Abstract
Objective
To determine the molecular characterization of Polymerase complex (PA, PB1 and PB2) genes of H9N2 avian influenza viruses and the genetic relationship of Iranian H9N2 viruses and other Asian viruses.
Methods
The Polymerase complex (PA, PB1 and PB2) genes from seven isolates of H9N2 viruses isolated from commercial chickens in Iran during 2008-2009 were amplified (by RT-PCR method) and sequenced. Nucleotide sequences (Open Reading Frame: orf) of the PA, PB1 and PB2 genes were used for phylogenetic tree construction.
Results
Most PB2 and PA genes of the H9N2 viruses isolated in 2008-2009 belonged to the unknown avian sublineage which grouped with the 2004 Pakistani H7N3 viruses. The PB1 genes of Iranian viruses indicated greater genetic diversity and shared a high level of similarity to PB1 genes from either H5 or H7 subtypes with compared to established H9N2 Eurasian sublineages.
Conclusions
Our findings demonstrated that the H9N2 viruses in Iran exhibit striking reassortment which has led to the generation of new genotypes.
Keywords: Genetic analysis, Polymerase complex, Avian influenza, Iran
1. Introduction
Influenza viruses belong to the Orthomyxoviridae family .They are enveloped viruses with a genome of single-stranded negative-sense RNA composed of eight genes segments encoding at least 10 proteins[1],[2]. According to the antigenic differences in their nucleoprotein and matrix protein, these viruses are divided into three major types A, B and C[2]. Influenza A virus can be further classified into subtypes on the basis of the antigenic properties of two surface glycoprotein's haemagglutinin (HA) and neuraminidase (NA). To date, 16 HA and 9 NA subtypes of influenza A virus have been recognized[3]. The entire genome of influenza A viruses are packaged into ribonucleoprotein particles (RNPs) including the nucleoprotein (NP) and polymerase complex. The polymerase complex consists of the PA, PB1 and PB2 subunits. Polymerase subunits are responsible for transcription and replication of viral RNA: During the initial steps, by using a cap-snatching method, they transcribe virally encoded genes; then, they replicate full-length viral RNA to produce first positive-strand complementary RNA and progeny viral RNA. The catalytic activity of the PB1 subunit cleaves host cell mRNA binding to the cap-binding PB2 subunit. PB1 residues implicated in the endonuclease and polymerase active sites have been recognized, although the position of the cap-binding site of PB2 remains controversial[1]. The role of the PA protein isn't exactly clear, but numerous studies indicated that it is essential for cap snatching and viral RNA promoter recognition[1],[4]. During 1999 to 2003, human cases of H9N2 virus infection have been reported in China[5]. Published surveys indicated that H9N2 viruses can infect humans and it is considered to be one of the potential public health risks[6]-[10]. These viruses are now enzootic in the Middle East countries and are associated with great economic losses[11],[12],[14],[15]. In this study, we investigated the phylogenetic patterns of the PA, PB1 and PB2 genes of H9N2 influenza viruses isolated from Commercial broiler chicken in the Iran between 1999 and 2009. We delineated the PA, PB1 and PB2 genes of these field isolates and established their phylogenetic relationship to the other Asian H9N2 viruses.
2. Materials and methods
2.1. Sampling and virus isolation
Samples collected from different parts of the country. Sample collection was performed according to the standard protocol[16]. During the period 2008 to 2009, lung and trachea Samples were submitted to the National Reference Laboratory. The samples were stored at -70 °C until used. They were treated with 2× phosphate buffer solution (PBS, pH 7.4) containing antibiotics and antifungal (Penicillin 10 000 unit/ml, Streptomycin 10 000 unit/ml and Nystatin 20 000 unit/ml). Initial viral isolation was performed by using ten days-old SPF (Specific Pathogen Free) embrocated chicken eggs (ECEs). Eggs candled daily, and embryos dying within 24-h post inoculation (PI) were discarded. Allantoic fluids were extracted from the eggs, and the presence of viruses was confirmed by haemagglutination test. Standard haemagglutination-inhibition (HI) and neuraminidase-inhibition (NI) tests were used for Subtype identification of the viruses[17].
