Abstract
Ten fragments of higher eucaryotic DNA were tested for upstream activation sequence activity in Saccharomyces cerevisiae by inserting them upstream of a CYC1::lacZ promoter lacking an upstream activation sequence. Fragments containing the 21-base-pair repeat region, the enhancer of simian virus 40 or both strongly stimulated beta-galactosidase synthesis, and three fragments from the polyomavirus enhancer region stimulated moderate levels. Three of the four controls of random DNA sequences failed to stimulate significant levels, and the fourth stimulated moderate levels. The stimulation in all cases was independent of the orientation of the inserted fragment. Two series of clones were examined in which between one and six tandemly arranged copies of a fragment were inserted into the XhoI site of the vector. Very interestingly, we detected an apparent exponential relationship between the number of copies of a fragment and the amount of beta-galactosidase produced. Southern analysis showed that increases in enzyme activity were not a result of increased plasmid copy number. Rather, quantitative S1 nuclease analysis demonstrated that the increases were correlated with steady-state levels of lacZ-specific mRNA. We suggest that there may be an evolutionary relationship between some transcriptional activation sequences in yeast cells and the higher eucaryotic regulatory elements that we tested.
Full text
PDF


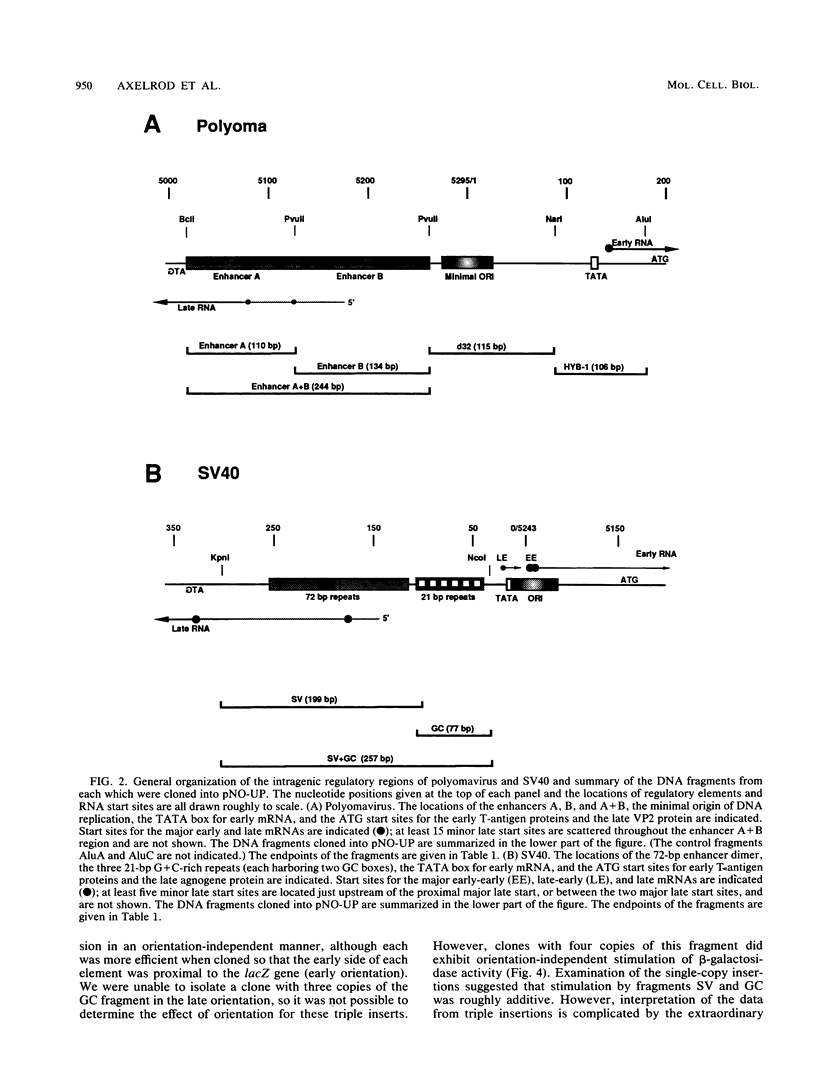







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arndt K., Fink G. R. GCN4 protein, a positive transcription factor in yeast, binds general control promoters at all 5' TGACTC 3' sequences. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8516–8520. doi: 10.1073/pnas.83.22.8516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayer D. E., Dynan W. S. Simian virus 40 major late promoter: a novel tripartite structure that includes intragenic sequences. Mol Cell Biol. 1988 May;8(5):2021–2033. doi: 10.1128/mcb.8.5.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji J., Rusconi S., Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981 Dec;27(2 Pt 1):299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- Benoist C., Chambon P. In vivo sequence requirements of the SV40 early promotor region. Nature. 1981 Mar 26;290(5804):304–310. doi: 10.1038/290304a0. [DOI] [PubMed] [Google Scholar]
- Benoist C., O'Hare K., Breathnach R., Chambon P. The ovalbumin gene-sequence of putative control regions. Nucleic Acids Res. 1980 Jan 11;8(1):127–142. doi: 10.1093/nar/8.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourachot B., Yaniv M., Herbomel P. Control elements situated downstream of the major transcriptional start site are sufficient for highly efficient polyomavirus late transcription. J Virol. 1989 Jun;63(6):2567–2577. doi: 10.1128/jvi.63.6.2567-2577.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady J., Radonovich M., Thoren M., Das G., Salzman N. P. Simian virus 40 major late promoter: an upstream DNA sequence required for efficient in vitro transcription. Mol Cell Biol. 1984 Jan;4(1):133–141. doi: 10.1128/mcb.4.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bram R. J., Kornberg R. D. Specific protein binding to far upstream activating sequences in polymerase II promoters. Proc Natl Acad Sci U S A. 1985 Jan;82(1):43–47. doi: 10.1073/pnas.82.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandl C. J., Struhl K. Yeast GCN4 transcriptional activator protein interacts with RNA polymerase II in vitro. