Abstract
Objective
To assess the antimicrobial and cytotoxic effects of Methylobacterium sp. isolated from soil sample of Doddabetta forest, Nilgiris, Western Ghats of Tamil Nadu.
Methods
Isolation of Methylobacterium was performed from soils by serial dilution plate technique. The strain was grown in modified nutrient gulucose agar (MNGA) medium to study the morphology and biochemical characteristics. Methylobacterium sp. was screened for its antimicrobial activity against pathogenic bacteria and fungi. The strain was subjected to 16S rRNA analysis and was identified as Methylobacterium sp. The nucleotide sequence of the 16S rRNA gene of the isolate exhibited close similarity with other Methylobacterium sp. and has been submitted to Genbank. The antibacterial substances were extracted using chloroform and ethyl acetate from MNGA medium in which ERI-135 had grown for 5 d at 30 °C. Cytotoxic effect was also studied. GC-MS analysis was carried out. The antimicrobial activity was assessed using broth micro dilution technique.
Results
Ethyl acetate extract showed activity against bacteria such as Bacillus subtilis, Klebsiella pneumoniae (K. pneumoniae), Pseudomonas aeruginosa, Salmonella typhimurium, Shigella flexneri, Enterobacter aerogenes, Staphylococcus aureu and Staphylococcus epidermidis (S. epidermidis) and fungi such as, Candida albicans and Trichophyton rubrum. The lowest minimum inhibitory concentrations were: 250 µg/mL against S. epidermidis and 250µg/mL against K. pneumonia. The isolate had the ability to produce enzymes such as protease. The exyract showed cytotoxic effect in human adenocarcinoma cancer cell line (A549). GC-MS analysis showed the presence of isovaleric acid (3.64%), 2-Methylbutanoic acid (5.03%), isobutyramide (5.05%), N,N-oimethylformamide-di-t-butylacetal (9.79%), benzeneacetamide (15.56%), octyl butyl phthalate (3.59%) and diisooctyl phthalate (5.79) in the extract.
Conclusions
Methylobacterium sp. (ERI-135) showed promising antibacterial and cytotoxic activity. This is the first report in the antimicrobial and cytotoxic effect of Methylobacterium sp.
Keywords: Methylobacterium sp, 16S rRNA, Antimicrobial, Cytotoxicity, GC-MS, Soil sample, Western Ghats
1. Introduction
Over the past 20 years there has been a lot of interest in the investigation of natural materials as sources of new antibacterial agents[1]. The genus Methylobacterium was first proposed in 1976 to accommodate Gram-negative bacteria that have the ability to utilize methane and other more complex organic compounds such as carbon and other energy sources. Methylobacterium sps are natural sources for the production of industrially important compounds and an alternative to current bacterial expression systems for the production of recombinant proteins. Furthermore, Methylobacterium is a promising microorganism for the commercial production of natural products, including polyhydroxybutyrate (PHB) and the very valuable co-polymer P (HB/HV). Particularly Methylobacterium sps possess one or more characteristics of plant-growth promoting bacteria (PGPB)[2]. Methylobacterium sp. have been recognized as common environmental isolates from such habitats as leaf surfaces, leaf nodules of plants, soil, water, grass, sewage, air, and rice grains[3]. Methylobacteium stimulated seed germination, plant development, contributed to plant flavour and inhibited the bacterial infection[4]. The present study was aimed at assessing the antimicrobial activity of a new Methylobacterium sp. isolate (ERI-135), obtained from Doddapetta forest soil, Nilgiris (Southern Western Ghats of Tamil Nadu), India.
2. Materials and methods
2.1. Sample collection
The soil samples were collected from the depth of 5-15 cm at Doddapetta forest, (southern Western Ghats), Tamil Nadu, India.
