Abstract
The expansion of the N-terminal poly-glutamine tract of the huntingtin (Htt) protein is responsible for Huntington disease (HD). A large number of studies have explored the neuronal phenotype of HD, but the molecular aethiology of the disease is still very poorly understood. This has hampered the development of an appropriate therapeutical strategy to at least alleviate its symptoms. In this short review, we have focused our attention on the alteration of a specific cellular mechanism common to all HD models, either genetic or induced by treatment with 3-NPA, i.e. the cellular dyshomeostasis of Ca2+. We have highlighted the direct and indirect (i.e. transcriptionally mediated) effects of mutated Htt on the maintenance of the intracellular Ca2+ balance, the correct modulation of which is fundamental to cell survival and the disturbance of which plays a key role in the death of the cell.
Keywords: Huntington disease, calcium, mitochondria, transcription, Huntington
Introduction
All patients affected by Huntington disease (HD) have as a common genetic defect, the expansion in the number of CAG triplets located in the N-terminal region of the protein huntingtin (Htt). Normally, the wild type protein is characterized by 15–35 CAG repeats, while in the Htt forms associated with HD the repeats increase up to 40–180. One of the hallmarks of HD is the presence, within the cell body, of insoluble inclusions, composed of aggregates of Htt fragments, produced by the cleavage of the protein by caspases, characterized by the trapping of very diverse proteins, among them calmodulin, transcription factors, components of the ubiquitin–proteasome system and polyubiquitin binding proteins, e.g., p62 that is involved in autophagy.1,2
Even if Htt is ubiquitously expressed, HD is characterized principally by specific motor and cognitive impairments, suggesting a precise role of Htt in certain tissues and brain domains. The exact function of Htt has not been clarified: however, many tasks for it have been proposed based on the results obtained in different models of HD that have highlighted impairment in organelle and vesicular trafficking, cholesterol biosynthesis and propensity to apoptosis.3,4 Numerous papers have also reported an imbalance in the generation and scavenging of reactive oxygen species (ROS),5,6 as well as defects in the respiratory chain complexes7,8 and in mitochondrial functions and morphology.9 A detailed discussion of these specific alterations, which have been already summarized elsewhere,4,10 is beyond the scope of this review, which will focus on the changes of intracellular Ca2+ homeostasis induced by mutated Htt.
The concentration of Ca2+ ([Ca2+]) within the cell is finely tuned by a series of mechanisms, since Ca2+ is a messenger that modulates different signal transduction pathways that are vital to cells: their disturbance can eventually even lead to cell death. Among the main actors involved in [Ca2+] handling there are proteins that act as Ca2+ buffers, proteins that export Ca2+ from the cytosol toward the extracellular medium (the plasma membrane Ca2+ pumps and Na+/ Ca2+ exchangers) or to the lumen of organelles and proteins that mediate Ca2+ entry in the cytoplasm: the influx of Ca2+ is mediated by a number of Ca2+ channels (among them those formed by the STIM/Orai proteins which are involved in the store operated Ca2+ entry (SOCE) process). Finally, there are proteins whose function is activated by binding to Ca2+, such as DREAM and calmodulin.11,12 The membrane of some organelles, e.g., that of mitochondria and the endoplasmic reticulum (ER), also contain systems that take up/extrude Ca2+, establishing a Ca2+ - linked crosstalk with neighboring organelles.13,14
Work on various animal models in which HD was either induced by the genetically produced presence of mutated Htt or by the treatment with 3-nitropropionic acid (3-NPA), an inhibitor of a component of the mitochondrial respiratory chain, complex II, that has been shown to induce a HD phenotype,15 has indeed suggested that one of the hallmarks of HD is the impairment of the intracellular [Ca2+] modulation. The presence of mutated Htt fragments has been associated to the altered expression of some genes involved in Ca2+ homeostasis both in human patients and in HD murine models.4,17 Direct binding of mutated Htt fragments to proteins involved in Ca2+ handling has also been reported.16
Both effects have been supported by convincing evidence. However, it is still unclear whether the transcriptional effects in HD neurons are a cell adaptation response to the variations of intracellular [Ca2+], which could be due to the direct interaction of mutated Htt (and/or Htt fragments) with Ca2+ binding/ Ca2+ transport proteins, or whether the changes of Ca2+ levels are instead a secondary consequence of primary effects of mutated Htt on the expression of regulatory proteins. It is of course still possible that transcriptional and non-transcriptional processes run in parallel and contribute jointly to the onset of HD.
