokernot RH, Smithburn KC, Weinbren MP. Neutralizing antibodies to arthropod-borne viruses in human beings and animals in the union of South Africa. J. Immunol. 1956;77:313–323. [PubMed] [Google Scholar]
- Kouri G, Mas P, Guzman MG, Soler M, Goyenechea A, Morier L. Dengue hemorrhagic fever in Cuba, 1981: rapid diagnosis of the etiologic agent. Bull Pan Am Health Organ. 1983;17(2):126–32. [PubMed] [Google Scholar]
- Kouri GP, Guzman MG, Bravo JR. Why dengue haemorrhagic fever in Cuba? 2. An integral analysis. Trans R Soc Trop Med Hyg. 1987;81(5):821–3. doi: 10.1016/0035-9203(87)90042-3. [DOI] [PubMed] [Google Scholar]
- Kouri GP, Guzman MG, Bravo JR, Triana C. Dengue haemorrhagic fever/dengue shock syndrome: lessons from the Cuban epidemic, 1981. Bull World Health Organ. 1989;67(4):375–80. [PMC free article] [PubMed] [Google Scholar]
- Kuhn RJ, Zhang W, Rossmann MG, Pletnev SV, Corver J, Lenches E, Jones CT, Mukhopadhyay S, Chipman PR, Strauss EG, Baker TS, Strauss JH. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell. 2002;108(5):717–25. doi: 10.1016/s0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuno G, Chang GJ, Tsuchiya KR, Karabatsos N, Cropp CB. Phylogeny of the genus Flavivirus. J. Virol. 1998;72(1):73–83. doi: 10.1128/jvi.72.1.73-83.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurane I, Ennis FE. Immunity and immunopathology in dengue virus infections. Sem. Immunol. 1992;4:121–127. [PubMed] [Google Scholar]
- Kusriastuti R, Sutomo S. Evolution of dengue Prevention and control programme in Indonesia. Dengue Bull. 2005;29:1–7. [Google Scholar]
- Lanciotti RS, Gubler DJ, Trent DW. Molecular evolution and phylogeny of dengue-4 viruses. J Gen Virol. 1997;78(Pt 9):2279–84. doi: 10.1099/0022-1317-78-9-2279. [DOI] [PubMed] [Google Scholar]
- Lanciotti RS, Lewis JG, Gubler DJ, Trent DW. Molecular evolution and epidemiology of dengue-3 viruses. J. Gen. Virol. 1994;75(Pt 1):65–75. doi: 10.1099/0022-1317-75-1-65. [DOI] [PubMed] [Google Scholar]
- Leichtenstern O. Influenza and Dengue. In: Nothnagel H, editor. Specielle Pathologie und Therapie. Vol. IV. Alfred Holder; Wien: 1896. pp. 133–226. [Google Scholar]
- Leitmeyer KC, Vaughn DW, Watts DM, Salas R, Villalobos I, de C, Ramos C, Rico-Hesse R. Dengue virus structural differences that correlate with pathogenesis. J Virol. 1999;73(6):4738–47. doi: 10.1128/jvi.73.6.4738-4747.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung D, Schroder K, White H, Fang NX, Stoermer MJ, Abbenante G, Martin JL, Young PR, Fairlie DP. Activity of recombinant dengue 2 virus NS3 protease in the presence of a truncated NS2B co-factor, small peptide substrates, and inhibitors. J Biol Chem. 2001;276(49):45762–71. doi: 10.1074/jbc.M107360200. [DOI] [PubMed] [Google Scholar]
- Lewis JA, Chang GJ, Lanciotti RS, Kinney RM, Mayer LW, Trent DW. Phylogenetic relationships of dengue-2 viruses. Virology. 1993;197(1):216–24. doi: 10.1006/viro.1993.1582. [DOI] [PubMed] [Google Scholar]
- Li H, Clum S, You S, Ebner KE, Padmanabhan R. The serine protease and RNA-stimulated nucleoside triphosphatase and RNA helicase functional domains of dengue virus type 2 NS3 converge within a region of 20 amino acids. J Virol. 1999;73(4):3108–16. doi: 10.1128/jvi.73.4.3108-3116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Lok SM, Yu IM, Zhang Y, Kuhn RJ, Chen J, Rossmann MG. The flavivirus precursor membrane-envelope protein complex: structure and maturation. Science. 2008;319(5871):1830–4. doi: 10.1126/science.1153263. [DOI] [PubMed] [Google Scholar]
- Lindenbach BD, Rice CM. Molecular biology of flaviviruses. Adv Virus Res. 2003;59:23–61. doi: 10.1016/s0065-3527(03)59002-9. [DOI] [PubMed] [Google Scholar]