2.2. Isolation of methylobacterium
Isolation of Methylobacterium was performed by serial dilution using dilution plate technique. One gram of soil was suspended in 9 mL of sterile distilled water. The dilution was carried out up to 10−6 dilutions. Aliquots (0.1 mL) of 10−2, 10−3, 10−4 and 10−5 were spread on the isolation plates containing Modified Nutrient agar. The plates were incubated at 28 °C for 5 d.
2.3. Cultural characterization
Active isolate (ERI-135) was characterized morphologically and physiologically following the directions given by the Bergey's Manual of Systematic Bacteriology[5]. Cultural characteristics of pure isolates were recorded after incubation for 5 d at 28 °C in different media. Active isolate (ERI-135) was identified using different pH levels, NaCl concentration, temperature and utilization of carbon sources following standard methods[6]. The ability to produce enzymes was also studied[7].
2.4. Extraction of bioactive metabolites
Culture inoculate of the isolate (ERI-135) was taken in 500 ml Erlenmeyer flasks containing 150 mL of modified nutrient gulucose agar (MNGA) medium and incubated at 30 °C in a shaker (200 rpm) for 5 d. After 5th day the culture broth was centrifuged at 10 000 rpm for 15 min to remove the biomass. Equal volumes of chloroform and ethyl acetate (1:1 v/v) were added. The organic solvent layer was transferred to a clean conical flask. The organic layer was concentrated using vacuum rotary evaporator at 40 °C. The extract was transferred to a 5 mL sterile vial.
2.5. Microbial organisms
The following bacteria and fungi were used for the experiment. Bacteria: Bacillus subtilis (B. subtilis) MTCC 441, Klebsiella pneumoniae (K. pneumoniae) MTCC 109, Pseudomonas aeruginosa (P. aeruginosa) MTCC 741, Salmonella typhimurium (S. typhimurium) MTCC 1251, Shigella flexneri (S. flexneri) MTCC 1457, Enterobacter aerogenes (E. aerogenes) MTCC 111, Staphylococcus aureus (S. aureus) MTCC 96 and Staphylococcus epidermidis (S. epidermidis) MTCC 3615; fungi: Candida albicans (C. albicans) MTCC 227 and Trichophyton rubrum (T. rubrum) 57/01. The reference cultures were obtained from Institute of Microbial Technology (IMTECH), Chandigarh, India-160 036. All the other cultures were obtained from the Department of Microbiology, Christian Medical College, Vellore, Tamil Nadu, India.
2.6. Antimicrobial assay
Antibacterial and antifungal activities were carried out using disc-diffusion method[8]. Petri plates were prepared with 20 mL of sterile Mueller Hinton agar (MHA) (Hi-media, Mumbai). The test cultures were swabbed on the top of the solidified media and allowed to dry for 10 min and a specific amount of crude extract 2 mg/disc was added to each disc separately. The loaded discs were placed on the surface of the medium and left for 30 min at room temperature for compound diffusion. Negative control was prepared using respective solvents. Streptomycin (10 µg/disc) was used as positive control against bacteria. Ketoconazole was used as positive control for fungi. The plates were incubated for 24 h at 37 °C for bacteria and for 48 h at 28 °C against fungi. Zones of inhibition were recorded in millimetres and the experiment was repeated twice.
2.7. Minimum inhibitory concentration (MIC)
Minimum inhibitory concentration studies of the crude extract were performed according to the standard reference methods for bacteria[9], for filamentous fungi[10] and yeasts[11]. The required concentrations (2 000 µg/mL, 1 000 µg/mL, 500 µg/mL, 250 µg/mL, 125 µg/mL, 62.5 µg/mL and 31.25 µg/mL) of the extract were dissolved in DMSO (2%), and diluted to give serial two-fold dilutions that were added to each medium in 96 well plates. An inoculum of 100 from each well was inoculated. The antifungal agents ketoconazole and fluconazole for fungi and the antibacterial agent streptomycin and ciprofloxacin for bacteria were included in the assays as positive controls.
