Abstract
Objective
To present an integrated molecular biology dedicated system for tuberculosis diagnosis.
Methods
One hundred and five sputum specimens from patients strongly suspected by clinical parameters of tuberculosis were studied by Ziehl-Neelsen staining, by cultivation on solid medium and by a balanced heminested fluorometric PCR system (Orange G3TB) that could preserve worker safety and produce a rather pure material free of potential inhibitors. DNA amplification was performed in a low cost tuberculosis termocycler-fluorometer. Produced double stranded DNA was flurometrically detected. The whole reaction was conducted in one single tube which would not be opened after adding the processed sample in order to minimize the risk of cross contamination with amplicons.
Results
The assay was able to detect 30 bacillus per sample mL with 99.8% interassay variation coefficient. PCR was positive in 23 (21.9%) tested samples (21 of them were smear negative). In our study it showed a preliminary sensitivity of 94.5% for sputum and an overall specificity of 98.7%.
Conclusions
Total run time of the test is 4 h with 2.5 real working time. All PCR positive samples are also positive by microbiological culture and clinical criteria. Results show that it could be a very useful tool to increase detection efficiency of tuberculosis disease in low bacilus load samples. Furthermore, its low cost and friendly using make it feasible to run in poor regions.
Keywords: Tuberculosis, Sputum, Molecular diagnosis, Low cost, Real-time PCR, Mycobacterium tuberculosis, Molecular biology technology, Sputum sample, Microbiological culture
1. Introduction
Tuberculosis (TB) is estimated to have caused the deaths of 1 billion people in the last 200 years[1]. TB is a serious and slowly developing bacterial infection that is caused by the bacterium Mycobacterium tuberculosis (M. tuberculosis). Despite being a global disease, the burden of TB is most severe in developing countries, particularly those in South America, Asia (South-East Asia and Western Pacific regions) and Africa[2]. Although the fact that TB is almost endemic in some regions, case detection rates remain very low, posing a significant obstacle for its control. The World Health Organization's estimation of the global burden of disease caused by TB in 2009 is listed as follows: 9.4 million incident cases (range, 8.9 million-9.9 million), 14 million prevalent cases (range, 12 million-16 million), 1.3 million deaths among HIV-negative people (range, 1.2 million-1.5 million) and 0.38 million deaths among HIV-positive people (range, 0.32 million-0.45 million)[3].
TB is out of control in developing countries[4]. Overcrowding is so common that people are likely to live in dark, unventilated rooms, and thus they are more likely to be infected by TB and to receive large doses of the bacilli[5]. Patients' resistance to the disease is reduced, particularly by malnutrition and other diseases, such as neglected tropical diseases[6],[7]. Extreme poverty and all its accompaniments-malnutrition, overcrowding or homelessness, addiction, and lack of access to health care are the major driving forces underlying the presence and spread of TB, including the current increases in tuberculosis caused by multiple drug-resistant and extensively drug resistant straits[8],[9]. In addition, the increase in TB in recent decades is directly connected with the HIV epidemic, and in countries with high HIV prevalence, the number of new TB cases has tripled in the past 15 years. Developing countries bear the brunt of the epidemics of AIDS and TB, and the management of patients with both diseases poses a particular challenge in these settings[3],[10]. The reemergence of tuberculosis as an important public health issue and the spread of drug-resistant tuberculosis have emphasized the need for rapid diagnosis. However, the standard culture methods currently in use are quite slow[11]. Detection of mycobacterial growth on conventional Löwenstein-Jensen medium requires 4 to 8 weeks. In addition, it requires technical skills and a high complexity bacteriology laboratory. The need for speed is also a factor when the results of tests have a positive impact to help infection control decisions regarding patient isolation and therapeutic management[12].
It is in these type of situations that molecular diagnostic methods can provide the data needed more rapidly, and in many cases it is more cost-effective than traditional culture methods[13],[14]. Four years ago, we decided to develop a system of TB diagnosis that could fulfill these requirements. In this article,we presented a novel low complexity/cost integrated molecular biology dedicated system for the rapid detection of TB.
2. Materials and methods
2.1. Study population
From January to July 2010 we conducted this study in Buenos Aires, Argentina. Sputum specimens (one or two spot samples obtained in the morning) were collected from 66 patients with clinical manifestations of pulmonary tuberculosis.
