SUMMARY
The effects of genomic changes in hepatitis B virus (HBV) on the occurrence of hepatocellular carcinoma (HCC) are still unclear, especially in relation to the genotype of HBV. In this study, we examined the effects of genomic changes in HBV of genotype C2 on the development of HCC. A total of 318 patients with HBV-associated HCC and 234 patients with chronic hepatitis B (CHB) were studied. All of HCC cases were diagnosed histologically and treated with surgical resection. The whole of the X, S, basal core promoter (BCP) and precore regions of the viral genome from sera or liver tissues were sequenced. All subjects had HBV of genotype C2. The prevalence of the T1653 mutation in the X region and the A1896 mutation in the precore region of HBV was significantly higher in the HCC group than in the control CHB group (22% vs 11%, P = 0.003; 50% vs 23%, P < 0.001, respectively). Moreover, the T1762/A1764 mutations in the BCP region in combination with either T1653 or A1896 were more common in the HCC compared with the CHB group (BCP+X1653: 18% vs 11%, P = 0.05; BCP+PC, 40% vs 15%, P < 0.001, respectively). In multivariate analysis, T1653 and A1896 were revealed to be independent risk factors for HCC development. G1896A in the precore region and C1653T mutation in the X region of genotype C2 HBV are important risk factors for HCC development. Also, the A1762T/G1764A double mutation may act in synergy with C1653T to increase the risk of HCC in patients chronically infected with HBV genotype C2.
Keywords: genomic mutation, genotype C2, hepatitis B virus, hepatocellular carcinoma
INTRODUCTION
More than 400 million people in the world have been infected chronically with hepatitis B virus (HBV) [1]. Chronic HBV infection is a major cause of hepatocellular carcinoma (HCC), and 50% to 80% of HCC cases world-wide are attributable to HBV [2, 3]. Accumulated evidence indicates that genotype and specific genetic mutations in the HBV genome are representative factors contributing to development of HCC [4–6].
According to phylogenetic analyses, HBV can be classified into eight genotypes, A to H [7–9]. Genotype B and C are the two main genotypes prevailing in Asia, a region contributing around 75% of the world’s population of chronic hepatitis B (CHB) [10]. It is well known that genotype C, especially genotype C2 has a high risk of development of HCC [11]. Korea has an extraordinarily high prevalence of genotype C HBV patients. Various studies have demonstrated that 95– 100% of HBV in Korea is genotype C [12, 13].
Genotypes may be associated with certain genomic changes that also serve as risk factors for HCC development in patients with chronic hepatitis B. Each HBV genotype displays unique patterns of mutations at different regions of the genome. It has been demonstrated in several studies that the HBV genotype C is associated with mutations in the core promoter region and these particular mutations have been shown to be independent risk factors for HCC development [10].
One of the most well known mutations in the HBV genome is the basal core promoter (BCP) double mutation that consists of the nucleotide substitution of A to T at position 1762 and of G to A at 1764 (A1762T/G1764A) in the core promoter region. This double mutation has been associated with higher occurrence of HCC perhaps because of its relationship with HBeAg seroconversion and persistent viral replication [4, 10, 14]. Another common mutation is the G to A transition at nucleotide 1896 (G1896A) in the precore (PC) region. This mutation terminates the translation of HBeAg by creating a premature stop codon at codon 28 and is detected frequently in HBeAg-negative asymptomatic carriers [15, 16]. But, the exact role of this mutation is still unknown as the mutation has been demonstrated to cause more severe liver disease yet. Several studies showed that two common mutations in the region encoding HBx, a nonstructural regulatory protein of HBV, include a nucleotide substitution from C to T at position 1653 (C1653T) of the enhancer II region and a substitution from T to V (C/A/G) at position 1753 (T1753V) of the core promoter region are associated with HCC development [6, 17]. Recent studies have also demonstrated the importance of HBV with deletions in the pre-S region in severe liver disease such as HCC. The pre-S deletion itself or the combination of the pre-S deletion and the BCP double mutations were significantly associated with the development of HCC [18, 19].
While the effects of individual HBV genomic mutations on HCC development have been extensively studied, the role of combined mutations on HCC occurrence has rarely been addressed. Therefore, we aimed to investigate the individual and synergistic roles of mutations in the HBV genome in the X, S, core promoter and precore regions in patients with HBV-associated HCC and CHB patients without HCC.