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2652–2656. doi: 10.1073/pnas.86.8.2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne B. J., Davis M. S., Yamaguchi J., Bergsma D. J., Subramanian K. N. Definition of the simian virus 40 early promoter region and demonstration of a host range bias in the enhancement effect of the simian virus 40 72-base-pair repeat. Proc Natl Acad Sci U S A. 1983 Feb;80(3):721–725. doi: 10.1073/pnas.80.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhnlein E., Chowdhury K., Gruss P. Functional analysis of the regulatory region of polyoma mutant F9-1 DNA. Nucleic Acids Res. 1985 Jul 11;13(13):4789–4809. doi: 10.1093/nar/13.13.4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhnlein E., Gruss P. Interaction of distinct nuclear proteins with sequences controlling the expression of polyomavirus early genes. Mol Cell Biol. 1986 May;6(5):1401–1411. doi: 10.1128/mcb.6.5.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill K. B., Carmichael G. G. Deletion analysis of the polyomavirus late promoter: evidence for both positive and negative elements in the absence of early proteins. J Virol. 1989 Sep;63(9):3634–3642. doi: 10.1128/jvi.63.9.3634-3642.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Struhl K. Yeast upstream activator protein GCN4 can stimulate transcription when its binding site replaces the TATA element. EMBO J. 1989 Jan;8(1):261–268. doi: 10.1002/j.1460-2075.1989.tb03372.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clare J., Farabaugh P. Nucleotide sequence of a yeast Ty element: evidence for an unusual mechanism of gene expression. Proc Natl Acad Sci U S A. 1985 May;82(9):2829–2833. doi: 10.1073/pnas.82.9.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynan W. S. Modularity in promoters and enhancers. Cell. 1989 Jul 14;58(1):1–4. doi: 10.1016/0092-8674(89)90393-0. [DOI] [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. Control of eukaryotic messenger RNA synthesis by sequence-specific DNA-binding proteins. 1985 Aug 29-Sep 4Nature. 316(6031):774–778. doi: 10.1038/316774a0. [DOI] [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. Isolation of transcription factors that discriminate between different promoters recognized by RNA polymerase II. Cell. 1983 Mar;32(3):669–680. doi: 10.1016/0092-8674(83)90053-3. [DOI] [PubMed] [Google Scholar]
- Everett R. D., Baty D., Chambon P. The repeated GC-rich motifs upstream from the TATA box are important elements of the SV40 early promoter. Nucleic Acids Res. 1983 Apr 25;11(8):2447–2464. doi: 10.1093/nar/11.8.2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faye G., Leung D. W., Tatchell K., Hall B. D., Smith M. Deletion mapping of sequences essential for in vivo transcription of the iso-1-cytochrome c gene. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2258–2262. doi: 10.1073/pnas.78.4.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell A. J., Cowie A., Legon S., Kamen R. Multiple 5' terminal cap structures in late polyoma virus RNA. Cell. 1979 Feb;16(2):357–371. doi: 10.1016/0092-8674(79)90012-6. [DOI] [PubMed] [Google Scholar]
- Fromental C., Kanno M., Nomiyama H., Chambon P. Cooperativity and hierarchical levels of functional organization in the SV40 enhancer. Cell. 1988 Sep 23;54(7):943–953. doi: 10.1016/0092-8674(88)90109-2. [DOI] [PubMed] [Google Scholar]