2.8. Cytotoxicity effect on A549 cancer cell lines
A549 human adenocarcinoma cell lines were maintained in complete tissue culture medium DMEM with 10% fetal Bovine serum and 2 mM L-Glutamine, along with antibiotics (about 100 IU/mL of penicillin, 100 µg/mL of streptomycin) with the pH adjusted to 7.2. The cytotoxicity was determined according to the method of Hsu et al[12]. The cytotoxicity against cancer cells was determined by measuring the absorbance of the converted dye at 570 nm in an ELISA reader. Cytotoxicity of each sample was expressed as IC50 value.
2.9. Gas chromatography-mass spectrometry (GC-MS)
Secondary metabolites were identified using GC-MS, GC-MS-QP 2010 SHIMADZU. For GC-MS analysis, a 30 m×0.25 mm TR-35MS capillary column with a film thickness of 0.25 µm was used. The carrier gas was helium maintained at a column flow of 1.51 mL/min (at a pressure of 105 kPa). A 1.0 µL sample of the extract was injected and the column temperature was maintained at 70 °C for 2 min followed by temperature programming at 10 °C/min to 200 °C for 2 min. This was raised to 240 °C at a rate of 5 °C/min for 2 min, and finally to 300 °C at a rate of 35 °C/min for 2 min (Scan range: 40 - 1 000 m/z). The mass spectrometer and transfer line were held at 290 °C.
2.10. Molecular identification
Extraction of genomic DNA of the isolate was performed according to the method[13]. The 16S ribosomal RNA gene was amplified using the PCR with Taq DNA polymerase and primers F (AGAGTTTGATCCTGGCTCAG) and R (ACGGCTACCTTGTTACGACTT). The conditions for thermal cycling were as follows: denaturation of the target DNA at 94 °C for four minutes followed by 35 cycles at 94 °C for one minute, primer annealing at 52 °C for one minute and primer extension at 72 °C for one minute. At the end of the cycling, the reaction mixture was held at 72 °C for 5 min and then cooled to 4 °C. The PCR product obtained was sequenced by an automated sequencer (Genetic Analyzer 3130, Applied Biosystem, USA).
2.11. Nucleotide sequence accession number
The partial 16S rRNA gene sequences of isolate (ERI-135) have been deposited in the GenBank database under accession number JQ350893. A phylogenetic tree was constructed using the neighbour-joining DNA distance algorithm using software MEGA (version 4.1)[14].
3. Results
The Methylobacterium sp. isolate (ERI-135) was recovered from Doddapetta forest soil Nilgiris, (Southern Western Ghats), Tamil Nadu, India. The isolate showed good growth on medium amended with sodium chloride up to 8%; no growth was seen at 10%. Permissive temperature for growth ranged from 28 °C to 37 °C with optimum of 28 °C and the pH range was 6-10 with optimum pH of 7. Utilization of various carbon sources by ERI-135 indicated a wide pattern of carbon source assimilation. According to the cultural characteristics, the culture produced good amount of protease enzymes. Primary screening revealed that MNGA medium was a very good base for the production of antibacterial compound. The growing cells of selected cultures were Gram-positive. Cell wall sugar analysis of ERI-135 revealed the presence of maltose, sucrose, glucose, starch, xylose and ribose. The isolate ERI-135 is susceptible to ciprofloxacin, gentamicin, ampicillin, cephaloridine, streptomycin, erythromycin, rifambicin and vancomycin.