2.2. Laboratory methods
2.2.1. Sample treatment and DNA extraction procedure
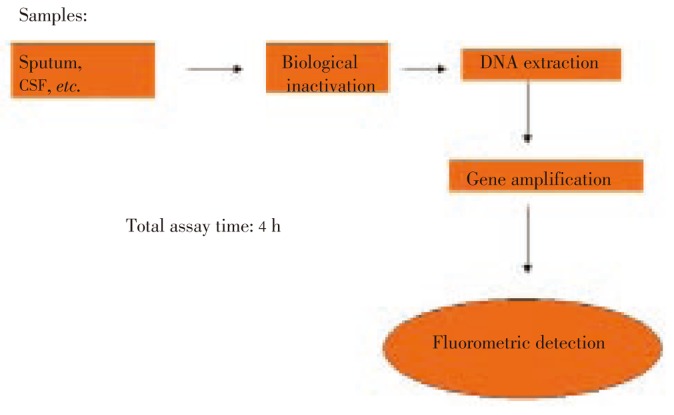
A sealed device for sample processing was designed to simultaneously preserve the bio-security of the operator and to exhibit nucleic acids for gene amplification. The schematic diagram was summarized in Figure 1.
Figure 1. Scheme of complete procedure for Orange G3TB tuberculosis diagnosis.
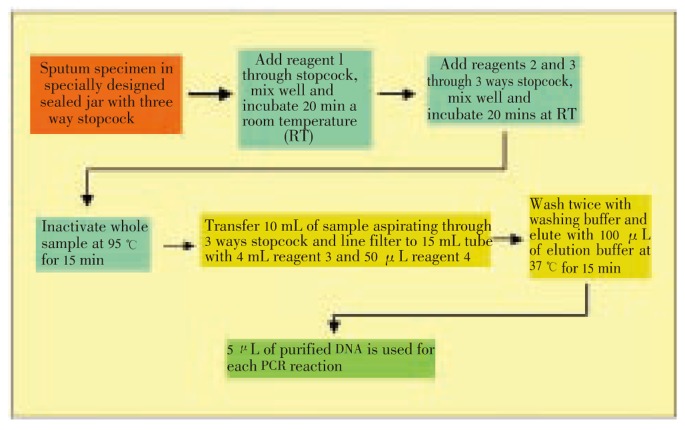
The system was designed in order to avoid any biohazard for the operator, to be easily manipulated by untrained personnel and to have efficient sample treatment to expose Mycobacterium DNA free of contaminants. Our system was based on a series of steps that included the liquefaction of the sample (in case of sputum material), the biological inactivation of the material and the purification of DNA. Everything was a standard system of syringes, filters and absorbents. Essentially it was a cycle completely closed without contact with the environment. In the last purification, all the genetic matter present in most of the time and the totality of the sample was obtained, which was used for amplification (Figure 2).
Figure 2. Scheme of DNA extraction procedure.
2.2.2. DNA amplification and M. tuberculosis detection
PCR procedure was conducted with a dedicated thermocycler equipment of low cost and with an incorporated and basic fluorometer. It was designed to be used in laboratories of very low complexity. Thus it avoided the edilicia and technological infrastructure of high complexity and it was easy to operate by non specialized personnel. It used a primitive but efficient heat exchanger. It also had a serial communication port via internet or telephone modem that allowed Public Health Department to monitor and to coordinate epidemiological data in real time (Figure 3). All reagents for amplification and fluorometric detection were included in PCR microtubes in a ready use form. Five microliters of previously sample purified DNA were used for each amplification, which took approximately 2 h. After amplification PCR products were evaluated by fluorometry in the dedicated fluometer and data were logged in memory and displayed on the screen. Produced double stranded DNA was end point and flurometrically detected in a simple but efficient low cost fluorometer designed “ad hoc”. The whole reaction was carried out in a single tube which was not opened after adding the processed sample to minimize the risk of cross contamination with amplicons. Time and temperature parameters for PCR reaction were inserted in microprocessor firmware to simplify the utilization of the equipment. The primer strategy design was based on Garcia-Quintanilla et al[15].
Figure 3. Low cost budget termocycler Orange G3TB for amplification of M. tuberculosis genome.

2.3. Statistical analysis
Assay parameters, sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV) and interassay coefficient were calculated. Results were expressed as proportion and 95% confidence interval was calculated. The differences between PCR and TB assays were evaluated and binomial McNemar's test was used.