The following seven viruses isolated in our study comprised: (A/chicken/Iran/RZ28/2008,A/chicken/Iran/RZ37/2008,A/chicken/Iran/RZ42/2009,A/chicken/Iran/RZ53/2008A/chicken/Iran/RZ71/2009,A/chicken/Iran/RZ75/2009, A/chicken/Iran/RZ77/2009).
2.2. RT-PCR and sequence analysis
The viral RNA was obtained directly from the allantoic fluid by applying the High Pure Viral Nucleic Acid Kit (Roche Germany). Purified genomic RNA was used to generate cDNA clones (RT-PCR) in accordance with the standard protocol[18]. The specific primers were used for genes amplification as described below:
-Primers used for PB2 amplification were: Forward primer (2052 bp):5′- AAAAGCAGGTCAATTATATTC-3′ Reverse primer (2052 bp): 5′- AAGGTCGTTTTTAAACTATTCA-3′ - Primers used for PB1 amplication were: Forward primer (2060 bp): 5′- GCAAAAGCAGGAGTGAAAATG-3′ Reverse primer (2060 bp): 5′- AGTCCTGAGCACAAATAACTGG-3′
- Primers used for PA amplication were: Forward primer (2133 bp):5′- AGCAAAAGCAGGTACTGAT-3′ Reverse primer (2133 bp):5′- AGTAGAAACAAGGTACTTTT-3′
High Pure Product Purification Kit (Roche Germany) was used for the PCR products purification and then purified products were used for direct sequencing (MWG co, Germany).
Nucleotide and deduced amino acid sequences of the three polymerase complex genes were edited with the Editseq (DNASTER Software package Version 5.2 )Nucleotide and deduced amino acid sequences were aligned by ClustalW, Version 1.4.
The phylogenetic tree construction was performed with the Meg Align program 2.8.