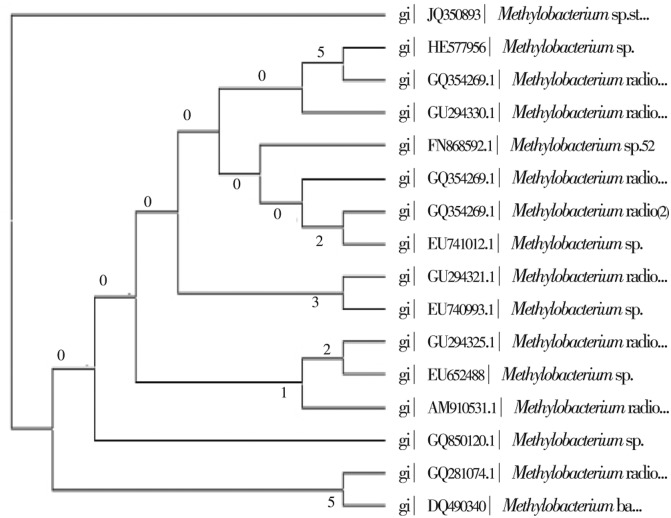
The result of the sequencing of ERI-135 was obtained in the form of rough electrophoregrams. The sequences have been chosen as reference sequences in which unidentified and unpublished sequences were not included. Alignment of this sequence through matching with reported 16S rRNA gene sequences in the Genbank showed high similarity (99%) to Methylobacterium sps 16S rRNA genes. The sequences were blasted via NCBI Blast software for comparison with the homologous sequences contained in the data bank (GenBank). The sequence results revealed that isolate ERI-135 was a Methylobacterium sp. Cluster analysis obtained by the MEGA4 method showed that isolate ERI-135 was taxonomically very close to Methylobacterium sp. (99%). The phylogenetic tree obtained by applying the neighbor joining method is illustrated in Figure 1. Based on preliminary screening results, ERI-135 was selected to study the antimicrobial property. The Methylobacterium sp. isolate (ERI-135) was extracted using chloroform and ethyl acetate solvents. The ethyl acetate extract (2 mg/mL) showed antibacterial and antifungal activities against bacteria and fungi (Table 1). The ethyl acetate extract produced maximum inhibitory zone when compared with chloroform extract. The MIC values are given in Table 2. The lowest minimum inhibitory concentrations were: 250 µg/mL against S. epidermidis and 250 µg/mL against K. pneumonia. The ethyl acetate extract showed maximum cytotoxicity against cancer cell lines at 2 000µg/mL concentration (Table 3). GC-MS analysis showed the presence of Isovaleric acid (3.64%), 2-Methylbutanoic acid (5.03%), Isobutyramide (5.05%), N,N-Dimethylformamide-di-t-butylacetal (9.79%), Benzeneacetamide (15.56%), Octyl butyl phthalate (3.59%) and Diisooctyl phthalate (5.79) (Table 4).
Figure 1. Phylogenetic tree derived from 16S rRNA gene sequences showing the relationship between isolate ERI-135 and species belonging to the genus Methylobacterium constructed using the neighbour-joining method. Bootstrap values were expressed as percentages of 1 000 replications.
Table 1. Antimicrobial activity of Methylobacterium sp. isolate (ERI-135) using disc diffusion method.
| Organism | Zone of inhibition (mm) |
||
| Chloroform extract | Ethyl acetate extract | Streptomycin (25 µg/mL) | |
| S. flexneri | - | 12 | 30 |
| E. aerogens | - | 10 | 22 |
| S. aureus | 8 | 10 | 14 |
| S. epidermidis | - | 12 | 14 |
| B. subtilis | - | 9 | 22 |
| K. pneumonia | 7 | 13 | 20 |
| P. aeruginosa | - | 11 | 30 |
| S. typhimurium | - | - | 24 |
| Fungi | Ketoconazole | ||
| C. albicans | - | 9 | 28 |
| T. rubrum | - | 8 | 30 |
Table 2. Minimum inhibitory concentrations of ethyl acetate extract of Methylobacterium sp. isolate (ERI-135) (µg/mL).
| Organism | Ethyl acetate extract | Streptomycin |
| S. flexneri | 500 | 30 |
| E. aerogens | 500 | 22 |
| S. aureus | 500 | 14 |
| S. epidermidis | 250 | 14 |
| B. subtilis | 1 000 | 22 |
| K. pneumonia | 250 | 20 |
| P. aeruginosa | 500 | 30 |
| S. typhimurium | - | 24 |
| Fungi | Ketoconazole | |
| C. albicans | 2 000 | 28 |
| T. rubrum | 2 000 | 30 |
-: No activity; Streptomycin - Standard antibacterial agent; Ketoconazole - standard antifungal agent.