3. Results
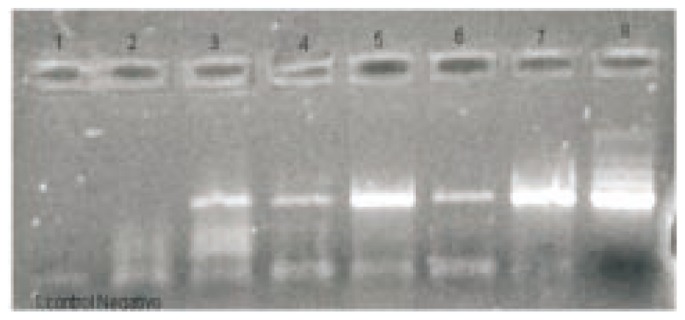
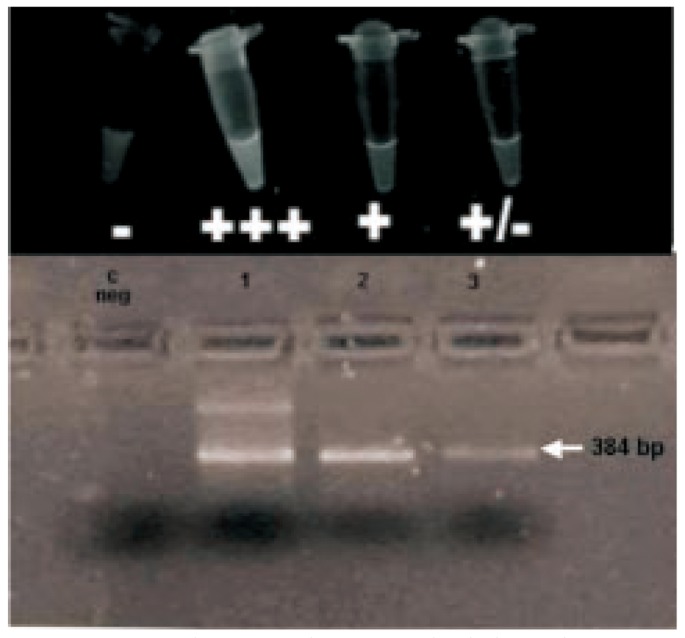
Sputums from 66 patients (105 samples) strongly suspected by clinical parameters of TB were studied by acid-fast bacillus smears direct observation (AFB), by culture and by a balanced heminested fluorometric PCR system (Orange G3TB) that included a specially developed syringe-attached membrane in order to preserve worker's safety and to produce a rather pure material free of potential inhibitors. DNA amplification was carried out in a low cost tuberculosis termocycler-fluorometer. It had a novel heat exchange system programmable controlled integrated circuit (PIC). The target of amplification was a segment of IS6110 insertion fragment. The assay was able to detect 30 bacillus per sample mL and it had a 99.8% interassay variation coefficient. Twenty-three samples from 19 patients were positive by Orange G3TB PCR assay and only two of them were positive by Ziehl Neelsen staining. All negative samples by AFB smear observation and confirmed by traditional culture methods were detected by PCR. In order to test simplicity of our procedure, samples proccesing assays were performed by duplicates including a team of laboratory technicians with only two weeks of training and clinical microbiologists. Results obtained by both teams were coincident. Samples proccesed by our improved extraction methodology were compared with standard TB DNA extraction method. Results were shown in Figure 4. It clearly showed that our DNA extraction system increased significantly the sensitivity of the molecular strategy for tuberculosis detection. The assay had an sensitivity of 94.5% for sputum specimens in our study, but only 10.5% in contrast with AFB. In addition, its overall specificity was 98.7%. Their corresponding NPV were 97.8 and 72.6 for Orange G3TB PCR and AFB direct observation, respectively. Total run time of the test was 4 h with 2.5 real working time. All TB detected by PCR specimens were also positive by microbiological culture and clinical criteria. In PCR positive samples, there were 2 bac pos and 17 bac neg, while only 47 bac neg without bac pos in PCR negative samples. PCR had a higher positive expression percentage (28.57%) than AFB-Ziehl-Neelsen (3.17%). And negative expression percentage in PCR was 71.43% and 96.83% was in AFB-Ziehl-Neelsen. Direct fluorescent detection and gel electrophoresis of a curve of positive control was shown in Figure 5.
Figure 4. Comparison of TB DNA extraction methods.
Lane 1: cont neg; lanes 2, 3: sample 1 and 2 extracted with the clasic procedure; lanes 4, 5: samples 1 and 2 processed with Orange G3TB extraction Kit; lanes 6, 7: samples 1 and 2 processed with Orange G3TB enhanced extraction Kit; lane 8: cont pos.