- Mangada MN, Igarashi A. Molecular and in vitro analysis of eight dengue type 2 viruses isolated from patients exhibiting different disease severities. Virology. 1998;244(2):458–66. doi: 10.1006/viro.1998.9093. [DOI] [PubMed] [Google Scholar]
- Medin CL, Fitzgerald KA, Rothman AL. Dengue virus nonstructural protein NS5 induces interleukin-8 transcription and secretion. J Virol. 2005;79(17):11053–61. doi: 10.1128/JVI.79.17.11053-11061.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer WB, Gubler DJ, Harris E, Sivananthan K, de Silva AM. Emergence and global spread of a dengue serotype 3, subtype III virus. Emerg Infect Dis. 2003;9(7):800–9. doi: 10.3201/eid0907.030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modis Y, Ogata S, Clements D, Harrison SC. Structure of the dengue virus envelope protein after membrane fusion. Nature. 2004;427(6972):313–9. doi: 10.1038/nature02165. [DOI] [PubMed] [Google Scholar]
- Monath TP. Dengue: the risk to developed and developing countries. Proc. Natl. Acad. Sci. USA. 1994;91(7):2395–400. doi: 10.1073/pnas.91.7.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monlun E, Zeller H, Traore-Lamizana M, Hervy JP, Adam F, Mondo M, Digoutte JP. Caracteres cliniques et epidemiologiques de la dengue 2 au Senegal. Med. Mal. Infect. 1992;22:718–721. [Google Scholar]
- More FW. Observations on dengue fever in Singapore. J. Malaya Branch Br. Med. Assoc. 1904;1:24–29. [Google Scholar]
- Munoz-Jordan JL, Sanchez-Burgos GG, Laurent-Rolle M, Garcia-Sastre A. Inhibition of interferon signaling by dengue virus. Proc Natl Acad Sci U S A. 2003;100(24):14333–8. doi: 10.1073/pnas.2335168100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myat Thu H, Lowry K, Jiang L, Hlaing T, Holmes EC, Aaskov J. Lineage extinction and replacement in dengue type 1 virus populations are due to stochastic events rather than to natural selection. Virology. 2005;336(2):163–72. doi: 10.1016/j.virol.2005.03.018. [DOI] [PubMed] [Google Scholar]
- Nogueira RM, de Araujo JM, Schatzmayr HG. Dengue viruses in Brazil, 1986-2006. Rev Panam Salud Publica. 2007;22(5):358–63. doi: 10.1590/s1020-49892007001000009. [DOI] [PubMed] [Google Scholar]
- Nogueira RM, Zagner SM, Martins IS, Lampe E, Miagostovich MP, Schatzmayr HG. Dengue haemorrhagic fever/dengue shock syndrome (DHF/DSS) caused by serotype 2 in Brazil. Mem Inst Oswaldo Cruz. 1991;86(2):269. doi: 10.1590/s0074-02761991000200018. [DOI] [PubMed] [Google Scholar]
- Novella IS, Hershey CL, Escarmis C, Domingo E, Holland JJ. Lack of evolutionary stasis during alternating replication of an arbovirus in insect and mammalian cells. J Mol Biol. 1999;287(3):459–65. doi: 10.1006/jmbi.1999.2635. [DOI] [PubMed] [Google Scholar]
- Nuegoonpipat A, Berlioz-Arthaud A, Chow V, Endy T, Lowry K, Mai le Q, Ninh TU, Pyke A, Reid M, Reynes JM, Su Yun ST, Thu HM, Wong SS, Holmes EC, Aaskov J. Sustained transmission of dengue virus type 1 in the Pacific due to repeated introductions of different Asian strains. Virology. 2004;329(2):505–12. doi: 10.1016/j.virol.2004.08.029. [DOI] [PubMed] [Google Scholar]
- Ong SH, Yip JT, Chen YL, Liu W, Harun S, Lystiyaningsih E, Heriyanto B, Beckett CG, Mitchell WP, Hibberd ML, Suwandono A, Vasudevan SG, Schreiber MJ. Periodic re-emergence of endemic strains with strong epidemic potential-a proposed explanation for the 2004 Indonesian dengue epidemic. Infect Genet Evol. 2008;8(2):191–204. doi: 10.1016/j.meegid.2007.12.005. [DOI] [PubMed] [Google Scholar]
- Ooi E-E, Goh K-T, Gubler DJ. Dengue prevention and 35 years vector control in Singapore. Emerg Infect Dis. 2006;12(6):887–893. doi: 10.3201/10.3201/eid1206.051210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PAHO Number of Reported Cases of Dengue & Dengue Hemorrhagic Fever (DHF), Region of the Americas (by country and subregion) 2007 http://www.paho.org/English/AD/DPC/CD/dengue-cases-2007.htm.