Table 3. MTT assay to determine the cytotoxicity of ethyl acetate extract of Methylobacterium sp. (ERI-135).
| Ethyl acetate extract (ERI-135) (µg/mL) | Optical density at 570 nm | Cytotoxicity (%) |
| 2 000.0 | 0.082 | 76.38 |
| 1 000.0 | 0.197 | 55.36 |
| 500.0 | 0.218 | 42.55 |
| 250.0 | 0.245 | 31.34 |
| 125.0 | 0.295 | 19.13 |
| 62.5 | 0.375 | 2.15 |
| Control | 0.385 | - |
Table 4. GC-MS analysis of ethyl acetate extract of Methylobacterium sp. isolate (ERI-135).
| Retention time | Area (%) | Molecules name |
| 4.246 | 3.64 | Isovaleric acid |
| 4.476 | 5.03 | 2-Methylbutanoic acid |
| 5.363 | 5.05 | Isobutyramide |
| 6.037 | 0.75 | Butanamide |
| 6.618 | 0.60 | 4-Methyl-Pent-3-Enoic acid |
| 7.052 | 9.79 | N,N-Dimethylformamide-di-t-butylacetal |
| 8.042 | 1.28 | 4-Methylhexanoic acid |
| 8.944 | 0.56 | Phenylethyl Alcohol |
| 9.015 | 1.20 | Hexanamide |
| 9.232 | 2.16 | 5-Acetyldihydro-2(3H)-furanone |
| 9.739 | 0.76 | 5,6-Dimethyltetrahydro-2H-Pyran-2-One |
| 10.021 | 2.24 | Benzoic acid |
| 10.375 | 0.55 | 4-(1-HYDROXY-ETHYL). Gamma. Butanolactone |
| 10.494 | 0.23 | 2H-Pyran-2-OL, Tetrahydro-, Acetate |
| 10.750 | 3.95 | gamma.-Methylvaleramide |
| 10.910 | 0.50 | 2-Coumaranone |
| 11.128 | 0.49 | alpha-Toluic acid |
| 11.870 | 0.47 | 2-Hydroxy-2-methyl-6-hepten-3-one |
| 11.968 | 0.34 | Styrene glycol |
| 12.390 | 0.65 | Benzamide |
| 12.897 | 0.37 | Methyl 2,3-dimethylbutanoate |
| 13.260 | 15.56 | Benzeneacetamide |
| 17.095 | 0.52 | 3-Pyrrolidin-2-YL-Propionic acid |
| 17.313 | 1.42 | dl-Alanyl-l-leucine |
| 17.395 | 1.01 | 3-Isopropyl-6-Methyl-2,5-Piperazinedione |
| 17.647 | 1.13 | Pyrrolidino[1,2-a]piperazine-3,6-dione |
| 17.714 | 0.35 | 4-MethylpentylS-2-(diisopropylamino)ethyl propylphosphonothiolate |
| 18.192 | 2.29 | 3-Isobutylhexahydropyrrolo[1,2-a]pyrazine-1,4-dione |
| 18.499 | 3.59 | Octyl butyl phthalate |
| 19.168 | 1.45 | 3-Isobutylhexahydropyrrolo[1,2-a]pyrazine-1,4-dione |
| 19.357 | 4.40 | 3-Isobutylhexahydropyrrolo[1,2-a]pyrazine-1,4-dione |
| 19.482 | 5.25 | 3-Isobutylhexahydropyrrolo[1,2-a]pyrazine-1,4-dione |
| 21.507 | 0.72 | 3-Benzyl-6-Methyl-2,5-Piperazinedione |
| 21.659 | 2.94 | 3,6-Diisobutyl-2,5- Piperazinedione |
| 23.118 | 2.70 | 3-Benzylhexahydropyrrolo[1,2-a]pyrazine-1,4-dione |
| 24.127 | 0.42 | n-Eicosane |
| 24.469 | 5.79 | Diisooctyl phthalate |
| 25.639 | 0.92 | 3-Benzyl-6-Isobutyl-2,5-Dioxo-Piperazine |
| 26.354 | 1.26 | Docosane |
| 27.044 | 1.11 | Tetratetracontane |
4. Discussion
A new Methylobacterium sp. isolate (ERI-135) was recovered from Nilgiris forest soil of Western Ghats of Tamil Nadu. Cultural characteristics and 16S rRNA studies strongly suggested that this isolate belonged to the genus Methylobacterium. The isolate (ERI-135) was identified as Methylobacterium sp. with homology of 99%. The universal primers seem to be sufficient for identifying the genus but not the species. The cultural and morphological characteristics of isolate ERI-135 was Gram-negative, aerobic, frequently branched and formed pink to red-pigmented colonies. The cultural and morphological characteristics and activities of different Methylobacterium isolates have been reported by several investigators[15]. The isolate ERI-135 showed good antimicrobial activity in MNGA medium. Our results indicated that the antimicrobial compounds were extracellular[16]. Most of the secondary metabolites and antibiotics were extracellular in nature and extra cellular products showed potent antimicrobial activities[17].