PATIENTS AND METHODS
Subjects
The study involved 318 HBV-related HCC and 234 CHB patients without HCC as controls. The HBV-related HCC cases underwent surgical treatment such as hepatic resection or liver transplantation and diagnosed histologically at the Asan Medical Center from 2005 to 2009. A total of 234 patients with CHB served as controls who were diagnosed between 1991 and 1998. There was no patient who developed HCC during the follow-up period of a median 105 months (range, 1–237). Patients were regularly monitored by serum biochemistry, serum alpha-fetoprotein (AFP) levels and imaging study such as ultrasonography or CT scan at 3–6 month intervals. This study was approved by the Institutional Review Board of the Asan Medical Center, Seoul, Korea.
DNA extraction from samples
Deoxyribonucleic acid (DNA) was extracted from serum or liver tissue samples using the QIAmp DNA extraction kit (Qiagen K.K., Tokyo, Japan). The serum samples were treated with Proteinase K. The DNA was then isolated by phenol and chloroform extraction followed by ethanol precipitation. The extracted DNA was dissolved in 50 µL of TE buffer.
Genotyping of HBV
The specific HBV genotypes were identified using the polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP) of the surface region of the HBV genome. Two fragments of the HBV genome between nucleotide position 2823 and 2845, and 61 and 80 were amplified using PCR and the products were treated with restriction enzymes, followed by 3% agarose gel electrophoresis. The six main genotypes (A-F) were identified by the restriction patterns of the DNA fragments.
Amplification and direct sequencing of genes to detect HBV mutations
The sequence was analysed by amplifying the entire X, core promoter, precore/core and S regions of the HBV genome. Nested PCR was performed for the amplification of these genes. For the first round, 25 µL of the reaction mixture containing 2 µL of the DNA sample, 1 X PCR buffer, 0.1 mM of each dNTP, 0.5 µmol of each outer primer and 1 U of Taq DNA polymerase was amplified in a thermal cycler in 35 cycles. Each cycle included a denaturation at 95° C for 30 s, primer annealing at varying degrees (52° C for X, 55° C for precore/core, 50° C and 57° C for pre-S) for 30 s and extension at 72° C for 45 s with an additional extension step at 72°C for 7 min. The second round of PCR required a reamplification of the PCR product for another 35 cycles with 0.5 µmol of each inner primer (Table 1).
Table 1.
Primers used for genotyping and amplification
| Region | Name | Sequence | Position | Tm |
|---|---|---|---|---|
| Genotyping | HBG1 | TCACCATATTCTTGGGAACAAGA | 2823–2845 | 53 |
| Genotyping | HBG2 | TTCCGGAACTGGAGCCACCA | 61–80 | 53 |
| preC/C_X | HB3 | GATCCATACTGCGGAACTCC | 1265–1284 | 52 |
| preC/C_X | HB9R | CGAGATTGAGATCTTCTGCG | 2425–2444 | 52 |
| preC/C_nested | HB5 | TGTGCTGCCAACTGGATCCT | 1389–1408 | 55 |
| preC/C_nested | HB8R | CTGACTACTAATTCCCTGGATGCTGGGTCT | 2133–2162 | 55 |
| preS | HB55 | GGGTCACCATATTCTTGGG | 2821–2830 | 50 |
| preS | HB42R | GACCCACAATTCTTTGACAT | 989–1008 | 50 |
| preS_nested | HB56 | TTCTTGGGAACAAGAGCTAC | 2833–2852 | 57 |
| preS_nested | HB58R | TTGAGAGAAGTCCACCACGA | 255–275 | 57 |
| preC/C_seq | HB15 | GTCTGCCGTTCCAGCCGACC | 1510–1520 | |
| preS_seq | HB57 | CTTGGGAACAAGAGCTACAGC | 2835–2855 |
Tm, melting temperature.
The PCR products were sequenced both in a forward and reverse direction using the inner primers of each region and the Taq DyeDeoxy Terminator Cycle Sequencing Kit (ABI; Applied Biosystems, Foster City, CA, USA).