2.3. Nucleotide sequence accession numbers
The sequences determined in this study are available in the GenBank under accession numbers: JX097026- JX097046.
3. Results
Phylogenetic analysis of H9N2 polymerase complex genes:
Phylogenetic analysis of three polymerase complex genes showed that they formed different Sub- lineages including: Beijing -like or Y280-like, G1-like, H5N1/01-like, unknown avian and three duck lineages (Dk1, Dk2, Dk3)
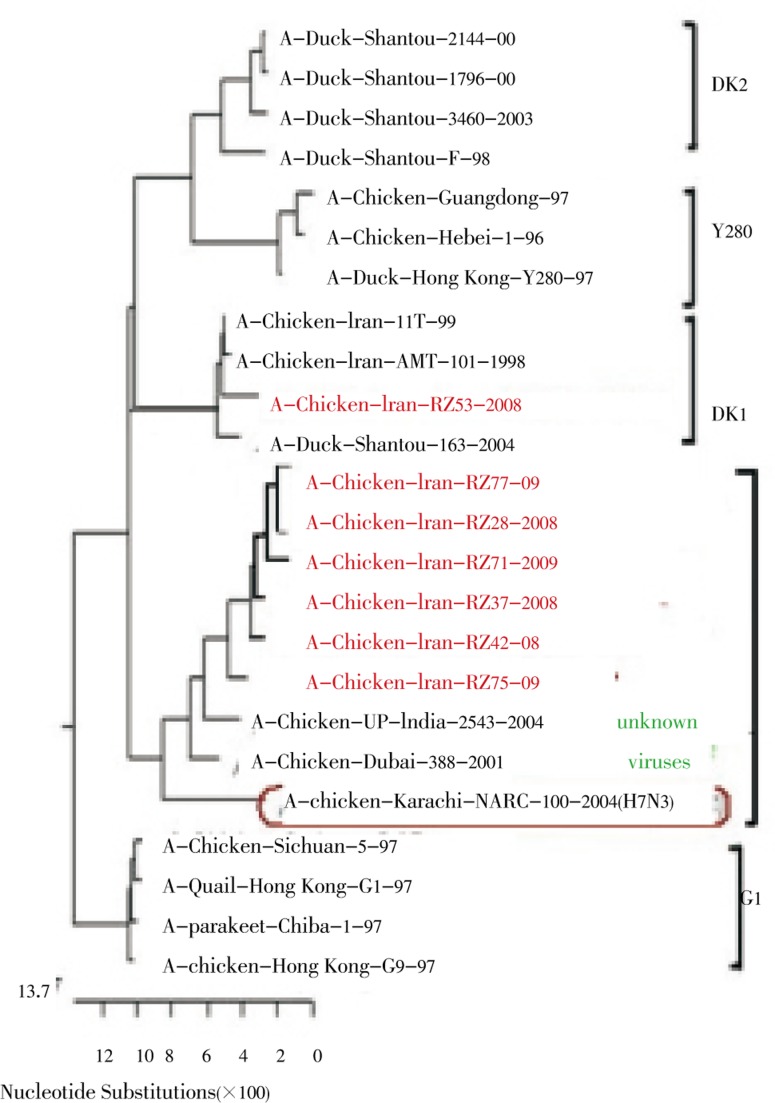
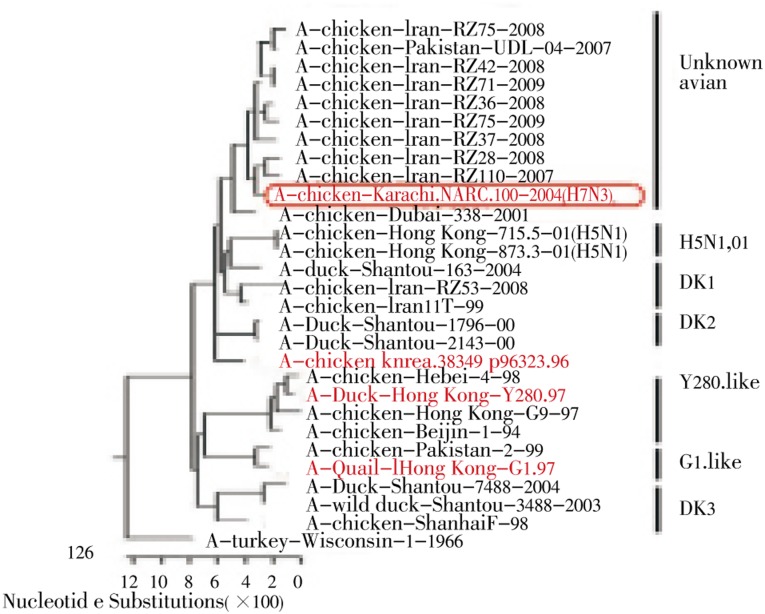
Phylogenetic analysis of the PB2 and PA genes showed that all of the Iranian H9N2 viruses fell in two groups: unknown avian and Dk1 (Figure 1 and Figure 2). All of the H9N2 viruses isolated in 2008-2009 except A/chicken/Iran/RZ53/2008 belonged to the unknown avian sublineage which grouped with the 2004 Pakistani H7N3 viruses. A/chicken/Iran/RZ53/2008 clustered with Dk1 sublineage that is most closely related to Dk/ST/163/04, which is isolated from migratory duck.
Figure 1. Phylogenetic analysis of the PB2 gene showed that all the PB2 genes of the Iranian H9N2 viruse fell in two groups, unknown avian and Dk1.
Figure 2. Phylogenetic analysis of the PA gene showed that all the PA genes of the Iranian H9N2 viruse fell in two groups, unknown avian and Dk1.
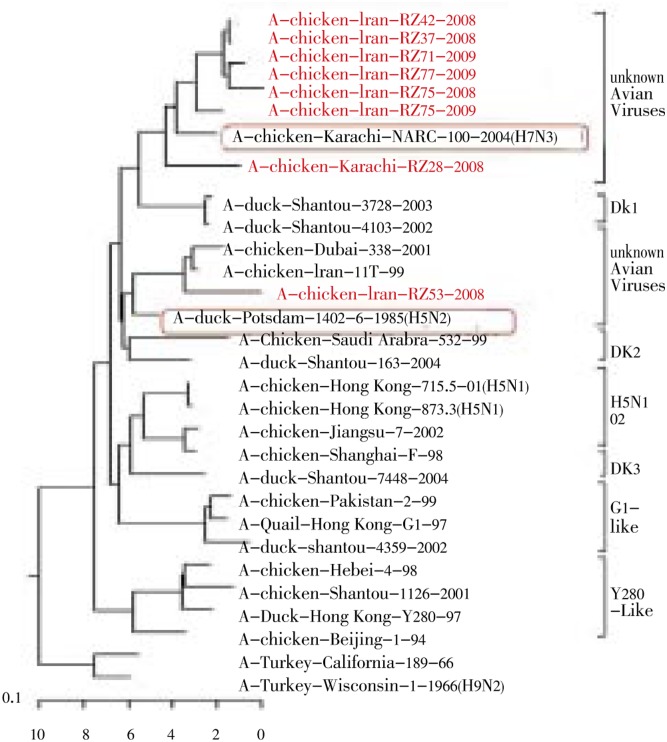
The PB1 genes of Iranian H9N2 viruses showed a high level of sequence diversity as compare with PA and PB2 genes.