The study of influence of different nutritional media and culture conditions on antimicrobial compound production indicated that the highest biological activities were obtained when YPG medium was used as a base. Our results indicated that the synthesis of antimicrobial metabolites depended on the medium constituents.
Our isolate ERI-135 produced ethyl acetate soluble extracellular products which were effective against pathogenic bacteria and fungi. It showed good antimicrobial activity in solid as well as in culture broth. It has been reported that the environmental factors like temperature, pH and incubation have profound influence on antibiotic production[18]. The antimicrobial compound from Methylobacterium sp. isolate (ERI-135) was recovered using ethyl acetate solvent. Normally most of the antimicrobial compounds are extracted using ethyl acetate[17]. It is evident from the present investigation that ethyl acetate extract of the isolate (ERI-135) showed significant cytotoxic effects on A549 cancer cell line. MTT assay is normally used to determine the cytotoxicity of potential drugs.
Acknowledgments
We are grateful to Entomology Research Institute, Loyola College, Chennai, Tamil Nadu, India, for the financial assistance (Grant No. ERI/2011/MB-05).
Footnotes
Foundation Project: Supported by Entomology Research Institute, Loyolacollege, Chennai, Tamil Nadu, India (Grant No. ERI/2011/MB-05).
Conflict of interest statement: We declare that we have no conflict of interest.
References
- 1.Zhang C, Ondeyka J, Dietrich L, Gailliot FP, Hesse M, Lester M, et al. Isolation, structure and biological activities of platencin A2-A4 from Streptomyces platensis. Bioorg Med Chem. 2010;18:2602–2610. doi: 10.1016/j.bmc.2010.02.030. [DOI] [PubMed] [Google Scholar]
- 2.Yim W, Poonguzhali S, Deka Boruah HP, Palaniappan P, Tong-Min S. Colonization pattern of gfp tagged Methylobacterium suomiens on rice and tomato plant root and leaf surfaces World Congress of Soil Science, Soil Solutions for a Changing World. 2010:1–6. [Google Scholar]
- 3.Rossetto PB, Dourado MN, Quecine MC, Andreote FD, Araújo WL, Azevedo JL, et al. Specific plant induced biofilm formation in Methylobacterium species. Braz J Microbiol. 2011;42:878–883. doi: 10.1590/S1517-83822011000300006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smejkalova H, Erb TJ, Fuchs G. Methanol assimilation in Methylobacterium extorquens AM1: Demonstration of all enzymes and their regulation. PLoS ONE. 2010;5:13001. doi: 10.1371/journal.pone.0013001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergeys manual of systematic bacteriology. Baltimore: Williams & Wilkins Company; 1989. Locci. pp. 2344–508. [Google Scholar]
- 6.Gottieb D. An evalution of criteria and procedures used in the description and characterization of Streptomyces. A co-operative study. Appl Microbiol. 1961;9:55. doi: 10.1128/am.9.1.55-65.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.National Committee for Clinical Laboratory Standards (NCCLS) Document M31-A performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. 1999. p. 57. Villanova: approved standard NCCLS.