Intracellular Ca2+ Dyshomeostasis in HD
Defects of intracellular Ca2+ homeostasis have been reported in the majority, if not all, the experimental models of HD. However, different alterations have been described depending on the model used. For instance, in medium spiny neurons from mice expressing exon 1 of the human HD gene (R6/2 mice) an increase in the basal cytosolic [Ca2+] has been detected that could induce an adaptation to excitotoxic stress.18 In clonal striatal progenitor cell lines obtained from knock-in mouse embryos carrying a 111 poly-glutamine repeat, we have reported an increased release of Ca2+ from the ER induced by the blockade of the SERCA pump with cyclopiazonic acid with respect to the wild type controls.9 Defects in the morphology of mitochondria have also been observed in the same cell line. The organelles appeared more fragmented: the deregulation of cytosolic Ca2+ contributed to the upregulation of the activity of the Ca2+-dependent phosphatase calcineurin that could dephosphorylate (and thus activate) the pro-fission dynamin related protein 1 (Drp1) leading to the fragmentation of mitochondria and increased propensity to apoptosis.9
Interestingly, the majority of the papers on Ca2+ dyshomeostasis in HD describe principally defects on Ca2+ uptake/extrusion by mitochondria, which are known to act as central regulators of intracellular Ca2+ levels. When the concentration of cytosolic [Ca2+] in the vicinity of mitochondria increases significantly, the low-affinity mitochondrial Ca2+ uniporter (MCU) becomes activated and mediates Ca2+ entry into the matrix. Mitochondria release Ca2+ by means of a Na+/Ca2+ exchanger13 or, in case of Ca2+ overload, through the opening of the permeability transition pore (PTP),14 which results in collapse of the membrane potential, rupture of the outer membrane and release of cytochrome c.14 Alterations in the Ca2+ buffering/handling activities of mitochondria have been reported in both early and late stages of HD, indicating a key role of these defects in its pathogenesis. Mitochondria isolated from cells obtained both from HD patients and HD murine models (carrying 72 or 150 poly-glutamine repeats) are more sensitive to Ca2+ loads, i.e., they have a lower [Ca2+] threshold for the depolarization of their inner membrane and higher propensity to PTP opening.19,20 Similar mitochondrial Ca2+ handling defects have been also reported in clonal striatal cells expressing an Htt form carrying 111Q.21
Results of this type could easily explain the higher susceptibility of HD cells to apoptosis; however, they have not been reproduced in all HD experimental models. For instance, striatal mitochondria from knock-in mouse models expressing different variants of Htt (with 20, 80, 92 or 111 glutamine stretches), from R6/2 mice (three months old) and from 12 months YAC128 mice (carrying a mutated Htt with 128 CAG triplets) were equally or even less susceptible to Ca2+ loads than their wild type counterparts.22,23 In addition, the sensitivity to Ca2+ of mitochondria from several HD models decreased in parallel with age and polyQ length,22 suggesting the intervention of compensatory mechanisms that would eventually protect these organelles from PTP opening.
In addition to interfering with mitochondrial Ca2+ handling, Htt also influences the release of Ca2+ from the ER and the function of SOCE channels. As to the ER, mutated Htt (but not the wild type protein) has been claimed to interact directly with the C-terminal portion of the inositol 1,4,5-trisphosphate (InsP3) receptor type 1 (InsP3R1),16 sensitizing it to InsP324 and thus modulating the efflux of Ca2+ from the ER. As suggested for other neurological disorders,25 this could in turn affect the ability of mitochondria to take up Ca2+, that is known to depend on the close proximity of mitochondria to the Ca2+ releasing ER. A recent finding has shown that InsP3R silencing reduces the aggregation of mutated Htt,26 underlining the importance of the interaction between the two proteins for the molecular pathogenesis of HD.
Interestingly, the association of Htt fragments with ER membranes has been recently confirmed and reported to be dependent on ER stress27: the translocation of Htt from the ER to the nucleus was induced by ER stress. Both the binding to the ER and the shuttling of the Htt fragments to the nucleus were affected by the length of the glutamine repeat.26,28,29
The capacitative, SOCE channels mediated entry of Ca2+ from the extracellular medium also appears to be affected in HD. Convincing experimental evidence has indeed shown that this process is enhanced in HD cells.30 Inhibitors of the capacitative Ca2+ entry slow down the progression/appearance of HD phenotypes in transgenic flies and exert protective effects on primary cultures of medium spiny neurons from YAC128 mice.30 SOCE channels could thus be a possible novel target for the design of new drugs to alleviate the symptoms of HD.
Full length and N-terminal fragments of Htt have also been reported to bind the synaptic region of N-type voltage dependent Ca2+ channels,31 possibly modulating their activity. Finally, mutated Htt can directly interact with numerous Ca2+ binding proteins.32 This ability should not be underestimated, since, as mentioned above, mutated Htt could sequester these proteins, preventing them from fulfilling their normal physiological tasks and leading to the impairment of signal transduction pathways.33 Notably, mutated Htt fragments have been found associated to Calmodulin in high molecular weight complexes,32 disruption of this interaction exerting neuroprotective effects.34,35 Another of the sequestered proteins is Calretinin, a calcium-binding protein with six EF-hand Ca2+ binding domains: the interaction between Calretinin and mutated Htt seems to have protective effects since Calretinin knockdown increases the susceptibility of HD cells to cell death, while its overexpression alleviates the [Ca2+] alterations associated to the presence of mutated Htt.36
One of the hallmarks of mutated Htt is its ability to aggregate, thus the question raises on whether Ca2+ dyshomeostasis is playing a role in the aggregation process or, in turn, whether the presence of Htt inclusions correlate with the appearance of Ca2+ deregulations. Actually the latter has been investigated in detail in a very elegant paper in which the authors overexpress in PC12 cells a truncated form of Htt (Httex1p) containing the polyglutamine repeat.37 Overexpression of Httex1p containing 97 glutamines caused both an alteration of the basal Ca2+ levels as well as an increase in the intracellular Ca2+ levels induced by glutamate. On the contrary, expression of the wild type Httex1p caused no variations in the resting Ca2+ levels, suggesting a key role for the expansion of the glutamine repeat on Ca2+ dyshomeostasis. As to the effect of aggregates per se on Ca2+ dyshomeostasis, it is worth mentioning that expression of mutants forms of Httex1p carrying either 97or 103 glutamines deprived of the first 17 amino acids (thus causing a reduction of their ability to aggregate) was sufficient to alter the basal Ca2+ levels, but not the response to glutamate stimulation.
