Abstract
Mammalian and fungal prion proteins form self-perpetuating β-sheet-rich fibrillar aggregates called amyloid. Prion inheritance is based on propagation of the regularly oriented amyloid structures of the prion proteins. All yeast prion proteins identified thus far contain aggregation-prone glutamine/asparagine (Gln/Asn)-rich domains, although the mammalian prion protein and fungal prion protein HET-s do not contain such sequences. In order to fill this gap, we searched for novel yeast prion proteins lacking Gln/Asn-rich domains via a genome-wide screen based on cross-seeding between two heterologous proteins and identified Mod5, a yeast tRNA isopentenyltransferase, as a novel non-Gln/Asn-rich yeast prion protein. Mod5 formed self-propagating amyloid fibers in vitro and the introduction of Mod5 amyloids into non-prion yeast induced dominantly and cytoplasmically heritable prion state [MOD+], which harbors aggregates of endogenous Mod5. [MOD+] yeast showed an increased level of membrane lipid ergosterol and acquired resistance to antifungal agents. Importantly, enhanced de novo formation of [MOD+] was observed when non-prion yeast was grown under selective pressures from antifungal drugs. Our findings expand the family of yeast prions to non-Gln/Asn-rich proteins and reveal the acquisition of a fitness advantage for cell survival through active prion conversion.
Keywords: prion, amyloid, Mod5, tRNA isopentenyltransferase, antifungal drug, cellular adaptation
Prions are protein-based genetic elements that are formed by structural conversion of a monomeric protein to β-sheet-rich fibrillar aggregates called amyloid.1-3 The prion phenomenon now includes yeast and fungal prion proteins,4,5 which share characteristics common to mammalian prions, including amyloid-based prion inheritance, a variety of heritable phenotypic strain variants and transmission barriers that limit prion infection between different species.6 In the budding yeast Saccharomyces cerevisiae, eight proteins (Sup35, Ure2, Rnq1, Swi1, Cyc8, Mot3, Sfp1 and Nup100) have been verified as yeast prion proteins2,4,5,7-12 and all of them contain a characteristic aggregation-inducing domain which is abundant in glutamine and/or asparagine (Gln/Asn) residues. The Gln/Asn-rich domain is critical to prion propagation as the fusion of Gln/Asn-rich domain to non-prion proteins can create novel prion proteins.13 Until now, both in silico and in vivo screening of yeast prion proteins have focused on Gln/Asn-rich proteins.11,14
In contrast to these yeast prion proteins identified to date, both mammalian prion protein and a filamentous fungal prion protein, HET-s, lack a Gln/Asn-rich domain but are capable of forming infectious heritable prion aggregates.15,16 Interestingly, a HET-s prion domain fused to GFP propagates as a prion in yeast, demonstrating that non-Gln/Asn-rich proteins were also able to act as yeast prions.16 Although these findings imply the possibility of non-Gln/Asn-rich yeast prion proteins, none has yet to be found.
The possible existence of a large number of yeast prion proteins11 suggests that prion conversion may play physiological roles in cells. Wickner and colleagues indicated that [PSI+], [URE3] and [PIN+] prion states might be deleterious to cells,1,17,18 whereas the prion state [PSI+] and [MOT+] could be beneficial for cells under certain conditions.19 A key question is that the conversion to prion states can be induced for cells to play physiological roles when the cells are faced with deleterious conditions. Tyedmers and colleagues reported that the [PSI+] prion state is induced to acquire a survival advantage under the selective pressure of environmental stresses such as hydrogen peroxide exposure and high concentrations of salt,20 indicating that prion conversion might help an organism adapt to environmental stress. However, it has been elusive whether cells actively utilize prion conversion to perform certain physiological functions or such observations have simply been consequences unrelated to physiological roles. Furthermore, our understanding of whether specific mechanisms exist to regulate the cellular fitness adaptation by active prion conversion has been limited.
Previous studies have demonstrated that several yeast prions act as “seeds” for the prion conversion of other prion proteins in yeast.8,21 Derkatch and colleagues previously performed a genome-wide genetic screen for PIN (inducible to [PSI+], a prion state resulting from Sup35 aggregation) factors that facilitate the de novo appearance of [PSI+] when overexpressed21 and identified Gln/Asn-rich Rnq1 as the causal factor for [PIN+].22 Overexpression of other Gln/Asn-rich proteins such as Ure2, Swi1 and Cyc8 were also shown to be PIN factors21 and subsequent studies demonstrated that these proteins are yeast prion proteins.2,9,10 Altogether, these studies suggest that screening of PIN proteins may be a reliable strategy to identify a new class of yeast prion proteins that lack Gln/Asn-rich domains.