- Pandey BD, Igarashi A. Severity-related molecular differences among nineteen strains of dengue type 2 viruses. Microbiol Immunol. 2000;44(3):179–88. doi: 10.1111/j.1348-0421.2000.tb02480.x. [DOI] [PubMed] [Google Scholar]
- Pandey BD, Morita K, Hasebe F, Parquet MC, Igarashi A. Molecular evolution, distribution and genetic relationship among the dengue 2 viruses isolated from different clinical severity. Southeast Asian J Trop Med Public Health. 2000;31(2):266–72. [PubMed] [Google Scholar]
- Parvin JD, Moscona A, Pan WT, Leider JM, Palese P. Measurement of the mutation rates of animal viruses: influenza A virus and poliovirus type 1. J Virol. 1986;59(2):377–83. doi: 10.1128/jvi.59.2.377-383.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper P. A note on David Bylon and dengue. Ann. Med. Hist. 1941;3:363–368. [PMC free article] [PubMed] [Google Scholar]
- Pires Neto RJ, Lima DM, de Paula SO, Lima CM, Rocco IM, Fonseca BA. Molecular epidemiology of type 1 and 2 dengue viruses in Brazil from 1988 to 2001. Braz J Med Biol Res. 2005;38(6):843–52. doi: 10.1590/s0100-879x2005000600005. [DOI] [PubMed] [Google Scholar]
- Podder G, Breiman RF, Azim T, Thu HM, Velathanthiri N, Mai le Q, Lowry K, Aaskov JG. Origin of dengue type 3 viruses associated with the dengue outbreak in Dhaka, Bangladesh, in 2000 and 2001. Am J Trop Med Hyg. 2006;74(2):263–5. [PubMed] [Google Scholar]
- Powell JR, Tabachnick WJ, Arnold J. Genetics and the origin of a vector population: Aedes aegypti, a case study. Science. 1980;208(4450):1385–7. doi: 10.1126/science.7375945. [DOI] [PubMed] [Google Scholar]
- Pryor MJ, Rawlinson SM, Butcher RE, Barton CL, Waterhouse TA, Vasudevan SG, Bardin PG, Wright PJ, Jans DA, Davidson AD. Nuclear localization of dengue virus nonstructural protein 5 through its importin alpha/beta-recognized nuclear localization sequences is integral to viral infection. Traffic. 2007;8(7):795–807. doi: 10.1111/j.1600-0854.2007.00579.x. [DOI] [PubMed] [Google Scholar]
- Raekiansyah M, Pramesyanti A, Bela B, Kosasih H, Ma’roef CN, Tobing SY, Rudiman PI, Alisjahbana B, Endi TP, Green S, Kalayanarooj S, Rothman AL, Sudiro TM. Genetic variations and relationship among dengue virus type 3 strains isolated from patients with mild or severe form of dengue disease in Indonesia and Thailand. Southeast Asian J Trop Med Public Health. 2005;36(5):1187–97. [PubMed] [Google Scholar]
- Regato M, Recarey R, Moratorio G, de Mora D, Garcia-Aguirre L, Gonzalez M, Mosquera C, Alava A, Fajardo A, Alvarez M, L DA, Dubra A, Martinez M, Khan B, Cristina J. Phylogenetic analysis of the NS5 gene of dengue viruses isolated in Ecuador. Virus Res. 2008;132(1-2):197–200. doi: 10.1016/j.virusres.2007.10.012. [DOI] [PubMed] [Google Scholar]
- Repik PM, Dalrymple JM, Brandt WE, McCown JM, Russell PK. RNA fingerprinting as a method for distinguishing dengue 1 virus strains. Am J Trop Med Hyg. 1983;32(3):577–89. doi: 10.4269/ajtmh.1983.32.577. [DOI] [PubMed] [Google Scholar]
- Rey FA, Heinz FX, Mandl C, Kunz C, Harrison SC. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature. 1995;375(6529):291–8. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- Rico-Hesse R. Molecular evolution and distribution of dengue viruses type 1 and 2 in nature. Virology. 1990;174(2):479–93. doi: 10.1016/0042-6822(90)90102-w. [DOI] [PubMed] [Google Scholar]
