Abstract
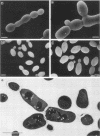
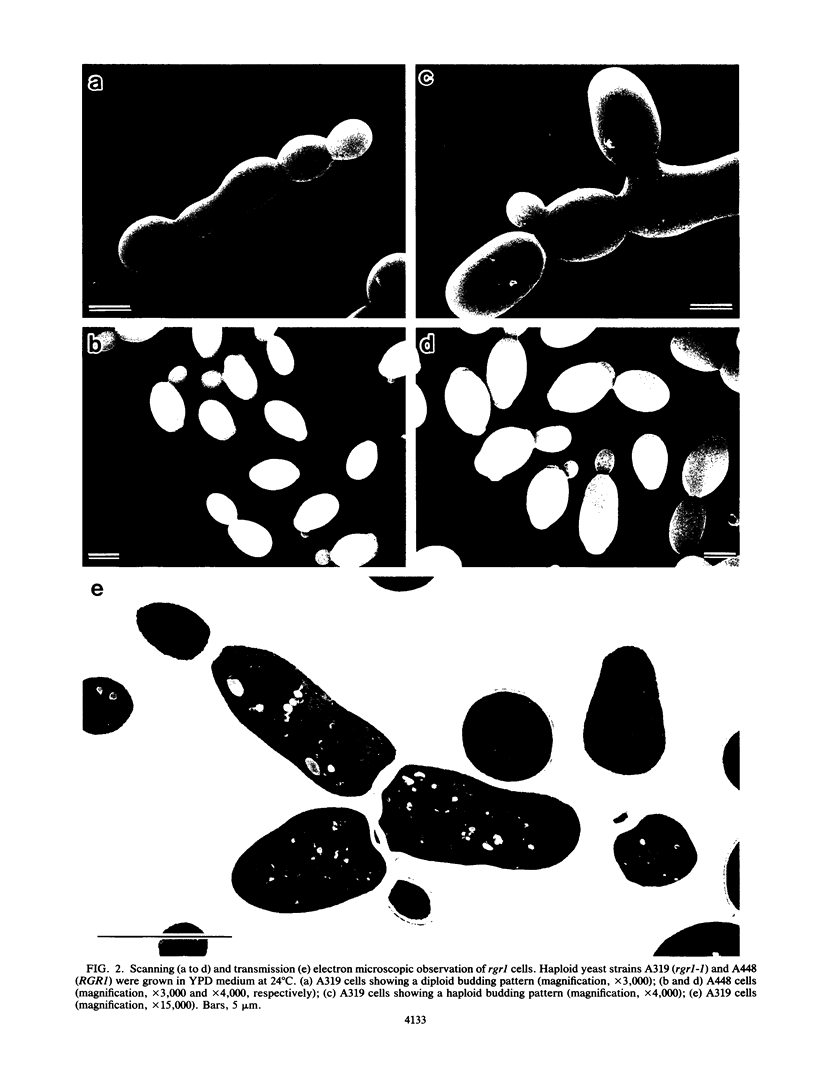
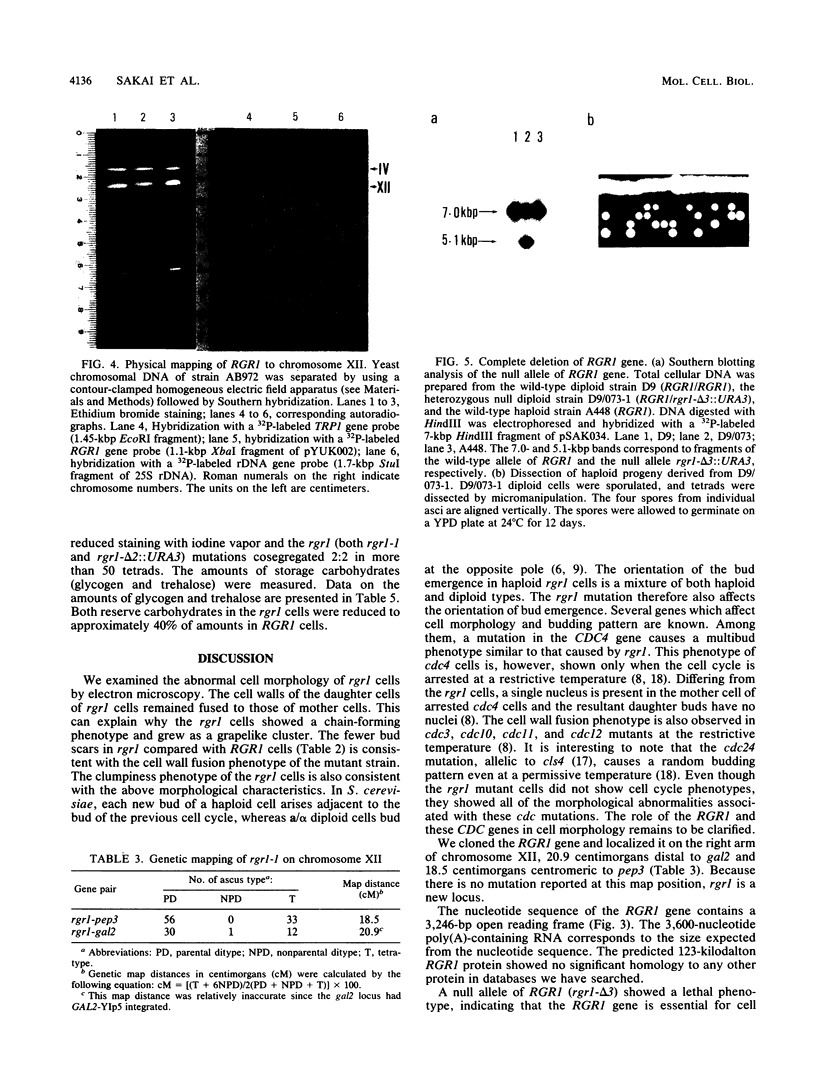
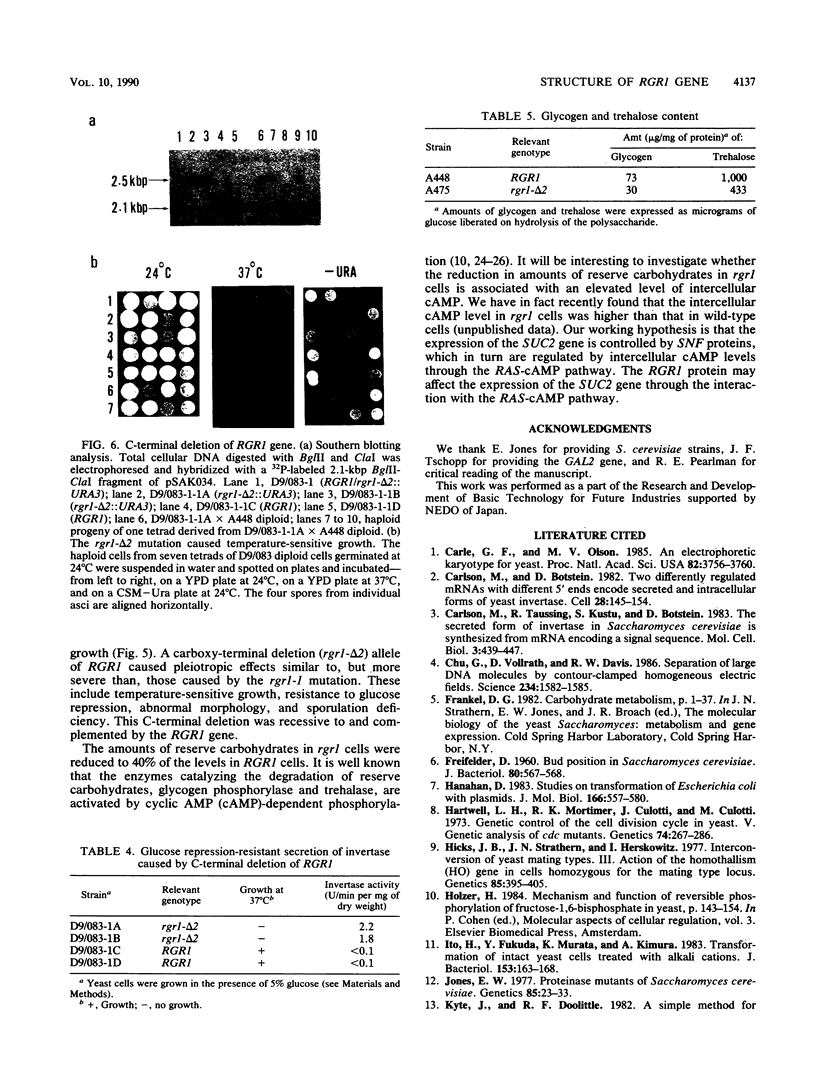
An RGR1 gene product is required to repress expression of glucose-regulated genes in Saccharomyces cerevisiae. The abnormal morphology of rgr1 cells was studied. Scanning and transmission electron microscopic observations revealed that the cell wall of the daughter cell remained attached to that of mother cell. We cloned the RGR1 gene by complementation and showed that the cloned DNA was tightly linked to the chromosomal RGR1 locus. The cloned RGR1 gene suppressed all of the phenotypes caused by the mutation and encoded a 3.6-kilobase poly(A)+ RNA. The RGR1 gene is located on chromosome XII, as determined by pulsed-field gel electrophoresis, and we mapped rgr1 between gal2 and pep3 by genetic analysis. rgr1 was shown to be a new locus. We also determined the nucleotide sequence of RGR1, which was predicted to encode a 123-kilodalton protein. The null mutation resulted in lethality, indicating that the RGR1 gene is essential for growth. On the other hand, a carboxy-terminal deletion of the gene caused phenotypes similar to but more severe than those caused by the original mutation. The amount of reserve carbohydrates was reduced in rgr1 cells. Possible functions of the RGR1 product are discussed.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Carle G. F., Olson M. V. An electrophoretic karyotype for yeast. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3756–3760. doi: 10.1073/pnas.82.11.