Transcriptional Deregulation of Ca2+ Handling Proteins in HD
Transcriptional deregulation has been described in almost every model of HD, suggesting a fundamental role for it in the molecular pathogenesis of the disease.38,39 The transcriptional effects have been ascribed to a variety of mechanisms: impairment of microRNA mediated gene expression,40,41 direct binding to the DNA and interaction with transcription factors both inside the nucleus and within the cytoplasm.39,42
The preferred mechanism seems to be direct binding: Htt fragments could behave like a “kidnapper,” sequestering negative and positive regulators of transcription in the Htt inclusion bodies. Among the molecules sequestered a number are indeed factors that regulate gene expression, some of which also contain glutamine stretches. This is the case of the CREB-binding protein (CBP),43 a coactivator of CREB (cAMP-Responsive Element Binding Protein). Interestingly, transient overexpression of CBP has protective role in cellular models expressing mutated Htt,43 and disruption of CREB function leads to a striatal phenotype reminiscent of HD.44 The impairment of CBP activity could have particularly detrimental consequences given its function as a master regulator of the peroxisome proliferator- activated receptor γ (PPARγ) coactivator-1a (PGC1α). PGC1α is primarily involved in mitochondria biogenesis: thus, impairment of its expression could contribute to the production of the mitochondria defects described above. The possible role of the impairment of PGC1α expression on the onset and progression of HD is supported by experimental evidence; the majority of HD models present changes in the level of PGC1α, and its overexpression has protective effects on striatal neurons expressing mutated Htt.44 Finally, PGC1α Knock out mice present lesions in the striatum and phenotype similarities with HD murine models.45,46
The genes that show transcriptional deregulation in HD are involved in numerous physiological processes. They range from those coding for molecules that act as neurotrophins (this is the case of the brain-derived neurotrophic factor (BDNF), whose mRNA levels are significantly lower in human and murine HD samples,47,48 for protein kinases (such as protein kinase C β II17)) or for cytosketetal proteins.17 Importantly, many genome-wide studies performed on various HD models have shown profound differences in the mRNA levels of genes coding for Ca2+ binding proteins, i.e., parvalbumin, calbindin or hippocalcin or proteins involved in the regulation of intracellular Ca2+ handling, i.e the ryanodine receptor type 1 or different voltage-dependent plasma membrane Ca2+ channels.17,38 To better illustrate this point, we have performed a simple bioinformatics analysis using results archived in public databases. Thus, several public gene expression studies performed with Affymetrix genechips (Table 1) were combined in one single file of logRatios for approximately 650 probes corresponding to genes related to Ca2+ homeostasis in neurons according to Affymetrix annotations to probesets included in their genechips. Four representative HD model systems were selected, the mouse Q7/Q111 knock-in striatal neurons, Q7 (wild type) neurons exposed to the mitochondrial toxin 3-NPA, the striatum from the YAC128 transgenic mice and lymphocytes from HD patients or pre-symptomatic mHtt carriers with their appropriate healthy controls (Table 1). A total of six individual genechip hybridization experiments were included in the analysis (Table 1).
Table 1. Transcriptomic experiments compared in the bioinformatics analysis.
| GEO ID | Organism (GeneChip) | Experimental (replicates) | Control (replicates) |
|---|---|---|---|
|
GSE3583 |
Mus musculus (MG430v2.0) |
Wild type Q7 cells 3NP treated (3) |
Wild Type Q7 cells untreated (3) |
| Mutant Q111 cells (3) |
Wild Type Q7 cells (3) |
||
|
GSE18551 |
Mus musculus (MG430v2.0) |
YAC128 mice 12 mo old (4) |
Wild Type mice 12 mo old (4) |
| YAC128 mice 24 mo old (6) |
Wild Type mice 24 mo old (4) |
||
| GSE1751 | Homo sapiens (HGU133A) | HD symptomatic (12) |
Healthy Controls (14) |
| Presymptomatic (5) | Healthy Controls (14) |
The Gene Expression Omnibus (NCBI-GEO, http://www.ncbi.nlm.nih.gov/geo/) ID of each experiment is shown on the right. In each case, the species and the experimental vs. the appropriate control are indicated. YAC128 is a transgenic mouse model for HD expressing a yeast artificial chromosome containing the mutated Huntingtin gene with 128 CAG repeats. Q7 and Q111 cells are neuroblasts cells derived from striatal neurons obtained from wild type or mHtt knock-in mice, respectively.
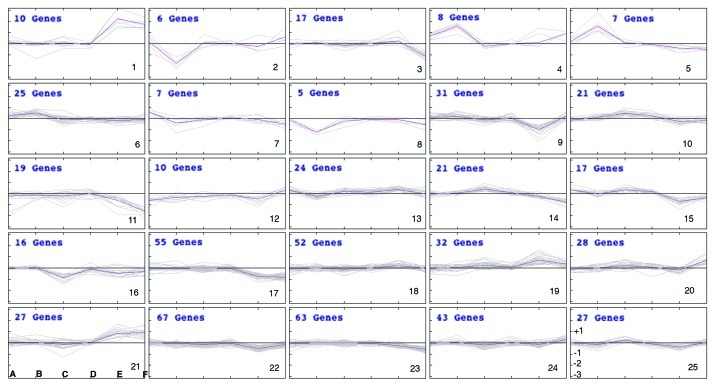
Prior to performing the bioinformatic analysis, to allow the comparison between human and mouse genechips, all probeset IDs from the human genechip (HGU133A) were converted to the orthologous probesets for the mouse Affymetrix GeneChip (MG430v2.0), by using the conversion tables provided by Affymetrix (www.affymetrix.com). Normalized log2Intensity values provided by GEO database (www.ncbi.nlm.nih.gov/geo) were averaged over all replicates (AvgLogIntensities). For each comparison LogRatios were calculated as: LogRatio = AvgLogIntensity (experiment) - AvgLogIntensity (control). Twenty-five clusters of similar logRatio patterns were generated by K-means algorithm (Fig. 1). Some of the most significant clusters from Figure 1 are commented below.