- Rico-Hesse R, Harrison LM, Nisalak A, Vaughn DW, Kalayanarooj S, Green S, Rothman AL, Ennis FA. Molecular evolution of dengue type 2 virus in Thailand. Am J Trop Med Hyg. 1998;58(1):96–101. doi: 10.4269/ajtmh.1998.58.96. [DOI] [PubMed] [Google Scholar]
- Rico-Hesse R, Harrison LM, Salas RA, Tovar D, Nisalak A, Ramos C, Boshell J, de Mesa MT, Nogueira RM, da Rosa AT. Origins of dengue type 2 viruses associated with increased pathogenicity in the Americas. Virology. 1997;230(2):244–51. doi: 10.1006/viro.1997.8504. [DOI] [PubMed] [Google Scholar]
- Rigau-Perez JG, Vorndam AV, Clark GG. The dengue and dengue hemorrhagic fever epidemic in Puerto Rico, 1994-1995. Am J Trop Med Hyg. 2001;64(1-2):67–74. doi: 10.4269/ajtmh.2001.64.67. [DOI] [PubMed] [Google Scholar]
- Robin Y, Cornet M, Heme G, Le Gonidec G. Isolement du virus dela dengue au Senegal. Ann. Virol (Institut Pasteur) 1980;131:149–154. [Google Scholar]
- Roche JC, Cordellier R, Hervy JP, Digoutte JP, Monteny N. Isolement de 96 souches de virus dengue 2 à partir de moustiques capturés en Côte d’Ivoire et Haute Volta. Ann. Virol. (Institut Pasteur) 1983;143:233–244. [Google Scholar]
- Rodier GR, Gubler DJ, Cope SE, Cropp CB, Soliman AK, Polycarpe D, Abdourhaman MA, Parra JP, Maslin J, Arthur RR. Epidemic dengue 2 in the city of Djibouti 1991-1992. Trans R Soc Trop Med Hyg. 1996;90(3):237–40. doi: 10.1016/s0035-9203(96)90228-x. [DOI] [PubMed] [Google Scholar]
- Roehrig JT, Bolin RA, Kelly RG. Monoclonal antibody mapping of the envelope glycoprotein of the dengue 2 virus, Jamaica. Virology. 1998;246(2):317–28. doi: 10.1006/viro.1998.9200. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19(12):1572–4. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Rudnick A. Studies of the ecology of dengue in Malaysia: a preliminary report. J. Med. Entomol. 1965;2(2):203–8. doi: 10.1093/jmedent/2.2.203. [DOI] [PubMed] [Google Scholar]
- Rudnick A. Ecology of Dengue virus. Asian J. Infect. Dis. 1978;2:156–160. [Google Scholar]
- Rudnick A. Proceedings of the International Conference on dengue/DHF; Kuala Lampur, Malaysia. 1984a. [Google Scholar]
- Rudnick A. The ecology of the dengue virus complex in Peninsular Malaysia. In: Pang T, Pathmanathan R, editors. Proceedings of the International Conference on Dengue/DHF; Kuala Lumpur: University of Malaysia Press; 1984b. p. 7. [Google Scholar]
- Rudnick A. Dengue virus ecology in Malaysia. Inst. Med. Res. Malays. Bull. 1986;23:51–152. [Google Scholar]
- Rudnick A, Lim TW. Dengue fever studies in Malaysia. Institute of Medical Research of Malaysia Bulletin. 1986;23:51–152. [Google Scholar]
- Rudnick A, Marchette NJ, Garcia R. Possible jungle dengue--recent studies and hypotheses. Jpn. J. Med. Sci. Biol. 1967;20:69–74. [PubMed] [Google Scholar]
- Rush AB. Medical Inquiries and Observations. Prichard and Hall; Philadelphia: 1789. An account of the bilious remitting fever, as it appeared in philadelphia in the summer and autumn of the year 1980; pp. 89–100. [Google Scholar]
- Russell PK, Buescher EL, McCown JM, Ordonez J. Recovery of dengue viruses from patients during epidemics in Puerto Rico and East Pakistan. Am J Trop Med Hyg. 1966;15(4):573–9. doi: 10.4269/ajtmh.1966.15.573. [DOI] [PubMed] [Google Scholar]