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M., Botstein D. Two differentially regulated mRNAs with different 5' ends encode secreted with intracellular forms of yeast invertase. Cell. 1982 Jan;28(1):145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Carlson M., Taussig R., Kustu S., Botstein D. The secreted form of invertase in Saccharomyces cerevisiae is synthesized from mRNA encoding a signal sequence. Mol Cell Biol. 1983 Mar;3(3):439–447. doi: 10.1128/mcb.3.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu G., Vollrath D., Davis R. W. Separation of large DNA molecules by contour-clamped homogeneous electric fields. Science. 1986 Dec 19;234(4783):1582–1585. doi: 10.1126/science.3538420. [DOI] [PubMed] [Google Scholar]
- FREIFELDER D. Bud position in Saccharomyces cerevisiae. J Bacteriol. 1960 Oct;80:567–568. doi: 10.1128/jb.80.4.567-568.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hartwell L. H., Mortimer R. K., Culotti J., Culotti M. Genetic Control of the Cell Division Cycle in Yeast: V. Genetic Analysis of cdc Mutants. Genetics. 1973 Jun;74(2):267–286. doi: 10.1093/genetics/74.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks J. B., Strathern J. N., Herskowitz I. Interconversion of Yeast Mating Types III. Action of the Homothallism (HO) Gene in Cells Homozygous for the Mating Type Locus. Genetics. 1977 Mar;85(3):395–405. doi: 10.1093/genetics/85.3.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones E. W. Proteinase mutants of Saccharomyces cerevisiae. Genetics. 1977 Jan;85(1):23–33. doi: 10.1093/genetics/85.