- Fromm M., Berg P. Simian virus 40 early- and late-region promoter functions are enhanced by the 72-base-pair repeat inserted at distant locations and inverted orientations. Mol Cell Biol. 1983 Jun;3(6):991–999. doi: 10.1128/mcb.3.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura F. K. Nuclear activity from F9 embryonal carcinoma cells binding specifically to the enhancers of wild-type polyoma virus and PyEC mutant DNAs. Nucleic Acids Res. 1986 Apr 11;14(7):2845–2861. doi: 10.1093/nar/14.7.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P. K., Lebowitz P. Simian virus 40 early mRNA's contain multiple 5' termini upstream and downstream from a Hogness-Goldberg sequence; a shift in 5' termini during the lytic cycle is mediated by large T antigen. J Virol. 1981 Oct;40(1):224–240. doi: 10.1128/jvi.40.1.224-240.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P. K., Reddy V. B., Swinscoe J., Lebowitz P., Weissman S. M. Heterogeneity and 5'-terminal structures of the late RNAs of simian virus 40. J Mol Biol. 1978 Dec 25;126(4):813–846. doi: 10.1016/0022-2836(78)90022-0. [DOI] [PubMed] [Google Scholar]
- Giniger E., Ptashne M. Cooperative DNA binding of the yeast transcriptional activator GAL4. Proc Natl Acad Sci U S A. 1988 Jan;85(2):382–386. doi: 10.1073/pnas.85.2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giniger E., Varnum S. M., Ptashne M. Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell. 1985 Apr;40(4):767–774. doi: 10.1016/0092-8674(85)90336-8. [DOI] [PubMed] [Google Scholar]
- Gong S. S., Subramanian K. N. Functional anatomy of the simian virus 40 late promoter. Virology. 1988 Apr;163(2):481–493. doi: 10.1016/0042-6822(88)90289-9. [DOI] [PubMed] [Google Scholar]
- Gruss P. Magic enhancers? DNA. 1984;3(1):1–5. doi: 10.1089/dna.1.1984.3.1. [DOI] [PubMed] [Google Scholar]
- Guarente L., Hoar E. Upstream activation sites of the CYC1 gene of Saccharomyces cerevisiae are active when inverted but not when placed downstream of the "TATA box". Proc Natl Acad Sci U S A. 1984 Dec;81(24):7860–7864. doi: 10.1073/pnas.81.24.7860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L., Mason T. Heme regulates transcription of the CYC1 gene of S. cerevisiae via an upstream activation site. Cell. 1983 Apr;32(4):1279–1286. doi: 10.1016/0092-8674(83)90309-4. [DOI] [PubMed] [Google Scholar]
- Guarente L. Yeast promoters: positive and negative elements. Cell. 1984 Apr;36(4):799–800. doi: 10.1016/0092-8674(84)90028-x. [DOI] [PubMed] [Google Scholar]
- Guarente L., Yocum R. R., Gifford P. A GAL10-CYC1 hybrid yeast promoter identifies the GAL4 regulatory region as an upstream site. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7410–7414. doi: 10.1073/pnas.79.23.7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen U., Sharp P. A. Sequences controlling in vitro transcription of SV40 promoters. EMBO J. 1983;2(12):2293–2303. doi: 10.1002/j.1460-2075.1983.tb01737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harshman K. D., Moye-Rowley W. S., Parker C. S. Transcriptional activation by the SV40 AP-1 recognition element in yeast is mediated by a factor similar to AP-1 that is distinct from GCN4. Cell. 1988 Apr 22;53(2):321–330. doi: 10.1016/0092-8674(88)90393-5. [DOI] [PubMed] [Google Scholar]
- Hartzell S. W., Byrne B. J., Subramanian K. N. Mapping of the late promoter of simian virus 40. Proc Natl Acad Sci U S A. 1984 Jan;81(1):23–27. doi: 10.1073/pnas.81.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearing P., Shenk T. The adenovirus type 5 E1A transcriptional control region contains a duplicated enhancer element. Cell. 1983 Jul;33(3):695–703. doi: 10.1016/0092-8674(83)90012-0. [DOI] [PubMed] [Google Scholar]