Figure 1. Representation of the 25 clusters of similar logRatio patterns generated by K-means algorithm. LogRatios were calculated as described, from the normalized intensities downloaded from GEO database for each experiment. They represent differential expresión of the genes in the altered samples (disease or transgenic) vs. the controls (healthy or wild type). LogRatios (from -3 to +3) are represented in the Y-axis in each cluster, as shown for cluster 25 at the bottom right corner. LogRatio equal to 0 is represented by the horizontal middle line in each cluster. Positive logRatios (above the horizontal middle line) indicate overexpression whereas negative logRatios (below the horizontal middle line) indicates under-expression. Clusters 1 to 25 are numbered in the bottom right corner. For each cluster the six experiments are labeled A to F (as shown for cluster 21, bottom left corner). (A) Q7–3NP vs Q7; (B) Q111 vs Q7; (C) YAC128–12 min vs WT-12 min; (D) YAC128–24 min vs WT-24 min; (E) HD vs Normal and (F) Pre-symptomatic HD vs Normal. Each cluster represents the coordinated changes in gene expression of a subset of calcium-related genes for each of the six experimental conditions (A to F) compared in this study.
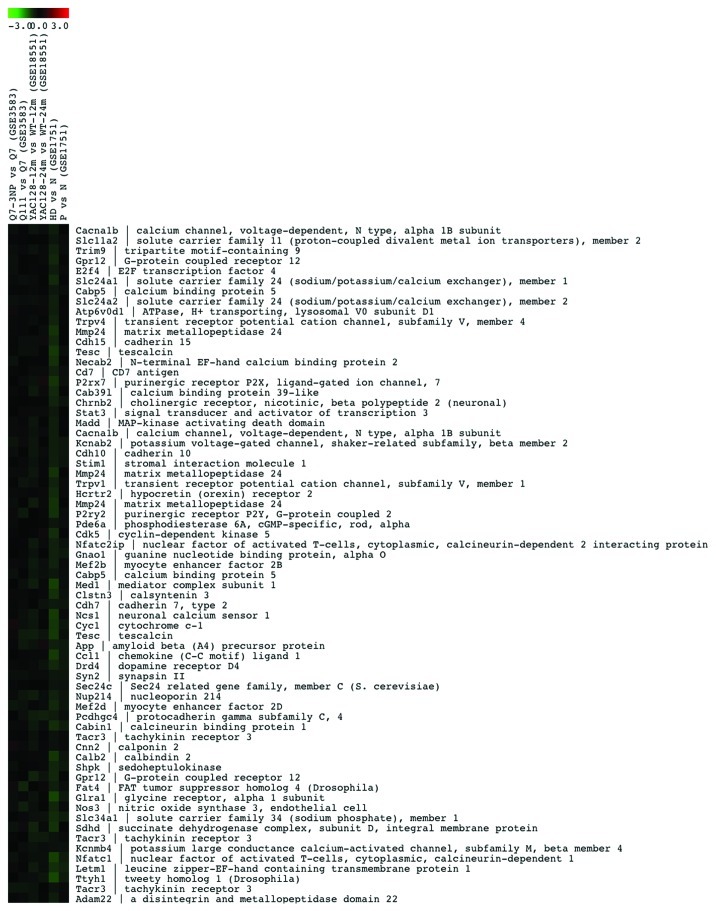
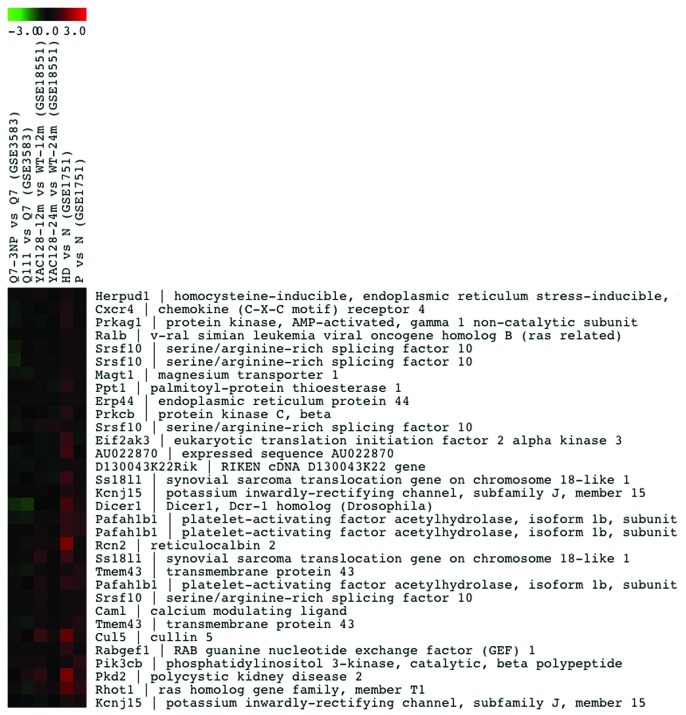
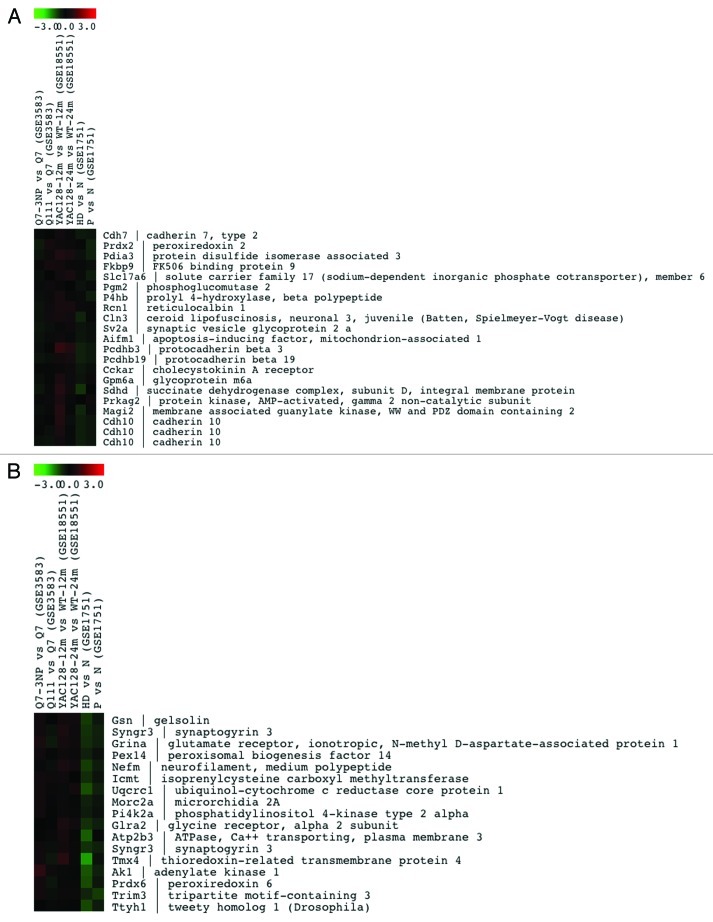
Genes related to calcium homeostasis that are downregulated in lymphocytes from HD patients, Yac128 mice and Q111 knock in cells (clusters 22 and 11) are shown in Figure 2 and in supplementary Figure 1A, respectively, while genes that are downregulated only in HD patients (cluster 17) or in HD patients and YAC128 mice (cluster 16) are shown in Figure S1B and C, respectively. Similarly, cluster 19 includes the most representative collection of genes related to calcium homeostasis that are induced in all four model-systems of HD (Fig. 3), while others in clusters 21 and 1 are mostly induced in human HD lymphocytes (Fig. S2A and B). Moreover, we have found that another significant number of genes change in opposite directions in the different model systems compared in this analysis. Thus, several genes included in clusters 10 and 15 are downregulated in human samples but are induced in non-human models (Fig. 4A and B). In summary, this preliminary bioinformatics analysis shows coordinated or divergent changes in the expression rate of many calcium homeostasis-related genes, that involve a wide variety of cellular functions related to calcium signaling, in HD. Several factors, however, preclude a more detailed commentary for each set of genes at this stage. For instance, the number of experiments introduced in the comparison is reduced specially when compared with the diversity of the experimental specimens that are compared. Thus, differences observed in the behavior of certain sets of genes could be related to the different tissues subjected to genome-wide analysis, human lymphocytes vs. neuronal tissue or cells or to the different approaches used to generate the HD model, i.e., the generally poor agreement between the transcriptional changes observed in striatal Q111 neurons and those present in Q7 cells after exposure to the 3-NPA toxin (Figs. 2–4). Further comparative bioinformatic analyses including more genome-wide studies would be required to complete the assessment of changes in calcium homeostasis in HD that are controlled at the transcriptional level.