- 8.Valan Arasu M, Duraipandiyan V, Agastian P, Ignacimuthu S. In vitro antimicrobial activity of Streptomyces spp. ERI-3 isolated from Western Ghats rock soil (India) J de Mycologie Médicale. 2009;19:22–28. [Google Scholar]
- 9.Duraipandiyan V, Ignacimuthu S. Antibacterial and antifungal activity of Flindersine isolated from the traditional medicinal plant, Toddalia asiatica (L.) Lam. J Ethnopharmacol. 2009;123:494–498. doi: 10.1016/j.jep.2009.02.020. [DOI] [PubMed] [Google Scholar]
- 10.Clinical and Laboratory Standards Institute (CLSI) Reference method for Broth dilution antifungal susceptibility testing of filamentous fungi; Approved standard second edition. CLSI document M38-A2. Pennsylvania: Clinical and Laboratory Standards Institute; 2008. [Google Scholar]
- 11.NCCLS . National Committee for Clinical Laboratory Standards. Reference method for broth dilution antifungal susceptibility testing of yeasts: proposed standard. Villanova: NCCLS; 2002. [Google Scholar]
- 12.Hsu H, Huang K, Lu K, Chiou S, Yen J, Chang C, et al. Typhonium blumei extract inhibits proliferation of human lung adenocarcinoma A549 cells via induction of cell cycle arrest and apoptosis. J Ethnopharmacol. 2011;135:492–500. doi: 10.1016/j.jep.2011.03.048. [DOI] [PubMed] [Google Scholar]
- 13.Moreira M, Noschang J, Neiva IF, Carvalho Y, Higuti IH, Vicente VA. Methodological Variations in the Isolation of Genomic DNA from Streptococcus Bacteria. Braz Arch Biol Technol. 2010;53:845–849. [Google Scholar]
- 14.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 15.Kim K, Madhaiyan M, Woo-Jong Yim, Chauhan PS, Tong-Min Sa. A novel pink-pigmented facultative Methylobacterium phyllosphaerae sp. nov. from phyllosphere of rice. 19th World Congress of Soil Science, Soil Solutions for a Changing World 1-6, 2010, Brisbane, Australia. Published on DVD.
- 16.Vital Pierangeli G, Rivera Windell L. Antimicrobial activity, cytotoxicity, and phytochemical screening of Voacanga globosa (Blanco) Merr. leaf extract (Apocynaceae) Asian Pac J Med. 2011;4(10):824–828. doi: 10.1016/S1995-7645(11)60202-2. [DOI] [PubMed] [Google Scholar]
- 17.Kavitha A, Prabhakar P, Vijayalakshmi M, Venkateswarlu Y. Purification and biological evaluation of the metabolites produced by Streptomyces sp. TK-VL_333. Res Microbiol. 2010;161:335–345. doi: 10.1016/j.resmic.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 18.Singh LS, Mazumder S, Bora TC. Optimisation of process parameters for growth and bioactive metabolite produced by a salt-tolerant and alkaliphilic actinomycete, Streptomyces tanashiensis strain A2D. J de Mycologie Médicale. 2009;19:225–223. [Google Scholar]