- Herbomel P., Bourachot B., Yaniv M. Two distinct enhancers with different cell specificities coexist in the regulatory region of polyoma. Cell. 1984 Dec;39(3 Pt 2):653–662. doi: 10.1016/0092-8674(84)90472-0. [DOI] [PubMed] [Google Scholar]
- Herr W., Clarke J. The SV40 enhancer is composed of multiple functional elements that can compensate for one another. Cell. 1986 May 9;45(3):461–470. doi: 10.1016/0092-8674(86)90332-6. [DOI] [PubMed] [Google Scholar]
- Hill D. E., Hope I. A., Macke J. P., Struhl K. Saturation mutagenesis of the yeast his3 regulatory site: requirements for transcriptional induction and for binding by GCN4 activator protein. Science. 1986 Oct 24;234(4775):451–457. doi: 10.1126/science.3532321. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. G., Lucchini G., Fink G. R. A synthetic HIS4 regulatory element confers general amino acid control on the cytochrome c gene (CYC1) of yeast. Proc Natl Acad Sci U S A. 1985 Jan;82(2):498–502. doi: 10.1073/pnas.82.2.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman C. S., Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57(2-3):267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. GCN4 protein, synthesized in vitro, binds HIS3 regulatory sequences: implications for general control of amino acid biosynthetic genes in yeast. Cell. 1985 Nov;43(1):177–188. doi: 10.1016/0092-8674(85)90022-4. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M., Dover J. Mutational analysis of the GAL4-encoded transcriptional activator protein of Saccharomyces cerevisiae. Genetics. 1988 Sep;120(1):63–74. doi: 10.1093/genetics/120.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakidani H., Ptashne M. GAL4 activates gene expression in mammalian cells. Cell. 1988 Jan 29;52(2):161–167. doi: 10.1016/0092-8674(88)90504-1. [DOI] [PubMed] [Google Scholar]
- Kamen R., Jat P., Treisman R., Favaloro J., Folk W. R. 5' termini of polyoma virus early region transcripts synthesized in vivo by wild-type virus and viable deletion mutants. J Mol Biol. 1982 Aug 5;159(2):189–224. doi: 10.1016/0022-2836(82)90493-4. [DOI] [PubMed] [Google Scholar]
- Kern F. G., Basilico C. Transcription from the polyoma late promoter in cells stably transformed by chimeric plasmids. Mol Cell Biol. 1985 Apr;5(4):797–807. doi: 10.1128/mcb.5.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern F. G., Dailey L., Basilico C. Common regulatory elements control gene expression from polyoma early and late promoters in cells transformed by chimeric plasmids. Mol Cell Biol. 1985 Aug;5(8):2070–2079. doi: 10.1128/mcb.5.8.2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury G., Gruss P. Enhancer elements. Cell. 1983 Jun;33(2):313–314. doi: 10.1016/0092-8674(83)90410-5. [DOI] [PubMed] [Google Scholar]
- Lee W., Haslinger A., Karin M., Tjian R. Activation of transcription by two factors that bind promoter and enhancer sequences of the human metallothionein gene and SV40. Nature. 1987 Jan 22;325(6102):368–372. doi: 10.1038/325368a0. [DOI] [PubMed] [Google Scholar]