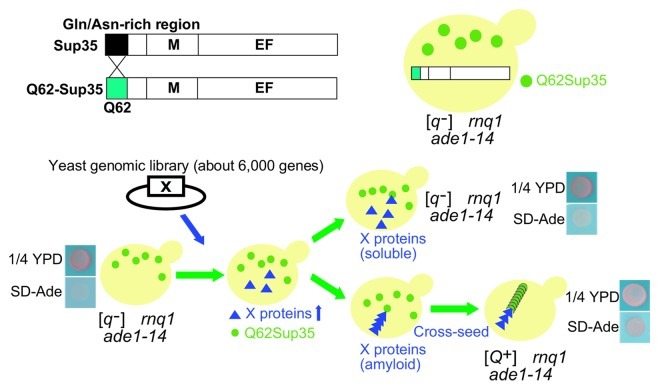
As such, we developed a genome-wide screen exploiting the [q−] yeast strain, a non-prion state of yeast expressing a chimera Sup35 protein containing 62 glutamine repeats (Q62-Sup35),23 and searched for novel QIN (inducible to a prion state [Q+]) factors (Fig. 1).24 [q−] yeast shows a lighter red color phenotype and a relatively higher frequency of spontaneous conversion from non-prion to prion states, compared with [psi−] yeast.23 This is consistent with the fact that expanded polyglutamine, which is responsible for polyglutamine diseases such as Huntington disease and Machado-Joseph disease, shows a high tendency to form amyloid. Accordingly, amyloid formation of proteins containing an expanded polyglutamine can be accelerated by the addition of aggregates of non-Gln/Asn-rich proteins such as bovine pancreas insulin and human immunoglobulin light-chain as well as Gln/Asn-rich Rnq1.25 Taken together, we reasoned that aggregates composed of not only Gln/Asn-rich but also non-Gln/Asn-rich proteins might act as seeds for “cross-seeding” with Q62-Sup35 and that non-Gln/Asn-rich proteins might be identified as novel QIN factors.
Figure 1. A procedure of genome-wide screen for identification of novel yeast prion proteins. A Q62-Sup35 chimera yeast strain was used to identify QIN factors (upper). A Sup35 protein consists of a N-terminal prion domain (N domain, residues 1–123), a highly charged middle domain (M domain, residues 124–253) and a C-terminal translation termination domain (EF domain, residues 254–685) The Gln/Asn-rich region in the SUP35 N domain (residues 1–40) was replaced by 62 glutamine repeats (Q62). The procedure for the genome-wide screen for novel QIN factors in the [Q+] yeast prion system is shown (lower). Expression plasmids from a yeast ORF library were introduced into [q-]Δrnq1 yeast cells, each yeast protein was overexpressed by galactose and de novo appearance of [Q+] was tested by growth on 1/4 YPD and SD-Ade plates. Color and growth phenotypes of [q-] and [Q+] yeasts on 1/4 YPD and SD-Ade plates are shown as negative and positive controls, respectively. (Reproduced from ref. 24.)
Among the 65 QIN factors identified in the screen, we focused on Mod5, a yeast isopentenyltransferase that catalyzes the transfer of an isopentenyl group to A37 in the anticodon loop,26 for several reasons. First, Mod5 does not contain any Gln/Asn-rich domains nor repeat sequences, but was found to act as both a QIN and PIN factor. Second, we took subcellular localization and a number of molecules per cell into account, as they might also be critical factors for yeast prions to propagate in a mitotically stable manner. Mod5 was located both in nucleus and cytoplasm, so the cytoplasmic localization of Mod5 would be suitable for cytoplasmic inheritance of prion states, although nuclear proteins such as Swi1 and Cyc8 and nucleoporin Nup100 have also been shown to act as yeast prion proteins.2,9,10,12 Mod5 is expressed at approximately 2,020 molecules per cell, which should be sufficient to support stable propagation since it is comparable to the number of molecules per cell for already known yeast prion proteins (e.g., 92 and 1,140 molecules per cell for Swi1 and Rnq1, respectively).27 These results suggested that Mod5 is a novel non-Gln/Asn-rich yeast prion protein candidate.