Figure 2. List of the 67 Ca2+-related genes in cluster 22 that are downregulated in the model systems of HD shown on the top. Color-coded LogRatio from -3 to +3 is shown on the top. For each gene in the cluster the acronym and the full name is shown. MultiExperiment Viewer software54 was used for generating and plotting the clusters.
Figure 3. List of the 32 Ca2+-related genes in cluster 19 that are upregulated in the model systems of HD shown on the top.
Figure 4. Lists of the 21 and 17 Ca2+-related genes in clusters 10 (A) and 15 (B), respectively, that are differentially regulated in the different model systems of HD shown on the top.
The transcriptional defects in the expression of proteins involved in the maintenance of intracellular Ca2+ homeostasis has certainly a role in the Ca2+ dyshomeostasis observed in HD models. The transcriptional defects could be further amplified by Ca2+ dependent transcriptional control mechanisms. This could occur by changes in the function of gene expression regulatory proteins modulated by Ca2+ binding. This is the case of the transcriptional repressor DREAM (downstream responsive element (DRE) Antagonist Modulator), that translocates to the nucleus following the increase of cytosolic [Ca2+] levels,49,50 or of the cofactor LMO4 (LIM domain only 4), the activity of which is induced by Ca2+ entry through voltage dependent Ca2+ channels.51
Conclusions
This short review has succintly summarized the present knowledge on the intracellular Ca2+ alterations that occur in HD models. Mutated Htt can directly bind to the InsP3R, thus modulating the ability of its channel to release Ca2+ and thus in turn the crosstalk between the ER and mitochondria and the active mitochondrial Ca2+ uptake.16,52,53 This could be a general mechanism in many neuropathological processes, since disturbances in the formation of peri-mitochondrial Ca2+ microdomains has been already reported to contribute to other disorders, such as for example Alzheimer disease.25
In parallel, strong evidence underlines the importance of transcriptional dysregulation in HD and of the ability of mutated Htt fragments to sequester or modulate proteins involved in gene expression. Interestingly, this is reflected in the changes of the expression of numerous genes related to Ca2+ homeostasis,17,38 raising the question of whether these two key processes that are affected in the disease run in parallel or in series.
A conciliatory hypothesis on the transcriptional and nontranscriptional effects of mutated Htt could be suggested by the interesting finding of Htt translocation from the ER to the nucleus.27-29 The expansion of the glutamine repeat in mutated Htt could affect the nuclear shuttling of Htt itself. At the same time it could cause the formation of inclusion bodies where transcription factors could be sequestered, in turn again influencing the expression of various genes, among them some coding for proteins of the Ca2+ homeostasis machinery.