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lillie S. H., Pringle J. R. Reserve carbohydrate metabolism in Saccharomyces cerevisiae: responses to nutrient limitation. J Bacteriol. 1980 Sep;143(3):1384–1394. doi: 10.1128/jb.143.3.1384-1394.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohya Y., Miyamoto S., Ohsumi Y., Anraku Y. Calcium-sensitive cls4 mutant of Saccharomyces cerevisiae with a defect in bud formation. J Bacteriol. 1986 Jan;165(1):28–33. doi: 10.1128/jb.165.1.28-33.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roncero C., Valdivieso M. H., Ribas J. C., Durán A. Effect of calcofluor white on chitin synthases from Saccharomyces cerevisiae. J Bacteriol. 1988 Apr;170(4):1945–1949. doi: 10.1128/jb.170.4.1945-1949.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Sakai A., Shimizu Y., Hishinuma F. Isolation and characterization of mutants which show an oversecretion phenotype in Saccharomyces cerevisiae. Genetics. 1988 Jul;119(3):499–506. doi: 10.1093/genetics/119.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevelein J. M. Cyclic-AMP content and trehalase activation in vegetative cells and ascospores of yeast. Arch Microbiol. 1984 May;138(1):64–67. doi: 10.1007/BF00425409. [DOI] [PubMed] [Google Scholar]
- Uno I., Matsumoto K., Adachi K., Ishikawa T. Genetic and biochemical evidence that trehalase is a substrate of cAMP-dependent protein kinase in yeast. J Biol Chem. 1983 Sep 25;258(18):10867–10872. [PubMed] [Google Scholar]
- Wingender-Drissen R., Becker J. U. Regulation of yeast phosphorylase by phosphorylase kinase and cAMP-dependent protein kinase. FEBS Lett. 1983 Oct 31;163(1):33–36. doi: 10.1016/0014-5793(83)81156-9. [DOI] [PubMed] [Google Scholar]