Statistics
Continuous data were expressed as median and range, and categorical data were expressed as number and percentage. Statistical analyses were performed using a chi-square test and Fischer’s exact test for categorical variables and Student’s t-test or Mann–Whitney U-test for continuous variables where appropriate. Multivariate analysis by logistic regression model was performed to assess the influence of each factor on the risk of HCC development. Significant associations identified in the multivariate analysis are presented as relative risk (95% confidence interval). A P value < 0.05 was considered as significant. All statistical procedures were performed using the SPSS 18.0 software (SPSS Inc., Chicago, IL, USA).
RESULTS
Baseline characteristics
The baseline characteristics for the 318 patients with HCC and 234 patients with CHB are summarized in Table 2. All of the 552 enrolled patients had genotype C2 HBV infection. The median age of patients with HCC was older than control patients (48 years vs 55 years, P < 0.001). Patients with HCC had higher serum AFP levels than patients with CHB (P = 0.018). On the other hand, the median serum ALT and albumin level were significantly lower in patients with HCC compared with control patients (P < 0.001). 29% of HCC and 84% of CHB patients were HBeAg positive, respectively (P < 0.001). However, serum HBV-DNA titre was not different between patients with HCC and controls.
Table 2.
Baseline characteristics
| HCC (n = 318) | CHB (n = 234) | |
|---|---|---|
| Age, years* | 55 (30–74)† | 48 (27–86)† |
| Gender, M/F, n (%) | 257/61 (81/19) | 195/39 (83/17) |
| ALT level, IU/L* | 37 (9–774)† | 150 (10–2170)† |
| Total bilirubin, mg/dL* | 1.0 (0.3–14.1) | 0.8 (0.3–15.4) |
| Albumin, g/dL* | 3.8 (1.3–4.7)† | 4.1 (2.2–5.2)† |
| Prothrombin time, %* | 89 (31–119)‡ | 88 (51–147)‡ |
| Platelet, × 103/mm3* | 159 (22–548)† | 176 (62–556)† |
| Serum AFP, ng/mL* | 51 (1–1.7 × 106)‡ | 5.6 (1–2150)‡ |
| Child-Pugh class, A/B/C, n (%) | 261/50/7 (82/16/2)† | 212/14/0 (94/6/0)† |
| HBeAg positivity, n (%) | 82 (29)† | 189 (84)† |