Supplementary Material
Acknowledgments
Work in JRN laboratory is supported by grants from CAM (Neurodegmodels); ERA-Net (SAF2008–03469); DGICYT (SAF2010–21784); FISS (CIBERNED); Fundación Reina Sofía (PI 006/09) and EU 6th Framework Program (NeuroNE, LSHG-CT-2006–037627).
Work in EC laboratory is supported by the same ERA-Net grant and by a Grant of the CARIPARO Foundation (Projects of Excellence 2008–2009).
Disclosure of Potential Conflict of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/prion/article/23581
References
- 1.Arrasate M, Finkbeiner S. Protein aggregates in Huntington’s disease. Exp Neurol. 2012;238:1–11. doi: 10.1016/j.expneurol.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nagaoka U, Kim K, Jana NR, Doi H, Maruyama M, Mitsui K, et al. Increased expression of p62 in expanded polyglutamine-expressing cells and its association with polyglutamine inclusions. J Neurochem. 2004;91:57–68. doi: 10.1111/j.1471-4159.2004.02692.x. [DOI] [PubMed] [Google Scholar]
- 3.Valenza M, Rigamonti D, Goffredo D, Zuccato C, Fenu S, Jamot L, et al. Dysfunction of the cholesterol biosynthetic pathway in Huntington’s disease. J Neurosci. 2005;25:9932–9. doi: 10.1523/JNEUROSCI.3355-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zuccato C, Valenza M, Cattaneo E. Molecular mechanisms and potential therapeutical targets in Huntington’s disease. Physiol Rev. 2010;90:905–81. doi: 10.1152/physrev.00041.2009. [DOI] [PubMed] [Google Scholar]
- 5.Bertoni A, Giuliano P, Galgani M, Rotoli D, Ulianich L, Adornetto A, et al. Early and late events induced by polyQ-expanded proteins: identification of a common pathogenic property of polYQ-expanded proteins. J Biol Chem. 2011;286:4727–41. doi: 10.1074/jbc.M110.156521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hands S, Sajjad MU, Newton MJ, Wyttenbach A. In vitro and in vivo aggregation of a fragment of huntingtin protein directly causes free radical production. J Biol Chem. 2011;286:44512–20. doi: 10.1074/jbc.M111.307587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tabrizi SJ, Workman J, Hart PE, Mangiarini L, Mahal A, Bates G, et al. Mitochondrial dysfunction and free radical damage in the Huntington R6/2 transgenic mouse. Ann Neurol. 2000;47:80–6. doi: 10.1002/1531-8249(200001)47:1<80::aid-ana13>3.3.co;2-b. [DOI] [PubMed] [Google Scholar]
- 8.Arenas J, Campos Y, Ribacoba R, Martín MA, Rubio JC, Ablanedo P, et al. Complex I defect in muscle from patients with Huntington’s disease. Ann Neurol. 1998;43:397–400. doi: 10.1002/ana.410430321. [DOI] [PubMed] [Google Scholar]
- 9.Costa V, Giacomello M, Hudec R, Lopreiato R, Ermak G, Lim D, et al. Mitochondrial fission and cristae disruption increase the response of cell models of Huntington’s disease to apoptotic stimuli. EMBO Mol Med. 2010;2:490–503. doi: 10.1002/emmm.201000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giacomello M, Hudec R, Lopreiato R. Huntington’s disease, calcium, and mitochondria. Biofactors. 2011;37:206–18. doi: 10.1002/biof.162. [DOI] [PubMed] [Google Scholar]
- 11.Saris NE, Carafoli E. A historical review of cellular calcium handling, with emphasis on mitochondria. Biochemistry (Mosc) 2005;70:187–94. doi: 10.1007/s10541-005-0100-9. [DOI] [PubMed] [Google Scholar]
- 12.Naranjo JR, Mellström B. Ca2+-dependent transcriptional control of Ca2+ homeostasis. J Biol Chem. 2012;287:31674–80. doi: 10.1074/jbc.R112.384982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carafoli E, Tiozzo R, Lugli G, Crovetti F, Kratzing C. The release of calcium from heart mitochondria by sodium. J Mol Cell Cardiol. 1974;6:361–71. doi: 10.1016/0022-2828(74)90077-7. [DOI] [PubMed] [Google Scholar]
- 14.Giacomello M, Drago I, Pizzo P, Pozzan T. Mitochondrial Ca2+ as a key regulator of cell life and death. Cell Death Differ. 2007;14:1267–74. doi: 10.1038/sj.cdd.4402147. [DOI] [PubMed] [Google Scholar]
- 15.Brouillet E, Hantraye P. Effects of chronic MPTP and 3-nitropropionic acid in nonhuman primates. Curr Opin Neurol. 1995;8:469–73. doi: 10.1097/00019052-199512000-00014. [DOI] [PubMed] [Google Scholar]
- 16.Tang TS, Tu H, Chan EY, Maximov A, Wang Z, Wellington CL, et al. Huntingtin and huntingtin-associated protein 1 influence neuronal calcium signaling mediated by inositol-(1,4,5) triphosphate receptor type 1. Neuron. 2003;39:227–39. doi: 10.1016/s0896-6273(03)00366-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luthi-Carter R, Hanson SA, Strand AD, Bergstrom DA, Chun W, Peters NL, et al. Dysregulation of gene expression in the R6/2 model of polyglutamine disease: parallel changes in muscle and brain. Hum Mol Genet. 2002;11:1911–26. doi: 10.1093/hmg/11.17.1911. [DOI] [PubMed] [Google Scholar]