- Sabin AB. Research on dengue during World War II. Am J Trop Med Hyg. 1952;1(1):30–50. doi: 10.4269/ajtmh.1952.1.30. [DOI] [PubMed] [Google Scholar]
- Sabin AB, Schlesinger MC. Production of immunity to dengue with virus modified by propagation in mice. Science. 1945;101:640–642. doi: 10.1126/science.101.2634.640. [DOI] [PubMed] [Google Scholar]
- Saleh AS, Hassan A, Scott RM, Mellick PW, Oldfield EC, 3rd, Podgore JK. Dengue in north-east Africa. Lancet. 1985;2(8448):211–2. doi: 10.1016/s0140-6736(85)91521-1. [DOI] [PubMed] [Google Scholar]
- Saluzzo JF, Cornet M, Castagnet P, Rey C, Digoutte JP. Isolation of dengue 2 and dengue 4 viruses from patients in Senegal. Trans R Soc Trop Med Hyg. 1986;80(1):5. doi: 10.1016/0035-9203(86)90182-3. [DOI] [PubMed] [Google Scholar]
- Sanchez IJ, Ruiz BH. A single nucleotide change in the E protein gene of dengue virus 2 Mexican strain affects neurovirulence in mice. J Gen Virol. 1996;77(Pt 10):2541–5. doi: 10.1099/0022-1317-77-10-2541. [DOI] [PubMed] [Google Scholar]
- Sanchez-Vargas I, Travanty EA, Keene KM, Franz AW, Beaty BJ, Blair CD, Olson KE. RNA interference, arthropod-borne viruses, and mosquitoes. Virus Res. 2004;102(1):65–74. doi: 10.1016/j.virusres.2004.01.017. [DOI] [PubMed] [Google Scholar]
- Sariol CA, Pelegrino JL, Martinez A, Arteaga E, Kouri G, Guzman MG. Detection and genetic relationship of dengue virus sequences in seventeen-year-old paraffin-embedded samples from Cuba. Am J Trop Med Hyg. 1999;61(6):994–1000. doi: 10.4269/ajtmh.1999.61.994. [DOI] [PubMed] [Google Scholar]
- Saugrain J, Rosen L, Outin-Fabre D, Moreau JP. [A recent epidemic due to arbovirus infections of the dengue type in Tahiti. Comparative study of the 1964 epidemic] Bull Soc Pathol Exot Filiales. 1970;63(6):636–42. [PubMed] [Google Scholar]
- Scholle F, Girard YA, Zhao Q, Higgs S, Mason PW. trans-Packaged West Nile virus-like particles: infectious properties in vitro and in infected mosquito vectors. J Virol. 2004;78(21):11605–14. doi: 10.1128/JVI.78.21.11605-11614.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh UB, Maitra A, Broor S, Rai A, Pasha ST, Seth P. Partial nucleotide sequencing and molecular evolution of epidemic causing Dengue 2 strains. J Infect Dis. 1999;180(4):959–65. doi: 10.1086/315043. [DOI] [PubMed] [Google Scholar]
- Sistayanarain A, Maneekarn N, Polprasert B, Sirisanthana V, Makino Y, Fukunaga T, Sittisombut N. Primary sequence of the envelope glycoprotein of a dengue type 2 virus isolated from patient with dengue hemorrhagic fever and encephalopathy. Southeast Asian J Trop Med Public Health. 1996;27(2):221–7. [PubMed] [Google Scholar]
- Sittisombut N, Sistayanarain A, Cardosa MJ, Salminen M, Damrongdachakul S, Kalayanarooj S, Rojanasuphot S, Supawadee J, Maneekarn N. Possible occurrence of a genetic bottleneck in dengue serotype 2 viruses between the 1980 and 1987 epidemic seasons in Bangkok, Thailand. Am J Trop Med Hyg. 1997;57(1):1008. doi: 10.4269/ajtmh.1997.57.100. [DOI] [PubMed] [Google Scholar]
- Smith CE. The history of dengue in tropical Asia and its probable relationship to the mosquito Aedes aegypti. J Trop Med Hyg. 1956;59(10):243–51. [PubMed] [Google Scholar]