- Martin M. E., Piette J., Yaniv M., Tang W. J., Folk W. R. Activation of the polyomavirus enhancer by a murine activator protein 1 (AP1) homolog and two contiguous proteins. Proc Natl Acad Sci U S A. 1988 Aug;85(16):5839–5843. doi: 10.1073/pnas.85.16.5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau P., Hen R., Wasylyk B., Everett R., Gaub M. P., Chambon P. The SV40 72 base repair repeat has a striking effect on gene expression both in SV40 and other chimeric recombinants. Nucleic Acids Res. 1981 Nov 25;9(22):6047–6068. doi: 10.1093/nar/9.22.6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller W. J., Dufort D., Hassell J. A. Multiple subelements within the polyomavirus enhancer function synergistically to activate DNA replication. Mol Cell Biol. 1988 Nov;8(11):5000–5015. doi: 10.1128/mcb.8.11.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller W. J., Mueller C. R., Mes A. M., Hassell J. A. Polyomavirus origin for DNA replication comprises multiple genetic elements. J Virol. 1983 Sep;47(3):586–599. doi: 10.1128/jvi.47.3.586-599.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. A., Tatchell K., Hall B. D., Astell C., Smith M. Physical analysis of mating-type loci in Saccharomyces cerevisiae. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 2):961–981. doi: 10.1101/sqb.1981.045.01.113. [DOI] [PubMed] [Google Scholar]
- Ondek B., Gloss L., Herr W. The SV40 enhancer contains two distinct levels of organization. Nature. 1988 May 5;333(6168):40–45. doi: 10.1038/333040a0. [DOI] [PubMed] [Google Scholar]
- Ostapchuk P., Diffley J. F., Bruder J. T., Stillman B., Levine A. J., Hearing P. Interaction of a nuclear factor with the polyomavirus enhancer region. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8550–8554. doi: 10.1073/pnas.83.22.8550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan T., Coleman J. E. Structure and function of the Zn(II) binding site within the DNA-binding domain of the GAL4 transcription factor. Proc Natl Acad Sci U S A. 1989 May;86(9):3145–3149. doi: 10.1073/pnas.86.9.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer K., Arcangioli B., Guarente L. Yeast HAP1 activator competes with the factor RC2 for binding to the upstream activation site UAS1 of the CYC1 gene. Cell. 1987 Apr 10;49(1):9–18. doi: 10.1016/0092-8674(87)90750-1. [DOI] [PubMed] [Google Scholar]
- Pfeifer K., Kim K. S., Kogan S., Guarente L. Functional dissection and sequence of yeast HAP1 activator. Cell. 1989 Jan 27;56(2):291–301. doi: 10.1016/0092-8674(89)90903-3. [DOI] [PubMed] [Google Scholar]
- Pfeifer K., Prezant T., Guarente L. Yeast HAP1 activator binds to two upstream activation sites of different sequence. Cell. 1987 Apr 10;49(1):19–27. doi: 10.1016/0092-8674(87)90751-3. [DOI] [PubMed] [Google Scholar]
- Piette J., Kryszke M. H., Yaniv M. Specific interaction of cellular factors with the B enhancer of polyoma virus. EMBO J. 1985 Oct;4(10):2675–2685. doi: 10.1002/j.1460-2075.1985.tb03987.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piette J., Yaniv M. Molecular analysis of the interaction between an enhancer binding factor and its DNA target. Nucleic Acids Res. 1986 Dec 22;14(24):9595–9611. doi: 10.1093/nar/14.24.9595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piette J., Yaniv M. Two different factors bind to the alpha-domain of the polyoma virus enhancer, one of which also interacts with the SV40 and c-fos enhancers. EMBO J. 1987 May;6(5):1331–1337. doi: 10.1002/j.1460-2075.1987.tb02372.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy V. B., Ghosh P. K., Lebowitz P., Piatak M., Weissman S. M. Simian virus 40 early mRNA's. I. Genomic localization of 3' and 5' termini and two major splices in mRNA from transformed and lytically infected cells. J Virol. 1979 Apr;30(1):279–296. doi: 10.1128/jvi.30.1.279-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassone-Corsi P., Wildeman A., Chambon P. A trans-acting factor is responsible for the simian virus 40 enhancer activity in vitro. Nature. 1985 Feb 7;313(6002):458–463. doi: 10.1038/313458a0. [DOI] [PubMed] [Google Scholar]