Figure 5. Direct fluorescent detection and gel electrophoresis of a curve of positive control.
c: negative (3×105, 3×103, 3×101 bacillus).
4. Discussion
New technology for rapid diagnosis of tuberculosis can be applied in developing countries where the prevalence of TB is high. It is based on methods that permit recognition of mycobacterial products in clinical specimens. We have developed a semi-automated PCR diagnostic test that can detect the presence of TB. A similar test, which is described in paper published in the New England Journal of Medicine[16], promises to help the public health sectors of low-income countries, where the occurrence of TB and multidrug-resistant pulmonary tuberculosis is high.
Previous TB diagnostic tests required skilled technicians and tools, and lacked either timeliness or sensitivity. Culture tests are highly sensitive, but these tests take as long as 2-6 weeks to produce results and demand specialized materials to support the virulent microbacteria in the culture. Although sputum smear test is quicker and produces results in about 30 min, it can only detect 10% to 75% of TB cases and requires trained microscopists. In developing countries, the technical expertise and tools needed to perform these tests are limited, and TB is often not diagnosed or treated early, which allows the disease to spread quickly in crowded living quarters and to build resistance to the drugs used in the treatment of the infection[17],[18].
Nucleic acid amplification (NAA) testing is useful for rapid identification of M. tuberculosis in respiratory samples, and results can be available within two to four hours. In addition, this technique has a higher sensitivity than sputum AFB smears. It can detect as few as one M. tuberculosis organism per 100 mL of sample[19].
Several techniques have been used for TB diagnosis, such as Gen-Probe MTD (Gen-Probe Incorporated, San Diego, CA). The enhanced MTD test (E-MTD) is an improved version of the Gen-Probe MTD that was approved in 1999 by the FDA for the diagnosis of TB in either smear-positive or smear-negative cases. A larger sample can be used and the processing time is shorter than that of the earlier MTD test, which requires three hours to complete. The Amplicor M. tuberculosis test kit (Roche Diagnostic Systems, Inc., Branchburg, NJ) is FDA-approved for testing smear positive respiratory samples[16],[20].
Here we presented the new heminested fluorometric PCR-based TB diagnostic test-called Orange G3TB, which is fast, sensitive and semi-automated. An accurate diagnosis can be obtained in less than 4 h. This novel technology increases the sensitivity and specificity for TB identification according to the results published by other authors[16],[18]. Our system is semi-automated and needs little technical training to perfom the assay. Orange G3TB also appears to have potential use for the determination of infectiousness. It is as specific as and more sensitive than serial AFB smear testing to screen for TB infectivity. This was illustrated in our prospective study (66 patients, total 105 samples). Two AFB smear-positive patients had also positive Orange G3TB test. In addition, another 21 samples from 16 patients with AFB smear-negative TB, were identified by Orange G3TB. The confirmation of AFB as M. tuberculosis within several hours can be valuable in the management of individual cases and permits the rapid mobilization of public health resources. Sputum specimens that are AFB positive and Orange G3TB positive always contain M. tuberculosis. Orange G3TB should be performed on all AFB smear-negative respiratory specimens. A positive Orange G3TB of an AFB smear-negative respiratory sample is diagnostic of tuberculosis.
A positive Orange G3TB result may be valuable in the early detection of the approximately 88 percent of active tuberculosis cases that are smear-negative. It is important to note that a relatively high clinical suspicion of TB is the driving factor leading to nucleic acid amplification testing in smear-negative cases. It is certainly not appropriate to test patients with AFB-negative smears in the absence of a strong clinical suspicion.
According to FDA data, the sensitivity of the tests for TB (compared with traditional culture) is approximately 95 percent in patients with a positive AFB smear, but only about 50 percent in smear-negative cases. However, in some patient population this sensitivity could be much smaller, which results in an important number of false negative. Specificity is greater than 95 percent in either smear-negative or smear-positive samples. When molecular methods are used in conjunction with classical epidemiology, their utility for TB control will be realized[18],[21].
In Argentina and in many other developing countries, the main approach for TB diagnosis is still the Ziehl Neelsen staining direct observation. AFB smear observation has a very poor sensitivity and a low negative predictive value in addition to exposing testing personnel to bacillus. Because of these technique parameter values, currently many patients are misdiagnosed and the epidemy is increasing[23].