- 18.Hansson O, Guatteo E, Mercuri NB, Bernardi G, Li XJ, Castilho RF, et al. Resistance to NMDA toxicity correlates with appearance of nuclear inclusions, behavioural deficits and changes in calcium homeostasis in mice transgenic for exon 1 of the huntington gene. Eur J Neurosci. 2001;14:1492–504. doi: 10.1046/j.0953-816x.2001.01767.x. [DOI] [PubMed] [Google Scholar]
- 19.Panov AV, Gutekunst CA, Leavitt BR, Hayden MR, Burke JR, Strittmatter WJ, et al. Early mitochondrial calcium defects in Huntington’s disease are a direct effect of polyglutamines. Nat Neurosci. 2002;5:731–6. doi: 10.1038/nn884. [DOI] [PubMed] [Google Scholar]
- 20.Choo YS, Johnson GV, MacDonald M, Detloff PJ, Lesort M. Mutant huntingtin directly increases susceptibility of mitochondria to the calcium-induced permeability transition and cytochrome c release. Hum Mol Genet. 2004;13:1407–20. doi: 10.1093/hmg/ddh162. [DOI] [PubMed] [Google Scholar]
- 21.Milakovic T, Quintanilla RA, Johnson GV. Mutant huntingtin expression induces mitochondrial calcium handling defects in clonal striatal cells: functional consequences. J Biol Chem. 2006;281:34785–95. doi: 10.1074/jbc.M603845200. [DOI] [PubMed] [Google Scholar]
- 22.Brustovetsky N, LaFrance R, Purl KJ, Brustovetsky T, Keene CD, Low WC, et al. Age-dependent changes in the calcium sensitivity of striatal mitochondria in mouse models of Huntington’s Disease. J Neurochem. 2005;93:1361–70. doi: 10.1111/j.1471-4159.2005.03036.x. [DOI] [PubMed] [Google Scholar]
- 23.Oliveira JM, Jekabsons MB, Chen S, Lin A, Rego AC, Gonçalves J, et al. Mitochondrial dysfunction in Huntington’s disease: the bioenergetics of isolated and in situ mitochondria from transgenic mice. J Neurochem. 2007;101:241–9. doi: 10.1111/j.1471-4159.2006.04361.x. [DOI] [PubMed] [Google Scholar]
- 24.Zhang H, Das S, Li QZ, Dragatsis I, Repa J, Zeitlin S, et al. Elucidating a normal function of huntingtin by functional and microarray analysis of huntingtin-null mouse embryonic fibroblasts. BMC Neurosci. 2008;9:38. doi: 10.1186/1471-2202-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kipanyula MJ, Contreras L, Zampese E, Lazzari C, Wong AK, Pizzo P, et al. Ca2+ dysregulation in neurons from transgenic mice expressing mutant presenilin 2. Aging Cell. 2012;11:885–93. doi: 10.1111/j.1474-9726.2012.00858.x. [DOI] [PubMed] [Google Scholar]
- 26.Bauer PO, Hudec R, Ozaki S, Okuno M, Ebisui E, Mikoshiba K, et al. Genetic ablation and chemical inhibition of IP3R1 reduce mutant huntingtin aggregation. Biochem Biophys Res Commun. 2011;416:13–7. doi: 10.1016/j.bbrc.2011.10.096. [DOI] [PubMed] [Google Scholar]
- 27.Atwal RS, Xia J, Pinchev D, Taylor J, Epand RM, Truant R. Huntingtin has a membrane association signal that can modulate huntingtin aggregation, nuclear entry and toxicity. Hum Mol Genet. 2007;16:2600–15. doi: 10.1093/hmg/ddm217. [DOI] [PubMed] [Google Scholar]
- 28.Benn CL, Landles C, Li H, Strand AD, Woodman B, Sathasivam K, et al. Contribution of nuclear and extranuclear polyQ to neurological phenotypes in mouse models of Huntington’s disease. Hum Mol Genet. 2005;14:3065–78. doi: 10.1093/hmg/ddi340. [DOI] [PubMed] [Google Scholar]
- 29.Cornett J, Cao F, Wang CE, Ross CA, Bates GP, Li SH, et al. Polyglutamine expansion of huntingtin impairs its nuclear export. Nat Genet. 2005;37:198–204. doi: 10.1038/ng1503. [DOI] [PubMed] [Google Scholar]
- 30.Wu J, Shih HP, Vigont V, Hrdlicka L, Diggins L, Singh C, et al. Neuronal store-operated calcium entry pathway as a novel therapeutic target for Huntington’s disease treatment. Chem Biol. 2011;18:777–93. doi: 10.1016/j.chembiol.2011.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swayne LA, Chen L, Hameed S, Barr W, Charlesworth E, Colicos MA, et al. Crosstalk between huntingtin and syntaxin 1A regulates N-type calcium channels. Mol Cell Neurosci. 2005;30:339–51. doi: 10.1016/j.mcn.2005.07.016. [DOI] [PubMed] [Google Scholar]
- 32.Bao J, Sharp AH, Wagster MV, Becher M, Schilling G, Ross CA, et al. Expansion of polyglutamine repeat in huntingtin leads to abnormal protein interactions involving calmodulin. Proc Natl Acad Sci U S A. 1996;93:5037–42. doi: 10.1073/pnas.93.10.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kreutz MR, Naranjo JR, Koch KW, Schwaller B. The Neuronal Functions of EF-Hand Ca2+-Binding Proteins. Front Mol Neurosci. 2012;5:92. doi: 10.3389/fnmol.2012.00092. [ì] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dudek NL, Dai Y, Muma NA. Neuroprotective effects of calmodulin peptide 76-121aa: disruption of calmodulin binding to mutant huntingtin. Brain Pathol. 2010;20:176–89. doi: 10.1111/j.1750-3639.2008.00258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dudek NL, Dai Y, Muma NA. Protective effects of interrupting the binding of calmodulin to mutant huntingtin. J Neuropathol Exp Neurol. 2008;67:355–65. doi: 10.1097/NEN.0b013e31816a9e60. [DOI] [PubMed] [Google Scholar]