Self-propagating β-sheet-rich conformation of amyloid constitutes the physical basis for prion propagation.28 Formation of self-perpetuating amyloid of non-Gln/Asn-rich Mod5 was confirmed by multiple biophysical methods. A key question was the identity of the core region of Mod5 amyloid because Mod5 does not have any Gln/Asn-rich domain and thus which amino acid regions are essential to amyloid formation is not readily apparent. Limited proteolysis followed by mass spectrometric analysis identified the amyloid core as the region consisting of residues 194–215. This region includes a sequence, FDTLFLWLY, which is rich in hydrophobic residues and the removal of this region dramatically decreased the amyloidogenicity of Mod5, thus confirming the crucial role of this region in amyloid formation.
Introduction of pure Mod5 amyloids into non-prion [mod−] yeast using a protein infection protocol29 induced de novo appearance of [MOD+], a prion state caused by aggregation of endogenous Mod5. [MOD+] yeast were expected to show phenotypes similar to Δmod5 yeast because aggregated Mod5 in the [MOD+] strain should be non or less functional. A previous study revealed that Δmod5 yeast are sensitive to 5-fluorouracil (5-FU)30 and we found that the [MOD+] yeast also exhibited the sensitivity to 5-FU. Thus, the 5-FU sensitivity assay allowed us to identify [MOD+] colonies from a pool composed mainly of [mod−] cells, providing us with a relatively simple method to detect [MOD+] yeast.
Importantly, the [MOD+] prion state was cured to the [mod−] non-prion state either by transient treatment of [MOD+] with 3mM guanidine hydrochloride, a specific inhibitor of an AAA+ ATPase chaperone Hsp104, or deletion of HSP104 in [MOD+] yeast.24 Overexpression of an ATPase-inactive dominant negative form of Hsp104 (Hsp104-KT) also converted [MOD+] back to the [mod−] non-prion state (data not shown). The Hsp104-dependent curing of prion states is characteristic to all known yeast prions,5 thereby confirming [MOD+] as a reversible prion state independent of any genomic changes. Subsequent mating and cytoduction experiments verified that [MOD+] yeast showed dominant and cytoplasmic inheritance. These results demonstrated that Mod5 was the first non-Gln/Asn-rich yeast prion protein and that, similar to mammalian and fungal prions, endogenous non-Gln/Asn-rich proteins can function as yeast prions.
Given that not only Gln/Asn-rich yeast proteins11 but also non-Gln/Asn-rich proteins have the potential to behave as cytoplasmically heritable elements, yeast may utilize prion conversion as a strategy to perform certain physiological roles by actively regulating reversible prion conversion. It is plausible that the prion conversion acts as a fast “molecular switch” to respond to new environments, whereas fitness adaptation by random genomic mutations and selection of favorable traits require a much longer time. If an organism were faced with environmental stresses that might threaten survival, rapid phenotypic changes to enhance cell survival would be particularly beneficial.
To this end, we found that [MOD+] yeast contains higher levels of ergosterol than [mod−] yeast and that they have enhanced resistance to antifungal drugs such as fluconazole and ketoconazole, which target ergosterol biosynthesis. More importantly, selective pressure of [mod−] yeast by these antifungal drugs increased the de novo appearance of [MOD+]. This result shows that yeast actively make use of Mod5 prion conversion to gain a fitness advantage for cell survival when faced with environmental stress. Furthermore, after removal of the antifungal drugs from the culture media, [MOD+] states were eventually lost over time and [mod−] yeast became dominant. Taken together, our findings suggested that active prion conversion in eukaryotes played critical roles in fast and on-demand cellular adaptation to harmful environments (Fig. 2).