In summary, the new heminested fluorometric PCR-based TB diagnostic test-Orange G3TB should be used in patients with an intermediate to high clinical suspicion of tuberculosis. It is helpful because the initial stains are negative in approximately 50 percent of cases. A negative NAA in this setting does not rule out tuberculosis, and additional examinations, such as bronchoscopy, are required[24],[26].
Orange G3TB is succefull for detecting TB directly in clinical samples, and for this reason, the “2010 International Science and Technology Meeting” is held in Buenos Aires. Argentina awarded it a “Technological Innovation Prize”. This system was conceptualized to be used in places of technical and economic limited resources to establish rapid TB diagnosis. Its advantages make it possible to be used in regions of low economic resources, where the incidence of the disease is often the greatest. In the whole experiment, there was no exhibition of biological material to the environment. This implies a dramatic reduction of false positive rates and avoids the possible contamination.
Our results showed that Orange G3TB integrated system is a very useful tool for detecting tuberculosis disease efficiently in low bacilus load samples. Furthermore, its relatively low cost and friendly using make it feasible to run in poor development regions. These preliminary results strongly suggest that this assay yield excellent predictive positive and negative values for diagnosis of TB.
Our objective now is to redesign this device of a form that the process is realized following a continuous flow within a column or injections. And it can be assembled on industrial scale with its original characteristics of easy operation and lowest cost.
We advocate to establish a collaboration in tuberculosis research between developed and developing nations in order to shed light on where it is most needed. Molecular epidemiology can be an important tool for understanding the current tuberculosis pandemic and for influencing the design of future policies in global TB control. Techniques based on nucleic acid detection have significantly changed pathogen disease diagnosis by improving specificity, sensitivity and accuracy. In the case of TB, a diagnostic technique based on this method would yield results in a much shorter time than the classical methods, which allow timely treatment of the disease and consequently decrease possible multi-drug resistance of TB The goal of innovative technologies and discoveries in the diagnostic field is to treat the right patient with right therapies and tools.
Acknowledgments
Partial results from this study were presented in a poster presentation at the 5th IAS Conference on HIV Pathogenesis, Treatment and Prevention in Cape Town, South Africa in July 2009 and at the III International Clinical Virology Symposium and advances in Buenos Aires, Argentina in November 2010.
Footnotes
Conflict of interest statement: We declare that we have no conflict of interest.
References
- 1.Ryan F. The forgotten plague: how the battle against tuberculosis was won and lost. Boston: Little, Brown and Co; 1992. p. 3. [Google Scholar]
- 2.Dye C, Watt J, Bleed M, Hosseini M, Raviglione C. Evolution of tuberculosis control and prospects for reducing tuberculosis incidence, prevalence, and deaths globally. JAMA. 2005;293:2767–2775. doi: 10.1001/jama.293.22.2767. [DOI] [PubMed] [Google Scholar]
- 3.WHO . Global tuberculosis control-epidemiology, strategy, financing: WHO report. Geneva: World Health Organization; 2009. (WHO/HTM/TB/2009.411.) [Google Scholar]
- 4.Kehinde O, Obaseki A, Cadmus I, Bakare A. Diagnosis of tuberculosis: urgent need to strengthen laboratory services. J Natl Med Assoc. 2005;97:394–396. [PMC free article] [PubMed] [Google Scholar]
- 5.Kimerling E. The Russian equation: an evolving paradigm in tuberculosis control. Int J Tuberc Lung Dis. 2000;4:160–167. [PubMed] [Google Scholar]
- 6.Marras K. Tuberculosis among tibetan refugee claimants in Toronto, 1998 to 2000. Chest. 2003;124:915–921. doi: 10.1378/chest.124.3.915. [DOI] [PubMed] [Google Scholar]