- Smith DR, Adams AP, Kenney JL, Wang E, Weaver SC. Venezuelan equine encephalitis virus in the mosquito vector Aedes taeniorhynchus: infection initiated by a small number of susceptible epithelial cells and a population bottleneck. Virology. 2008;372(1):176–86. doi: 10.1016/j.virol.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton AT. The mosquitoes of the Far East ports with special referenceto the prevalence of Stegomyia fasciata. Bull. Ent. Res. 1920;10:333–346. [Google Scholar]
- Steadman GW. Some account of an anomalous disease which raged in the islands of St. Thomas and Santa Cruz, in the West Indies, during the months of September, October, November, December, and January 1827-8. Edinb. Med. and Surg. Journ. 1828;30(97):227–248. [PMC free article] [PubMed] [Google Scholar]
- Steinhauer DA, Domingo E, Holland JJ. Lack of evidence for proofreading mechanisms associated with an RNA virus polymerase. Gene. 1992;122(2):281–8. doi: 10.1016/0378-1119(92)90216-c. [DOI] [PubMed] [Google Scholar]
- Tabachnick WJ, Powell JR. A world-wide survey of genetic variation in the yellow fever mosquito, Aedes aegypti. Genet Res. 1979;34(3):215–29. doi: 10.1017/s0016672300019467. [DOI] [PubMed] [Google Scholar]
- Thomas SJ, Strickman D, Vaughn DW. Dengue epidemiology: virus epidemiology, ecology, and emergence. Adv Virus Res. 2003;61:235–89. doi: 10.1016/s0065-3527(03)61006-7. [DOI] [PubMed] [Google Scholar]
- Thu HM, Lowry K, Myint TT, Shwe TN, Han AM, Khin KK, Thant KZ, Thein S, Aaskov J. Myanmar dengue outbreak associated with displacement of serotypes 2, 3, and 4 by dengue 1. Emerg Infect Dis. 2004;10(4):593–7. doi: 10.3201/eid1004.030216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolou HJ, Couissinier-Paris P, Durand JP, Mercier V, de Pina JJ, de Micco P, Billoir F, Charrel RN, de Lamballerie X. Evidence for recombination in natural populations of dengue virus type 1 based on the analysis of complete genome sequences. J Gen Virol. 2001;82(Pt 6):1283–90. doi: 10.1099/0022-1317-82-6-1283. [DOI] [PubMed] [Google Scholar]
- Trent DW. Antigenic characterization of flavivirus structural proteins separated by isoelectric focusing. J Virol. 1977;22(3):608–18. doi: 10.1128/jvi.22.3.608-618.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trent DW, Grant JA, Monath TP, Manske CL, Corina M, Fox GE. Genetic variation and microevolution of dengue 2 virus in Southeast Asia. Virology. 1989;172(2):523–35. doi: 10.1016/0042-6822(89)90195-5. [DOI] [PubMed] [Google Scholar]
- Trent DW, Manske CL, Fox GE, Chu MC, Kliks SC, Monath TP. The molecular epidemiology of dengue viruses: genetic variation and microevolution. Applied Virology Research. 1990;2:293–315. [Google Scholar]
- Twiddy SS, Farrar JJ, Vinh Chau N, Wills B, Gould EA, Gritsun T, Lloyd G, Holmes EC. Phylogenetic relationships and differential selection pressures among genotypes of dengue-2 virus. Virology. 2002;298(1):63–72. doi: 10.1006/viro.2002.1447. [DOI] [PubMed] [Google Scholar]
- Twiddy SS, Holmes EC, Rambaut A. Inferring the rate and time-scale of dengue virus evolution. Mol Biol Evol. 2003;20(1):122–9. doi: 10.1093/molbev/msg010. [DOI] [PubMed] [Google Scholar]
- Twiddy SS, Woelk CH, Holmes EC. Phylogenetic evidence for adaptive evolution of dengue viruses in nature. J Gen Virol. 2002;83(Pt 7):1679–89. doi: 10.1099/0022-1317-83-7-1679. [DOI] [PubMed] [Google Scholar]