Nucleotide sequence comparisons showed that two isolates (A/chicken/Iran/RZ53/2008 and A/chicken/Iran/T11/1399) contained a PB1 gene closely related to H5N2 viruses from Germany (A/duck/Potsdam/1402-6/1986; 95.4-96.1%). The other Iranian isolates were similar to a H7N3 chicken isolate from Pakistan and formed a distinct subclass compared to Eurasian sublineages (G1, Korean- and Y280-like) (Figure 3).
Figure 3. The PB1 genes of Iranian viruses indicated greater genetic diversity and shared a high level of similarity to PB1 genes from either H5 or H7 subtypes.
3.1. Molecular characterization
To analyze the molecular characteristics of Iranian H9N2 viruses, the deduced amino acid sequences of the PA, PB1 and PB2 proteins were aligned and compared with other H9N2 viruses.
The Iranian isolates did not indicate deletions or insertions within three polymerase complex genes with compare to the prototype, A/turkey/winconsin/66, but rather several point mutations were registered.
These viruses characterized by eight amino acid substitutions in PB1 gene at the following positions: 87 (V to I), 270 (S to C), 271 (T to A) 280 (N to S), 285 (R to K), 301(H to k), 416 (M to L).
One of the Iranian isolates (A/chicken/Iran/RZ71/2009) carry amino acid substitution K 615 R in the PA gene. Arginine to Lysine (Arg to Lys) substitution at position 615, has been associated with the adaptation of H9N2 avian influenza virus in human and mice.
3.2. Genotyping
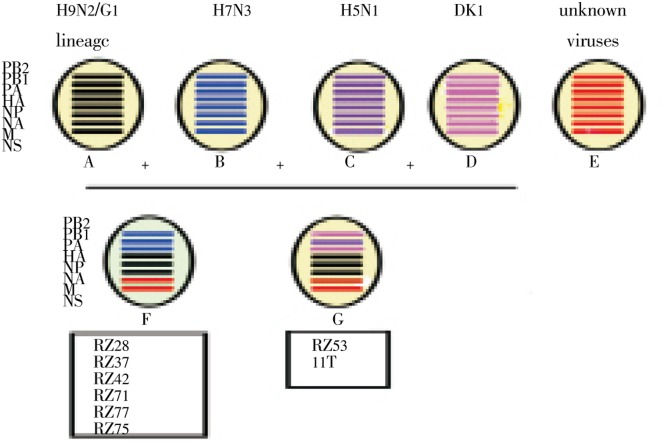
Based on sequence comparison and our previous studies[18]-[20] we recognized at least two different genotypes, designated F and G among these seven viruses (Figure 4).
Figure 4. newly identified genotype of H9N2 viruses. Phylogenetic analysis of the Iranian Polymerase complex genes revealed at least two different genotypes.
The polymerase genes of the recent H9N2 viruses originated from three sublineages. The Polymerase complex genes (PB1, PB2, and PA) of A/chicken/Iran/RZ53/2008 (genotype G) belonged to the Dk1 sublineage or closely related to German H5N2 viruses. However, most PB1, PB2 and PA genes of the H9N2 viruses isolated in 2008-2009 (genotypes F) were similar to the Pakistani H7N3 viruses.
4. Discussion
Avian influenza H9N2 viruses circulated widely in the Middle East (Iran, the UAE and Israel) and were associated with economic losses in poultry[11],[12],[14],[15]. In this study, we have reported the first genetic analysis of the Polymerase complex genes (PB1, PB2 and PA) of H9N2 avian influenza viruses and founded that Iranian viruses had undergone genetic reassortment. The molecular basis of host-range restriction and adaptation of influenza A viruses to a new host species has not exactly been determined. Previous studies revealed that mutation of the polymerase complex is necessary for adaptation to a new host and may increase replication and transcription of the adapted virus in mammalian species[5],[21].