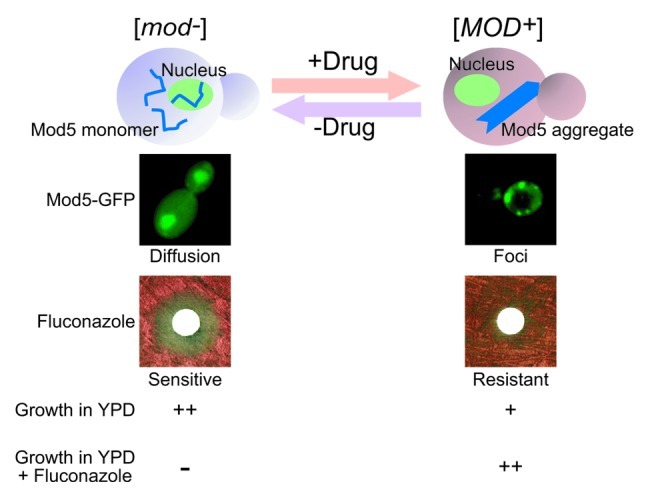
Figure 2. Prion conversion as a molecular switch to respond to environmental stress. [mod−] yeast contains the Mod5 protein as a soluble and functional monomer while [MOD+] yeast contains the Mod5 protein in its aggregated non/less functional form (top, schematic illustration). Mod5-GFP exhibits nuclear and cytoplasmic diffusible localization in [mod−] yeast cells, while Mod5-GFP forms cytoplasmic foci in [MOD+] yeast cells (middle images). [mod−] yeast show the sensitivity to antifungal drugs such as fluconazole and ketoconazole whereas [MOD+] yeast are resistant against them (bottom images). Prion conversion between [mod-] and [MOD+] occurs at very low frequency. However, in the presence of environmental stress such as antifungal drugs, [MOD+] yeast cells become dominant because of the newly acquired drug resistant phenotype. Once cells are released from the environmental stress, [mod−] yeast cells rapidly become dominant due to their growth advantage under normal conditions. Thus, the prion conversion of Mod5 acts as a “molecular switch” for enhanced survival under different conditions.
As shown for the antifungal resistance of [MOD+] yeast, the reversible prion conversion in eukaryotes may contribute to adaptation to the other environmental stresses such as oxidative stress and changes in pH, temperature and ionic strength outside cells. To further this hypothesis, perhaps amyloid formation in some neurodegenerative diseases such as Alzheimer disease and Parkinson disease could act to reduce the amount of potentially toxic oligomeric and/or monomeric species.28 More broadly, amyloid formation as well as prion conversion may represent a positive adaptation strategy for cell survival in response to environmental stresses such as those upon cellular aging.
Many of the yeast prion proteins that have been fully identified to date are transcriptional or translational regulators. Mod5 is a unique yeast prion protein not only in that it is a non-Gln/Asn-rich protein but also it catalyzes modification of tRNA. While the increase in ergosterol levels in [MOD+] yeast would provide the prion-state yeast with the antifungal resistance, the decreased isopentenylation activity at A37 in the anticodon loop of tRNA by Mod5 aggregation may also partly contribute to the enhanced survival of [MOD+] yeast. A previous study revealed that cells could respond to oxidative stress by altering tRNA modification levels.31 It would be of interest to investigate whether the change in tRNA modification in [MOD+] yeast are linked to translation of specific mRNAs, leading to the acquisition of new genetic traits.
We demonstrated that yeast cells actively induce [MOD+] prion states to gain a survival advantage under the stressful condition. Halfmann and colleagues found that one-third of approximately 700 wild strains of Saccharomyces harbored unknown prion elements that created diverse and beneficial phenotypes.19 Furthermore, recent studies suggest that a large number of proteins have the potential to form amyloid fibrils that would be required for prion propagation.32 These findings imply that Mod5 is not a solely non-Gln/Asn-rich yeast prion protein. By developing novel screening methods that do not rely on the aggregation property of Gln/Asn-rich proteins, other unknown non-Gln/Asn-rich yeast prion proteins could be found and they might also positively play physiological roles by utilizing prion conversion.
Mod5 is the first prototypical member of a novel class of non-Gln/Asn-rich yeast prion proteins. In light of this uniqueness of Mod5, it would be worth addressing in the future whether the molecular basis of in vitro amyloid formation and in vivo prion propagation of non-Gln/Asn-rich Mod5 is similar to or different from that of Gln/Asn-rich yeast prion proteins such as Sup35. Previous studies revealed that a prion-forming fragment of Sup35, Sup35NM, forms oligomers under specific conditions and their formation driven by non-native interactions leads to the production of highly infectious prion conformations.33,34 Future studies will examine whether Mod5 can also form oligomeric species that prefer specific amyloid conformations and misfold into different amyloid conformations that lead to phenotypically distinct heritable prion states. It would also be critical to determine whether Hsp40 and Hsp70 family proteins such as Sis1 and Ssa1 modulate the propagation of [MOD+] states as was observed for [PSI+].5 By taking advantage of yeast genetics, further efforts will clarify the detailed mechanisms of both prion propagation and curing in vitro and in vivo. It should be noted that Mod5 and mammalian prion protein do not contain any Gln/Asn-rich domains. Therefore, these studies in yeast might contribute to a better understanding of the pathogenic mechanism of mammalian prion disease and provide further insights into the development of novel therapeutic strategies for prion disease and the other neurodegenerative disorders.