| Serum HBV-DNA, copies/mL* | 4.0 × 103 (ND – 2.3 × 109) | 4.2 × 106 (ND-1.1 × 109) |
HCC, hepatocellular carcinoma; CHB, chronic hepatitis B; ALT, alanine aminotransferase; AFP, α-fetoprotein; ND, not-detected.
Median (range).
P < 0.001.
P < 0.05.
Prevalence of HBV genomic mutations in patients with HCC and in patients with CHB
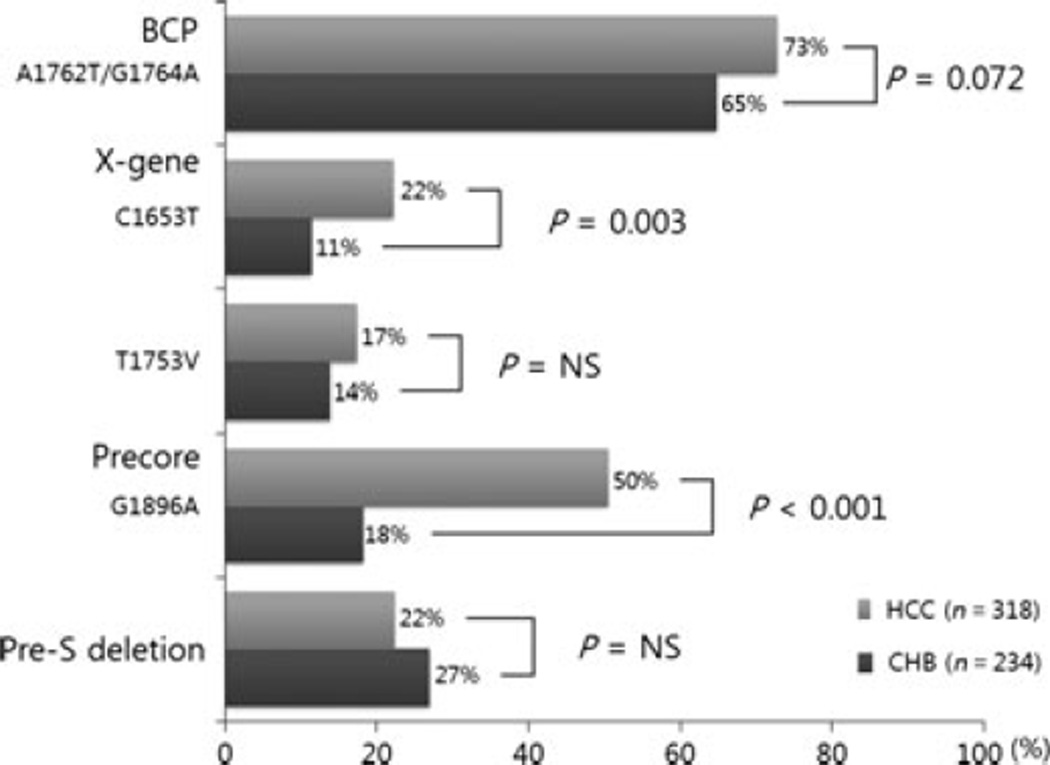
Figure 1 shows the prevalence of each HBV genomic mutation in the HCC and control groups. The BCP double mutation A1762T/G1764A was found in 73% (202/278) of patients with HCC and 65% (132/204) of patients with CHB, but this difference was not statistically significant (P = 0.072). The C1653T mutation in the X region was found in 22% (62/281) and 11% (24/210) of the HCC and CHB groups, respectively (P = 0.003). Another point mutation, the T1753V in the X region did not show significant difference in prevalence between patients with HCC and CHB [17% (49/284) vs 14% (29/210), P > 0.05].
Fig. 1.
The frequencies of genomic changes in HBV: comparison between patients with HCC and CH controls.
The G1896A mutation in the PC region was more frequently seen in patients with HCC than in patients with CHB [50% (142/282) vs 23% (48/210), P < 0.001]. The pre-S deletion was seen in 22% (64/287) and 27% (63/ 234) of the patients with HCC and patients with CHB, respectively (P > 0.05).
Synergistic effect of HBV mutations on HCC development
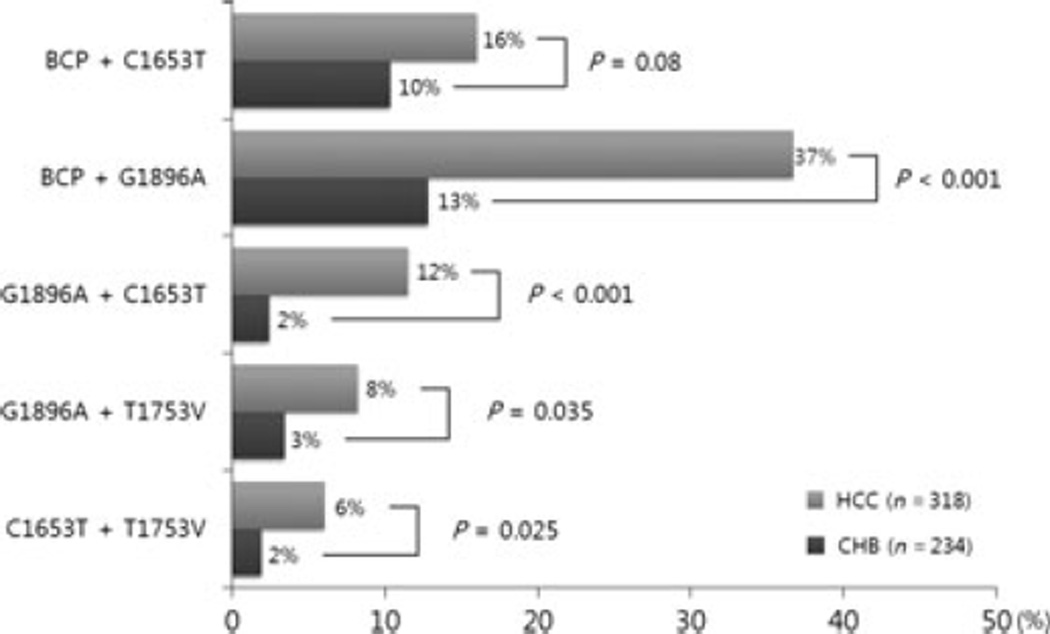
To evaluate any synergistic effect between the HBV mutations, the differences in frequencies of combined mutations between the HCC and CHB groups were assessed (Fig. 2). The combined BCP double mutation, A1762T/G1764A and the G1896A in the PC region, were more common in the HCC group compared with the CHB group [n = 102 (37%) vs n = 26 (13%), P < 0.001]. The combination of the BCP double mutation, A1762T/G1764A and C1653T in the X region, tended to occur more frequently in patients with HCC compared with the controls [n = 44 (16%) vs n = 21 (10%), P = 0.08]. However, combinations of A1762T/G1764A and T1753V or pre-S deletions did not differ significantly between the two groups.