Earlier reports concentrated on the key role of the HA, M1 and NS1 proteins in host-range restriction and adaptation[22],[23]. But at the present, numerous studies indicate that the virulence of influenza viruses is probably to be a multigenic trait[5],[21]. Analysis of protein sequences showed Arg to Lys ( 615) substitution in the PA gene in one of the Iranian isolate . K 615 R substitution in the PA gene is a crucial determinant for influenza virus pathogenicity and host specificity. Gabriel et al (2005) have suggested that the K 615 R substitution may be essential for adaptation of avian viruses to mammalian hosts[4]. It seems that the substitution K615R observed here in Iranian viruses may also lead to increased pathogenicity and replicative efficiency of H9N2 influenza viruses in mammalian hosts.
Instead of Lysine (K) at position 615 within the PA protein, Argnine (R) was existed in human H1N1, H5N1, and H9N2 isolates including A/HK/483/97, A/HK/485/97 and A/HK/1073/99 which confirm the relevance of PA 615 Arg for host change[5]. Previous studies have shown that the Eurasian lineage consists of at least three sublineages represented by their prototype strains: A/chicken/Korea/38349-p96323/96 (Korean-like), A/duck/Hong Kong/Y280/97 (Y280-like), and A/quail/Hong Kong/G1/97 (G1- like)[22]. As reported by Xu et al (2007), our result also showed that Polymerase complex genes of H9N2 viruses formed different sublineages including G1-like , Ck/Beijing -like( or Y280-like) , three duck lineages (Dk1, Dk2,Dk3) and unknown avian[25].
Our previous studies[18]-[20] indicated that Iranian surface glycoprotein genes (HA and NA) and one internal gene (NP) were similar to G1-like virus represented by Qa/HK/G1/97, whereas the PA,PB1 and PB2 genes of the Iranian H9N2 viruses, formed a distinct group compared to G1-, Korean- and Y280-like sublineage.
polymerase complex genes sequence homologies of the Iranian isolates showed more similarity with a H7N3 chicken isolate from Pakistan (A/Chicken/Karachi/NARC-100/2004( 92.5-95.5%) compared to Qa/HK/G1/97 (85.3-86.6%), Dk/HK/Y280/97 (84.7-86.9%) and Ck/Korea/323/96 (88.2-89.9%) .Furthermore, the PB1 gene of Iranian isolates were more similar to a H5N2 duck isolate from Germany (Dk/Potsdam/2216-4/84; 95.3-95.4%) compared to Eurasian sublineage.
Based on the genetic similarities and phylogenetic analysis, our results suggested that the Iranian viruses had undergone genetic reassortment with other influenza subtypes including H7 and H5 viruses.
Like the Iranian isolates, reassortment between H9N2 and the highly pathogenic avian influenza virus H7N3 subtype was reported in Pakistan[26]. It is also noted that the viruses from Dubai and Pakistan shared an out group relationship with the Iranian viruses in the PA gene tree suggesting that these viruses are derived from the same gene pool.
Phylogenetic analysis of the Iranian polymerase complex genes revealed at least two different genotypes. Our identification of novel genotypes of H9N2 viruses in 2008-2009 was markedly similar to those of a recent study conducted by Igbal et al in Pakistan[27]. This finding suggests a high degree of diversity among the H9N2 viruses in the regions of the Middle East and Indian sub-continent.
In recent years, novel genotypes of H9N2 avian influenza viruses from domestic poultry in China, Korea, Vietnam, India and Pakistan have been identified and well characterized[27]-[34]. In February 2006, highly pathogenic H5N1 virus was isolated from wild birds in Northern provinces of Iran[28]. It seems that the association of highly pathogenic H5N1 viruses and H9N2 cases raised the probability of novel genotypes in Iran. In view of this situation, we would expect to isolate additional novel genotypes with unique combinations of genes.