- Schirm S., Jiricny J., Schaffner W. The SV40 enhancer can be dissected into multiple segments, each with a different cell type specificity. Genes Dev. 1987 Mar;1(1):65–74. doi: 10.1101/gad.1.1.65. [DOI] [PubMed] [Google Scholar]
- Schöler H. R., Gruss P. Specific interaction between enhancer-containing molecules and cellular components. Cell. 1984 Feb;36(2):403–411. doi: 10.1016/0092-8674(84)90233-2. [DOI] [PubMed] [Google Scholar]
- Struhl K., Brandl C. J., Chen W., Harbury P. A., Hope I. A., Mahadevan S. Transcriptional activation by yeast GCN4, a functional homolog to the jun oncoprotein. Cold Spring Harb Symp Quant Biol. 1988;53(Pt 2):701–709. doi: 10.1101/sqb.1988.053.01.080. [DOI] [PubMed] [Google Scholar]
- Struhl K. Genetic properties and chromatin structure of the yeast gal regulatory element: an enhancer-like sequence. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7865–7869. doi: 10.1073/pnas.81.24.7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Promoters, activator proteins, and the mechanism of transcriptional initiation in yeast. Cell. 1987 May 8;49(3):295–297. doi: 10.1016/0092-8674(87)90277-7. [DOI] [PubMed] [Google Scholar]
- Tyndall C., La Mantia G., Thacker C. M., Favaloro J., Kamen R. A region of the polyoma virus genome between the replication origin and late protein coding sequences is required in cis for both early gene expression and viral DNA replication. Nucleic Acids Res. 1981 Dec 11;9(23):6231–6250. doi: 10.1093/nar/9.23.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman G. M., Lupton S., Kamen R. Polyomavirus enhancer contains multiple redundant sequence elements that activate both DNA replication and gene expression. Mol Cell Biol. 1985 Apr;5(4):649–658. doi: 10.1128/mcb.5.4.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigneron M., Barrera-Saldana H. A., Baty D., Everett R. E., Chambon P. Effect of the 21-bp repeat upstream element on in vitro transcription from the early and late SV40 promoters. EMBO J. 1984 Oct;3(10):2373–2382. doi: 10.1002/j.1460-2075.1984.tb02142.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiher H., König M., Gruss P. Multiple point mutations affecting the simian virus 40 enhancer. Science. 1983 Feb 11;219(4585):626–631. doi: 10.1126/science.6297005. [DOI] [PubMed] [Google Scholar]
- Wildeman A. G., Zenke M., Schatz C., Wintzerith M., Grundström T., Matthes H., Takahashi K., Chambon P. Specific protein binding to the simian virus 40 enhancer in vitro. Mol Cell Biol. 1986 Jun;6(6):2098–2105. doi: 10.1128/mcb.6.6.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenke M., Grundström T., Matthes H., Wintzerith M., Schatz C., Wildeman A., Chambon P. Multiple sequence motifs are involved in SV40 enhancer function. EMBO J. 1986 Feb;5(2):387–397. doi: 10.1002/j.1460-2075.1986.tb04224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Villiers J., Olson L., Tyndall C., Schaffner W. Transcriptional 'enhancers' from SV40 and polyoma virus show a cell type preference. Nucleic Acids Res. 1982 Dec 20;10(24):7965–7976. doi: 10.1093/nar/10.24.7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Villiers J., Schaffner W. A small segment of polyoma virus DNA enhances the expression of a cloned beta-globin gene over a distance of 1400 base pairs. Nucleic Acids Res. 1981 Dec 11;9(23):6251–6264. doi: 10.1093/nar/9.23.6251. [DOI] [PMC free article] [PubMed] [Google Scholar]