- Uchil PD, Kumar AV, Satchidanandam V. Nuclear localization of flavivirus RNA synthesis in infected cells. J Virol. 2006;80(11):5451–64. doi: 10.1128/JVI.01982-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usuku S, Castillo L, Sugimoto C, Noguchi Y, Yogo Y, Kobayashi N. Phylogenetic analysis of dengue-3 viruses prevalent in Guatemala during 1996-1998. Arch Virol. 2001;146(7):1381–90. doi: 10.1007/s007050170098. [DOI] [PubMed] [Google Scholar]
- Uzcategui NY, Camacho D, Comach G, Cuello de Uzcategui R, Holmes EC, Gould EA. Molecular epidemiology of dengue type 2 virus in Venezuela: evidence for in situ virus evolution and recombination. J Gen Virol. 2001;82(Pt 12):2945–53. doi: 10.1099/0022-1317-82-12-2945. [DOI] [PubMed] [Google Scholar]
- Uzcategui NY, Comach G, Camacho D, Salcedo M, Cabello de Quintana M, Jimenez M, Sierra G, Cuello de Uzcategui R, James WS, Turner S, Holmes EC, Gould EA. Molecular epidemiology of dengue virus type 3 in Venezuela. J Gen Virol. 2003;84(Pt 6):1569–75. doi: 10.1099/vir.0.18807-0. [DOI] [PubMed] [Google Scholar]
- Vasilakis N, Durbin A, Travassos da Rosa APA, Munoz-Jordan JL, Tesh RB, Weaver SC. Antigenic relationships between sylvatic and endemic dengue viruses. Am J Trop Med Hyg. 2008;79(1):128–132. [PubMed] [Google Scholar]
- Vasilakis N, Holmes EC, Fokam EB, Faye O, Diallo M, Sall AA, Weaver SC. Evolutionary processes among sylvatic Dengue-2 viruses. Journal of Virology. 2007a;81:9591–9595. doi: 10.1128/JVI.02776-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilakis N, Shell EJ, Fokam EB, Mason PW, Hanley KA, Estes DM, Weaver SC. Potential of ancestral sylvatic dengue-2 viruses to re-emerge. Virology. 2007b;358(2):402–12. doi: 10.1016/j.virol.2006.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilakis N, Tesh RB, Weaver SC. Sylvatic dengue virus type 2 activity in humans, Nigeria, 1966. Emerg Infect Dis. 2008;14(3):502–4. doi: 10.3201/eid1403.070843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vezza AC, Rosen L, Repik P, Dalrymple J, Bishop DH. Characterization of the viral RNA species of prototype dengue viruses. Am J Trop Med Hyg. 1980;29(4):643–52. doi: 10.4269/ajtmh.1980.29.643. [DOI] [PubMed] [Google Scholar]
- Wang E, Ni H, Xu R, Barrett AD, Watowich SJ, Gubler DJ, Weaver SC. Evolutionary relationships of endemic/epidemic and sylvatic dengue viruses. J Virol. 2000;74(7):3227–34. doi: 10.1128/jvi.74.7.3227-3234.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WK, Chao DY, Lin SR, King CC, Chang SC. Concurrent infections by two dengue virus serotypes among dengue patients in Taiwan. J Microbiol Immunol Infect. 2003;36(2):89–95. [PubMed] [Google Scholar]
- Wang WK, Sung TL, Lee CN, Lin TY, King CC. Sequence diversity of the capsid gene and the nonstructural gene NS2B of dengue-3 virus in vivo. Virology. 2002;303(1):181–91. doi: 10.1006/viro.2002.1635. [DOI] [PubMed] [Google Scholar]
- Weaver SC, Brault AC, Kang W, Holland JJ. Genetic and fitness changes accompanying adaptation of an arbovirus to vertebrate and invertebrate cells. J Virol. 1999;73(5):4316–26. doi: 10.1128/jvi.73.5.4316-4326.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver SC, Coffey LL, Nussenzveig R, Ortiz D, Smith D. Vector Competence. In: Gillespie SH, Smith GL, Osbourn A, editors. Microbe–vector Interactions in Vector-borne Diseases. Cambridge University Press; Cambridge: 2004. pp. 139–180. [Google Scholar]