Recently, it has been demonstrated that not only aggregates of mammalian prion protein but also many causative proteins for neurodegenerative disorders such as polyglutamine, α-synuclein and SOD1 show prion-like cell-to-cell propagation.35 This finding is consistent with the fact that neurodegeneration in the associated intractable diseases often spreads from one region to another in the affected brains. Interestingly, some of the causal proteins for such diseases do not contain Gln/Asn-rich domains but their aggregates can propagate in a manner similar to that observed for yeast Mod5. The molecular basis of this prion-like aggregate propagation in mammalian cells has yet to be clarified. We propose that analysis of non-Gln/Asn-rich Mod5 will be helpful to better our understanding of the mechanism of prion-like cell-to-cell transmission of non-Gln/Asn-rich causal proteins for neurodegenerative diseases.
In summary, Mod5, a yeast tRNA modification enzyme lacking Gln/Asn-rich domains, was identified as a novel yeast prion protein and its active prion conversion regulates the sterol biosynthetic pathway and provides cellular resistance against antifungal drugs. These new findings expand the yeast prion world to include non-Gln/Asn-rich proteins and revealed that active prion conversion could play essential roles for cell survival.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Kelvin Hui for his comments on the manuscript. Funding was provided by JST PRESTO, Grants from MEXT (Priority Area on Protein Society), the Next Program, the Sumitomo Foundation and the Novartis Foundation (Japan) for the Promotion of Science (M.T.).
Footnotes
Previously published online: www.landesbioscience.com/journals/prion/article/22685
References
- 1.Wickner RB, Edskes HK, Shewmaker F, Nakayashiki T. Prions of fungi: inherited structures and biological roles. Nat Rev Microbiol. 2007;5:611–8. doi: 10.1038/nrmicro1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tuite MF, Serio TR. The prion hypothesis: from biological anomaly to basic regulatory mechanism. Nat Rev Mol Cell Biol. 2010;11:823–33. doi: 10.1038/nrm3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Toyama BH, Weissman JS. Amyloid structure: conformational diversity and consequences. Annu Rev Biochem. 2011;80:557–85. doi: 10.1146/annurev-biochem-090908-120656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wickner RB. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264:566–9. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- 5.Liebman SW, Chernoff YO. Prions in yeast. Genetics. 2012;191:1041–72. doi: 10.1534/genetics.111.137760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tessier PM, Lindquist S. Unraveling infectious structures, strain variants and species barriers for the yeast prion [PSI+] Nat Struct Mol Biol. 2009;16:598–605. doi: 10.1038/nsmb.1617. [PSI+] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sondheimer N, Lindquist S. Rnq1: an epigenetic modifier of protein function in yeast. Mol Cell. 2000;5:163–72. doi: 10.1016/S1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- 8.Osherovich LZ, Weissman JS. Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI(+)] prion. Cell. 2001;106:183–94. doi: 10.1016/S0092-8674(01)00440-8. [DOI] [PubMed] [Google Scholar]
- 9.Du Z, Park KW, Yu H, Fan Q, Li L. Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat Genet. 2008;40:460–5. doi: 10.1038/ng.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patel BK, Gavin-Smyth J, Liebman SW. The yeast global transcriptional co-repressor protein Cyc8 can propagate as a prion. Nat Cell Biol. 2009;11:344–9. doi: 10.1038/ncb1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–58. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halfmann R, Wright JR, Alberti S, Lindquist S, Rexach M. Prion formation by a yeast GLFG nucleoporin. Prion. 2012;6:391–9. doi: 10.4161/pri.20199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li L, Lindquist S. Creating a protein-based element of inheritance. Science. 2000;287:661–4. doi: 10.1126/science.287.5453.661. [DOI] [PubMed] [Google Scholar]
- 14.Michelitsch MD, Weissman JS. A census of glutamine/asparagine-rich regions: implications for their conserved function and the prediction of novel prions. Proc Natl Acad Sci U S A. 2000;97:11910–5. doi: 10.1073/pnas.97.22.11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Legname G, Baskakov IV, Nguyen HO, Riesner D, Cohen FE, DeArmond SJ, et al. Synthetic mammalian prions. Science. 2004;305:673–6. doi: 10.1126/science.1100195. [DOI] [PubMed] [Google Scholar]