Homayounimehr et al (2010) and Soltanialvar et al (2010) have shown that the Iranian isolates possessed amino acid leucine (L) at position 226 instead of glutamine (Q) at the receptor binding site of haemagglutinins (HA) which is similar to A/Quail/HongKong/G1/97 and two human isolates: A/HK/1073/99, A/HK/1074/99[35]-[37],[18]. Amino acid differences in the receptor binding sites of HAs have been shown to be associated with differences in receptor binding specificity[22]. So Iranian H9N2 isolates can bind to α (2, 6) receptors. This feature suggested the pandemic potential of the H9N2 avian influenza virus and emphasizes the need for continuous surveillance in Iran, which has been continuing since 2000[39]-[41].
Acknowledgments
This study was supported by grant NO 8254 from The Islamic Azad University, Shoushtar Branch. The authors thank the excellent technical support provided by Mrs. Akbari. The sequences determined in this study are available in the GenBank under accession numbers: JX097026- JX097046.
Footnotes
Fundation Project: Supported by The Islamic Azad University, Shoushtar Branch (grant No. 8254).
Conflict of interest statement: We declare that we have no conflict of interest.
References
- 1.Bouvier NM, Plese P. The biology of influenza viruses. Vaccine. 2008;26:49–53. doi: 10.1016/j.vaccine.2008.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee CW, Saif YM. Avian influenza virus. Comp Immun Micro Infec Dis. 2009;32:301–310. doi: 10.1016/j.cimid.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 3.Fouchier R, Munster V, Wallensten A, Bestebroer TM, Herfst S, Smith D, et al. Characterization of a novel influenza A virus hemagglutinin subtype (HI6) obtained from black-headed gulls. J of Virol. 2005;79:2814–2822. doi: 10.1128/JVI.79.5.2814-2822.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gabriel G, Dauber B, Wolff T, Planz O, Klenk HD, Stech J. The viral polymerase mediates adaptation of an avian influenza virus to a mammalian host. Proc Natl Acad Sci USA. 2005;102(51):18590–18595. doi: 10.1073/pnas.0507415102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butt KM, Smith GJ, Chen H, Zhang LJ, Leung YH, Xu KM, et al. Human infection with an avian H9N2 influenza A virus in Hong Kong in 2003. J Clin Mic. 2005;43:5760–5767. doi: 10.1128/JCM.43.11.5760-5767.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuiken T, Taubenberger JF. Pathology of human influenza revisited. Vaccine. 2008;26S:D59–D66. doi: 10.1016/j.vaccine.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lupiani B, Reddy SM. The history of avian influenza. Comp Immun Mic Infec Dis. 2009;32:311–323. doi: 10.1016/j.cimid.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 8.Wit ED, Kawaoka Y, Jong MDE, Fouchier RAM. Pathogenicity of highly pathogenic avian influenza virus in mammals. Vaccine. 2008;26:54–58. doi: 10.1016/j.vaccine.2008.07.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalthoff D, Globig A, Beer M. (Highly pathogenic) avian influenza as a zoonotic agent. Vet Microb. 2010;140:237–245. doi: 10.1016/j.vetmic.2009.08.022. [DOI] [PubMed] [Google Scholar]
- 10.Brianda S, Tama JS, Blocha M, Mumforda E. Avian and pandemic influenza threats: The current situation. Proc Vaccino. 2010:159–165. [Google Scholar]
- 11.Aamir UB, Wernery U, Ilyushina N, Webster RG. Characterization of avian H9N2 influenza viruses from United Arab Emirates 2000-2003. J Virol. 2007;361:45–55. doi: 10.1016/j.virol.2006.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alexander DJ. An overview of the epidemiology of avian influenza. Vaccine. 2007;25:5637–5644. doi: 10.1016/j.vaccine.2006.10.051. [DOI] [PubMed] [Google Scholar]