- 7.Lukács J, Tubak V, Mester J, David S, Bártfai Z, Kubica T, et al. Conventional and molecular epidemiology of tuberculosis in homeless patients in budapest, hungary. J Clin Microbiol. 2004;42:5931–5934. doi: 10.1128/JCM.42.12.5931-5934.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reyes H, Coninx R. Pitfalls of tuberculosis programmes in prisons. BMJ. 1997;315:1447–1450. doi: 10.1136/bmj.315.7120.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elbers C, Lanjouw O, Lanjouw P. Micro-level estimation of poverty and inequality. Econometrica. 2002;71:355–364. [Google Scholar]
- 10.Evans T. Challenging inequities in health: from ethics to action. Oxford: University Press; 2001. pp. 144–149. [Google Scholar]
- 11.Noordhoek T, Kaan A, Mulder S, Wilke H, Kolk H. Routine application of the polymerase chain reaction for detection of Mycobacterium tuberculosis in clinical samples. J Clin Pathol. 1995;48:810–814. doi: 10.1136/jcp.48.9.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watterson A, Drobniewski A. Modern laboratory diagnosis of mycobacterial infections. J Clin Pathol. 2000;53:727–732. doi: 10.1136/jcp.53.10.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Urdea M, Penny A, Olmsted S. Requirements for high impact diagnostics in the developing world. Nature. 2006;444:73–79. doi: 10.1038/nature05448. [DOI] [PubMed] [Google Scholar]
- 14.Raja S, Ching J, Xi L. Technology for automated, rapid, and quantitative PCR or reverse transcription-PCR clinical testing. Clin Chem. 2005;51:882–890. doi: 10.1373/clinchem.2004.046474. [DOI] [PubMed] [Google Scholar]
- 15.García-Quintanilla A, Garcia L, Tudó G, Navarro M, González JT, Jiménez de Anta T. Single-tube balanced heminested PCR for detecting Mycobacterium tuberculosis in smear-negative samples. J Clin Microbiol. 2000;38:1166–1169. doi: 10.1128/jcm.38.3.1166-1169.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boehme C, Nabeta P, Hillemann D, Nicol P, Shenai S, Krapp F, et al. Rapid molecular detection of tuberculosis and rifampin resistance. N Engl J Med. 2009;363:1005–1015. doi: 10.1056/NEJMoa0907847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helb D, Jones M, Story E, Boehme C, Wallace E, Ken H. Rapid detection of Mycobacterium tuberculosis and rifampin resistance by use on on-demand, near patient technology. J Clin Microbiol. 2010;48:237–239. doi: 10.1128/JCM.01463-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blakemore R, Story E, Helb D, Kop A, Banada P, Owens R, et al. Evaluation of the analytical preformance of the Xpert MTB/RIF assay. J Clin Microbiol. 2010;48:2495–2501. doi: 10.1128/JCM.00128-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dahle R, Eldholm V, Winje A, Mannsaker T, Heldal E. Impact of immigration on the molecular epidemiology of Mycobacterium tuberculosis in a low-incidence country. Am J Respir Crit Care Med. 2007;176:930–935. doi: 10.1164/rccm.200702-187OC. [DOI] [PubMed] [Google Scholar]
- 20.Barnes F. Rapid diagnostic tests for tuberculosis. Progress, but not gold standard. Am J Respir Crit Care Med. 1997;155:1497–1498. doi: 10.1164/ajrccm.155.5.9154847. [DOI] [PubMed] [Google Scholar]
- 21.Mathema B, Kurepina E, Bifani J, Kreiswirth N. Molecular epidemiology of tuberculosis: current insights. Clin Microbiol Rev. 2006;19:658–685. doi: 10.1128/CMR.00061-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Banada B, Sivasubramani K, Blakemore R, Boehme C, Perkins M, Fennely K, et al. Containment of bioaerosol infection risk by the Xpert MTB/RIF assay and its applicability to point-of-care settings. J Clin Microbiol. 2010;48:3351–3357. doi: 10.1128/JCM.01053-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bradley P, Reed L, Catanzaro A. Clinical efficacy of the amplified Mycobacterium tuberculosis direct test for the diagnosis of pulmonary tuberculosis. Am J Respir Crit Care Med. 1996;153:1606. doi: 10.1164/ajrccm.153.5.8630609. [DOI] [PubMed] [Google Scholar]
- 24.Soham G, Vishnu PS, Indira B, Muralidharan S. Diagnostic efficacy of Ziehl-Neelsen method against fluorescent microscopy in detection of acid fast bacilli. Asian Pac J Trop Med. 2010;3(4):328–329. [Google Scholar]
- 25.Prakash S, Sasikala SL, Aldous V, Huxley J. Isolation and identification of MDR-Mycobacterium tuberculosis and screening of partially characterised antimycobacterial compounds from chosen marine micro algae. Asian Pac J Trop Med. 2010;3(8):655–661. [Google Scholar]
- 26.Neeraj T, Vineeta K, Ajoy T,K, Ram MR, Tolia VS. Investigation of tuberculosis clusters in Dehradun city of India. Asian Pac J Trop Med. 2010;3(6):486–490. [Google Scholar]