- 16.Taneja V, Maddelein ML, Talarek N, Saupe SJ, Liebman SW. A non-Q/N-rich prion domain of a foreign prion, [Het-s], can propagate as a prion in yeast. Mol Cell. 2007;27:67–77. doi: 10.1016/j.molcel.2007.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakayashiki T, Kurtzman CP, Edskes HK, Wickner RB. Yeast prions [URE3] and [PSI+] are diseases. Proc Natl Acad Sci U S A. 2005;102:10575–80. doi: 10.1073/pnas.0504882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGlinchey RP, Kryndushkin D, Wickner RB. Suicidal [PSI+] is a lethal yeast prion. Proc Natl Acad Sci U S A. 2011;108:5337–41. doi: 10.1073/pnas.1102762108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halfmann R, Jarosz DF, Jones SK, Chang A, Lancaster AK, Lindquist S. Prions are a common mechanism for phenotypic inheritance in wild yeasts. Nature. 2012;482:363–8. doi: 10.1038/nature10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tyedmers J, Madariaga ML, Lindquist S. Prion switching in response to environmental stress. PLoS Biol. 2008;6:e294. doi: 10.1371/journal.pbio.0060294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: the story of [PIN(+)] Cell. 2001;106:171–82. doi: 10.1016/S0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- 22.Patel BK, Liebman SW. “Prion-proof” for [PIN+]: infection with in vitro-made amyloid aggregates of Rnq1p-(132-405) induces [PIN+] J Mol Biol. 2007;365:773–82. doi: 10.1016/j.jmb.2006.10.069. [PIN+] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Osherovich LZ, Cox BS, Tuite MF, Weissman JS. Dissection and design of yeast prions. PLoS Biol. 2004;2:E86. doi: 10.1371/journal.pbio.0020086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzuki G, Shimazu N, Tanaka M. A yeast prion, Mod5, promotes acquired drug resistance and cell survival under environmental stress. Science. 2012;336:355–9. doi: 10.1126/science.1219491. [DOI] [PubMed] [Google Scholar]
- 25.Derkatch IL, Uptain SM, Outeiro TF, Krishnan R, Lindquist SL, Liebman SW. Effects of Q/N-rich, polyQ, and non-polyQ amyloids on the de novo formation of the [PSI+] prion in yeast and aggregation of Sup35 in vitro. Proc Natl Acad Sci U S A. 2004;101:12934–9. doi: 10.1073/pnas.0404968101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dihanich ME, Najarian D, Clark R, Gillman EC, Martin NC, Hopper AK. Isolation and characterization of MOD5, a gene required for isopentenylation of cytoplasmic and mitochondrial tRNAs of Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:177–84. doi: 10.1128/mcb.7.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–41. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 28.Eisenberg D, Jucker M. The amyloid state of proteins in human diseases. Cell. 2012;148:1188–203. doi: 10.1016/j.cell.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature. 2004;428:323–8. doi: 10.1038/nature02392. [DOI] [PubMed] [Google Scholar]
- 30.Gustavsson M, Ronne H. Evidence that tRNA modifying enzymes are important in vivo targets for 5-fluorouracil in yeast. RNA. 2008;14:666–74. doi: 10.1261/rna.966208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan CT, Pang YL, Deng W, Babu IR, Dyavaiah M, Begley TJ, et al. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat Commun. 2012;3:937. doi: 10.1038/ncomms1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goldschmidt L, Teng PK, Riek R, Eisenberg D. Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc Natl Acad Sci U S A. 2010;107:3487–92. doi: 10.1073/pnas.0915166107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Serio TR, Cashikar AG, Kowal AS, Sawicki GJ, Moslehi JJ, Serpell L, et al. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science. 2000;289:1317–21. doi: 10.1126/science.289.5483.1317. [DOI] [PubMed] [Google Scholar]
- 34.Ohhashi Y, Ito K, Toyama BH, Weissman JS, Tanaka M. Differences in prion strain conformations result from non-native interactions in a nucleus. Nat Chem Biol. 2010;6:225–30. doi: 10.1038/nchembio.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brundin P, Melki R, Kopito R. Prion-like transmission of protein aggregates in neurodegenerative diseases. Nat Rev Mol Cell Biol. 2010;11:301–7. doi: 10.1038/nrm2873. [DOI] [PMC free article] [PubMed] [Google Scholar]