- 13.Mosleh N, Dadras H, Mohammadi A. Molecular quantitation of H9N2 avian influenza virus in various organs of broiler chickens using TaqMan real time PCR. J Mole Genet Med. 2009;3:152–157. doi: 10.4172/1747-0862.1000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perk S, Golender N, Banet C, Shihmanter E, Pirak M, Tendler Y, et al. Phylogenetic analysis of hemagglutinin, neuraminidase, and nucleoprotein genes of H9N2 avian influenza viruses isolated In Israel during the 2000-2005 epizootic, Compara Immun. Mic Infec Dis. 2009;32:221–238. doi: 10.1016/j.cimid.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 15.Swayne DE. A laboratory manual for the isolation and identification of avian pathogens. 4th ed. Pennsylvania, USA: American Association of Avian Pathologist; 1998. p. 311. [Google Scholar]
- 16.Alexander DJ, Spackman D. Characterization of influenza A viruses isolated from turkeys during March-May 1979. Avia Path. 1981;10:281–293. doi: 10.1080/03079458108418477. [DOI] [PubMed] [Google Scholar]
- 17.Lee MS, Chang PC, Shien JH, Cheng MC, Shieh SK. Identification and subtyping of avian influenza viruses by reverse transcription PCR. J Virol Meth. 2001;97:13–22. doi: 10.1016/s0166-0934(01)00301-9. [DOI] [PubMed] [Google Scholar]
- 18.Soltanialvar M, Shoshtari H, Bozorgmehrifard M, Charkhkar S, Eshratabadi F. Molecular characterization of hemagglutinin, and neuraminidase Ggenes of H9N2 avian influenza viruses isolated from commercial broiler chicken in Iran. J Biolo Sci. 2010;10:145–150. [Google Scholar]
- 19.Soltanialvar M, Shoshtari H, Morovati M, Daliranni A, Akbarnejad F. Sequence and phylogenetic analysis of nucleoprotein gene in Iranian H9N2 avian influenza viruses. Modar J Med Sci Path. 2011;13:43–51. [Google Scholar]
- 20.Soltanialvar M, Shoshtari H, Bozorgmehrifard M, Charkhkar S, Akbarnejad F. Sequence and phylogenetic analysis of the neuraminidase genes of H9N2 avian influenza viruses isolated from commercial chickens in Iran. Trop Anim Health Prod. 2012;44:419–425. doi: 10.1007/s11250-011-9913-2. [DOI] [PubMed] [Google Scholar]
- 21.Wu R, Zhang H, Yang K, Liang W, Xiong Z, Yang X, et al. Multiple amino acid substitutions are involved in the adaptation of H9N2 vian influenza virus to mice. Vet Microbiol. 2009;138:85–91. doi: 10.1016/j.vetmic.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 22.Wan H, Perez DR. Amino acid 226 in the hemagglutinin of H9N2 influenza viruses determines cell tropism and replication in human air way epithelial cells. J Virol. 2007;81:5181–5191. doi: 10.1128/JVI.02827-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackson D, Hossain M J, Hickman D, Perez DR, Lamb RA. A new influenza virus virulence determinant: the NS1 protein four C-terminal residues modulate pathogenicity. Proc Natl Acad Sci USA. 2008;105:4381–4386. doi: 10.1073/pnas.0800482105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matrosovich MN, Krauss S, Webster RG. H9N2 influenza A viruses from poultry in Asia have human virus-like receptor specificity. Virol. 2001;281:156–162. doi: 10.1006/viro.2000.0799. [DOI] [PubMed] [Google Scholar]
- 25.Xu KM, Smith GJD, Bahl J, Duan L, Tai H, Vijaykrishna D, et al. The genesis and evolution of H9N2 influenza viruses in poultry from southern China, 2000 to 2005. J Virol. 2007:10389–10401. doi: 10.1128/JVI.00979-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abbas MA, Spackman DE, Swayne Z, Ahmed L, Sarmento D, et al. Sequence and phylogenetic analysis of H7N3 avian influenza viruses isolated from poultry in Pakistan 1995-2004. Virol J. 2010;7:137–146. doi: 10.1186/1743-422X-7-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iqbal M, Yaqub T, Reddy K, McCauley JW. Novel genotypes of H9N2 influenza A viruses isolated from poultry in Pakistan containing NS genes similar to highly pathogenic H7N3 and H5N1 viruses. Plos One. 2009;4:5788–5788. doi: 10.1371/journal.pone.0005788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun Y, Pu J, Jiang Z, Guan T, Xia Y, Xu Q, et al. Genotypic evolution and antigenic drift of H9N2 influenza viruses in China from 1994 to 2008. Vet Microbiol. 2010;146:215–225. doi: 10.1016/j.vetmic.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 29.Tosh C, Nagarajan S, Behera P, Rajukumar K, Purohit V, Kamal RP, et al. Genetic analysis of H9N2 avian influenza viruses isolated from India. Arch Virol. 2008;153:1433–1439. doi: 10.1007/s00705-008-0131-9. [DOI] [PubMed] [Google Scholar]