- 36.Dong G, Gross K, Qiao F, Ferguson J, Callegari EA, Rezvani K, et al. Calretinin interacts with huntingtin and reduces mutant huntingtin-caused cytotoxicity. J Neurochem. 2012;123:437–46. doi: 10.1111/j.1471-4159.2012.07919.x. [DOI] [PubMed] [Google Scholar]
- 37.Rockabrand E, Slepko N, Pantalone A, Nukala VN, Kazantsev A, Marsh JL, et al. The first 17 amino acids of Huntingtin modulate its sub-cellular localization, aggregation and effects on calcium homeostasis. Hum Mol Genet. 2007;16:61–77. doi: 10.1093/hmg/ddl440. [DOI] [PubMed] [Google Scholar]
- 38.Hodges A, Strand AD, Aragaki AK, Kuhn A, Sengstag T, Hughes G, et al. Regional and cellular gene expression changes in human Huntington’s disease brain. Hum Mol Genet. 2006;15:965–77. doi: 10.1093/hmg/ddl013. [DOI] [PubMed] [Google Scholar]
- 39.Seredenina T, Luthi-Carter R. What have we learned from gene expression profiles in Huntington’s disease? Neurobiol Dis. 2012;45:83–98. doi: 10.1016/j.nbd.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 40.Savas JN, Makusky A, Ottosen S, Baillat D, Then F, Krainc D, et al. Huntington’s disease protein contributes to RNA-mediated gene silencing through association with Argonaute and P bodies. Proc Natl Acad Sci U S A. 2008;105:10820–5. doi: 10.1073/pnas.0800658105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee ST, Chu K, Im WS, Yoon HJ, Im JY, Park JE, et al. Altered microRNA regulation in Huntington’s disease models. Exp Neurol. 2011;227:172–9. doi: 10.1016/j.expneurol.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 42.Hogel M, Laprairie RB, Denovan-Wright EM. Promoters are differentially sensitive to N-terminal mutant huntingtin-mediated transcriptional repression. PLoS One. 2012;7:e41152. doi: 10.1371/journal.pone.0041152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nucifora FC, Jr., Sasaki M, Peters MF, Huang H, Cooper JK, Yamada M, et al. Interference by huntingtin and atrophin-1 with cbp-mediated transcription leading to cellular toxicity. Science. 2001;291:2423–8. doi: 10.1126/science.1056784. [DOI] [PubMed] [Google Scholar]
- 44.Mantamadiotis T, Lemberger T, Bleckmann SC, Kern H, Kretz O, Martin Villalba A, et al. Disruption of CREB function in brain leads to neurodegeneration. Nat Genet. 2002;31:47–54. doi: 10.1038/ng882. [DOI] [PubMed] [Google Scholar]
- 45.Cui L, Jeong H, Borovecki F, Parkhurst CN, Tanese N, Krainc D. Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127:59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 46.Lin J, Wu PH, Tarr PT, Lindenberg KS, St-Pierre J, Zhang CY, et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119:121–35. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 47.Gambazzi L, Gokce O, Seredenina T, Katsyuba E, Runne H, Markram H, et al. Diminished activity-dependent brain-derived neurotrophic factor expression underlies cortical neuron microcircuit hypoconnectivity resulting from exposure to mutant huntingtin fragments. J Pharmacol Exp Ther. 2010;335:13–22. doi: 10.1124/jpet.110.167551. [DOI] [PubMed] [Google Scholar]
- 48.Zuccato C, Marullo M, Conforti P, MacDonald ME, Tartari M, Cattaneo E. Systematic assessment of BDNF and its receptor levels in human cortices affected by Huntington’s disease. Brain Pathol. 2008;18:225–38. doi: 10.1111/j.1750-3639.2007.00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ledo F, Kremer L, Mellström B, Naranjo JR. Ca2+-dependent block of CREB-CBP transcription by repressor DREAM. EMBO J. 2002;21:4583–92. doi: 10.1093/emboj/cdf440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Savignac M, Pintado B, Gutierrez-Adan A, Palczewska M, Mellström B, Naranjo JR. Transcriptional repressor DREAM regulates T-lymphocyte proliferation and cytokine gene expression. EMBO J. 2005;24:3555–64. doi: 10.1038/sj.emboj.7600810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kashani AH, Qiu Z, Jurata L, Lee SK, Pfaff S, Goebbels S, et al. Calcium activation of the LMO4 transcription complex and its role in the patterning of thalamocortical connections. J Neurosci. 2006;26:8398–408. doi: 10.1523/JNEUROSCI.0618-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Giacomello M, Drago I, Bortolozzi M, Scorzeto M, Gianelle A, Pizzo P, et al. Ca2+ hot spots on the mitochondrial surface are generated by Ca2+ mobilization from stores, but not by activation of store-operated Ca2+ channels. Mol Cell. 2010;38:280–90. doi: 10.1016/j.molcel.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 53.Csordás G, Várnai P, Golenár T, Roy S, Purkins G, Schneider TG, et al. Imaging interorganelle contacts and local calcium dynamics at the ER-mitochondrial interface. Mol Cell. 2010;39:121–32. doi: 10.1016/j.molcel.2010.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, et al. TM4 microarray software suite. Methods Enzymol. 2006;411:134–93. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.