- Weaver SC, Rico-Hesse R, Scott TW. Genetic diversity and slow rates of evolution in New World alphaviruses. Curr Top Microbiol Immunol. 1992;176:99–117. doi: 10.1007/978-3-642-77011-1_7. [DOI] [PubMed] [Google Scholar]
- White RS. Three years mosquito control work in Calcutta. Bull. Ent. Res. 1934;25:551–573. [Google Scholar]
- WHO . In: Trend of dengue case and CFR in SEAR countries. W. R. O. f. S. E. Asia, editor. Vol. 2008. WHO; 2006. [Google Scholar]
- Winkler G, Maxwell SE, Ruemmler C, Stollar V. Newly synthesized dengue-2 virus nonstructural protein NS1 is a soluble protein but becomes partially hydrophobic and membrane-associated after dimerization. Virology. 1989;171(1):302–5. doi: 10.1016/0042-6822(89)90544-8. [DOI] [PubMed] [Google Scholar]
- Wittke V, Robb TE, Thu HM, Nisalak A, Nimmannitya S, Kalayanrooj S, Vaughn DW, Endy TP, Holmes EC, Aaskov JG. Extinction and rapid emergence of strains of dengue 3 virus during an interepidemic period. Virology. 2002;301(1):148–56. doi: 10.1006/viro.2002.1549. [DOI] [PubMed] [Google Scholar]
- Woelk CH, Holmes EC. Reduced positive selection in vector-borne RNA viruses. Mol Biol Evol. 2002;19(12):2333–6. doi: 10.1093/oxfordjournals.molbev.a004059. [DOI] [PubMed] [Google Scholar]
- Wolfs TF, de Jong JJ, Van den Berg H, Tijnagel JM, Krone WJ, Goudsmit J. Evolution of sequences encoding the principal neutralization epitope of human immunodeficiency virus 1 is host dependent, rapid, and continuous. Proc Natl Acad Sci U S A. 1990;87(24):9938–42. doi: 10.1073/pnas.87.24.9938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Dong H, Shi N, Liu S, Zhou A, Cheng Z, Chen G, Liu J, Fang T, Zhang H, Gu C, Tan X, Ye J, Xie S, Cao G. An outbreak of dengue virus serotype 1 infection in Cixi, Ningbo, People’s Republic of China, 2004, associated with a traveler from Thailand and high density of Aedes albopictus. Am J Trop Med Hyg. 2007;76(6):1182–8. [PubMed] [Google Scholar]
- Yu IM, Zhang W, Holdaway HA, Li L, Kostyuchenko VA, Chipman PR, Kuhn RJ, Rossmann MG, Chen J. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science. 2008;319(5871):1834–7. doi: 10.1126/science.1153264. [DOI] [PubMed] [Google Scholar]
- Zanotto PM, Gould EA, Gao GF, Harvey PH, Holmes EC. Population dynamics of flaviviruses revealed by molecular phylogenies. Proc Natl Acad Sci U S A. 1996a;93(2):548–53. doi: 10.1073/pnas.93.2.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanotto PM, Gould EA, Gao GF, Harvey PH, Holmes EC. Population dynamics of flaviviruses revealed by molecular phylogenies [see comments] Proc. Natl. Acad. Sci. U.S.A. 1996b;93(2):548–53. doi: 10.1073/pnas.93.2.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Mammen MP, Jr., Chinnawirotpisan P, Klungthong C, Rodpradit P, Monkongdee P, Nimmannitya S, Kalayanarooj S, Holmes EC. Clade replacements in dengue virus serotypes 1 and 3 are associated with changing serotype prevalence. J Virol. 2005;79(24):15123–30. doi: 10.1128/JVI.79.24.15123-15130.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]