- 30.Wu R, Sui ZW, Zhang HB, Chen QJ, Liang WW, Yang KL, et al. Characterization of a pathogenic H9N2 influenza A virus isolated from central China in 2007. Arch Virol. 2008;153:1549–1555. doi: 10.1007/s00705-008-0139-1. [DOI] [PubMed] [Google Scholar]
- 31.Moon HJ, Song MS, Cruz DJM, Park KJ, Pascua PQ, Lee JH, et al. Active reassortment of H9 influenza viruses between wild birds and live-poultry markets in Korea. Arch Virol. 2010;155:229–241. doi: 10.1007/s00705-009-0577-4. [DOI] [PubMed] [Google Scholar]
- 32.Bi Y, Lu L, Li J, Yin Y, Zhang Y, Gao H, et al. Novel genetic reassortants in H9N2 influenza A viruses and their diverse pathogenicity to mice. Virol J. 2011;8:505. doi: 10.1186/1743-422X-8-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nomura N, Sakoda Y, Endo M, Yoshida H, Yamamoto N, Okamatsu M, et al. Characterization of avian influenza viruses isolated from domestic ducks in Vietnam in 2009 and 2010. Arch Virol. 2012;157(2):247–257. doi: 10.1007/s00705-011-1152-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou JP, Ge FF, Liu J, Ju HB, Yang DQ, Wang J, et al. Epidemiological survey and genetic evolution of H9 subtype influenza viruses in Shanghai, China, from 2006 to 2010. Arch Virol. 2012;157(6):1193–1198. doi: 10.1007/s00705-012-1266-2. [DOI] [PubMed] [Google Scholar]
- 35.Shoushtari AH, Habl-Alvarid MH, Vascellari M, Hedayati A. Mortality of wild swans associated with naturally infection with highly pathogenic H5N1 avian influenza virus in Iran. Arch Razi Institute. 2007;62:207–213. [Google Scholar]
- 36.Khun M, Heng C, Harun-Or-Rashid Md, Kasuya H, Sakamoto J. Knowledge, attitudes and practices towards avian influenza A (H5N1) among Cambodian women: A cross-sectional study. Asian Pac J Trop Med. 2012;5(9):727–734. doi: 10.1016/S1995-7645(12)60115-1. [DOI] [PubMed] [Google Scholar]
- 37.Gugong VT, Ajogi I, Junaidu K, Okolocha EC, Ngbede EO, Abraham MN, et al. Avian influenza in village chickens, its awareness and presence of potential risk practices among rural dwellers. Asian Pac J Trop Dis. 2012;2(4):282–285. [Google Scholar]
- 38.Homayounimehr AR, Dadras H, Shoushtari A, Pourbakhsh SA. Seuence and phylogenetic analysis of the haemagglutinin genes of H9N2 avian influenza viruses isolated from commercial chickens in Iran. Trop Anim Heal Prod. 2010;42:1291–1297. doi: 10.1007/s11250-010-9565-7. [DOI] [PubMed] [Google Scholar]
- 39.Rahimian A, Shoshtari AH, Pourbakhsh SA, Momayez R, Rahimi E, Mehrabanpour MJ. Serological and molecular survey of avian influenza H9N2 in human poultry farm industries. Med J Mashhad Univ Med Sci. 2009;52:133–140. [Google Scholar]
- 40.Pazani J, Karimi V, Bozorgmehri Fard MH, Ghalyanchi Langeroudi A, Barin A. Phylogenetic analysis of PB2 gene of H9N2 subtype of avian influenza viruses isolated from commercial chickens in Tehran province of Iran during 1998-2001. Iran J Veter Res Shiraz Univ. 2011;12:324–331. [Google Scholar]
- 41.Vatandour S, Bozorgmehrifard M, Shoushtari H, Charkhkar S, Bakhtiari S. Molecular characterization and phylogenetic analysis of neuraminidase gene of avian Influenza H9N2 viruses isolated from commercial broiler chicken in Iran during a period of 1998-2007. Afric J Microbiol Res. 2011;5:4182–4189. [Google Scholar]