Fig. 2.
The synergistic effect of genomic changes in HBV on the development of HCC.
The combination of G1896A and C1653T was observed in 12% of the HCC group and in just 2% of the control group. This difference was statistically significant (P < 0.001). Similarly, combination of the G1896A and T1753V in the X region was associated more frequently with HCC rather than CHB [n = 23 (8%) vs n = 7 (3%), P = 0.035]. In addition, coexistence of two X gene mutations such as C1653T and T1753V was more frequent in the HCC than in the CHB group [n = 17 (6%) vs n = 4 (2%), P = 0.025]. On the other hand, other combinations of mutations such as G1896A and pre-S deletion, pre-S deletion and C1653T, and pre-S deletion and T1753V did not show significant differences between groups.
Multivariate analysis for HBV mutations affecting the development of HCC
The presence of the G1896A in the PC region was independently associated with the development of HCC [odds ratio (OR), 2.725; 95% confidence interval (CI), 1.260– 5.894; P = 0.011]. The presence of C1653T in the X region (OR, 5.278; 95% CI, 1.182–23.575; P = 0.029) and older age (OR, 1.094; 95% CI, 1.069–1.120; P < 0.001) were also significant risk factors for HCC development. Moreover, the coexistence of A1762T/G1764A and C1653T was another risk factor for HCC, which had an OR of 5.396 (95% CI, 1.106–26.319, P = 0.037). However, the BCP double mutation or other combinations of mutations did not show a significant association with HCC development in multivariate analysis (Table 3).
Table 3.
Multivariate analysis of HBV mutations affecting HCC development
| Variables | OR | 95% CI | P value |
|---|---|---|---|
| Older age, years | 1.094 | 1.069–1.120 | <0.001 |
| Male | 1.306 | 0.752–2.267 | 0.344 |
| G1896A | 2.725 | 1.260–5.894 | 0.011 |
| C1653T | 5.278 | 1.182–23.575 | 0.029 |
| BCP | 1.502 | 0.821–2.746 | 0.186 |
| BCP + C1653T | 5.396 | 1.106–26.319 | 0.037 |
| BCP + G1896A | 1.326 | 0.503–3.495 | 0.569 |
| G1896A + C1653T | 1.505 | 0.400–5.658 | 0.545 |
| G1896A + T1753V | 1.220 | 0.426–3.495 | 0.712 |
| C1653T + T1753V | 2.669 | 0.701–10.163 | 0.150 |
OR, odds ratio; CI, confidence interval; BCP, basal core promoter A1762T/G1764A double mutation.
DISCUSSION
The importance of HBV genotype and genomic mutations has been emphasized in relation to the development of HCC. Genotype C of HBV is predominant in Asia including Korea and known to be associated with increased risk of HCC development [11, 20]. In this study, all the enrolled patients were infected with genotype C HBV, especially subtype C2, which is similar to previous reports; Korean patients are almost always infected with C2 genotype [12, 13]. Thus, some of the findings in this report could be applicable just to CHB patients infected with HBV genotype C2.
According to previous studies, the HBV genomic mutations affecting HCC occurrence include the BCP double mutation, the precore G1896A, the C1653T and T1753V in the HBx region and pre-S deletions. The present study is the largest which examined the role of HBV genomic mutations on the development of HCC. Our study did not show an association between the BCP A1762T/G1764A, T1753V or pre-S deletion and HCC development. However, the present study showed that the G1896A and C1653T were independent risk factors for HCC. Moreover, the synergistic effect of A1762T/G1764A and C1653T on HCC occurrence was also demonstrated in this study.
The G1896A mutation creates a stop codon that prevents translation of the PC protein and abolishes the production of HBeAg. However, these patients continue to synthesize HBV-DNA at sufficient levels to cause continual liver damage with progression to cirrhosis [18, 21, 22]. In the current study, the presence of PC mutants was detected more frequently in patients with HCC than in controls. This result would support the previous finding that the G1896A mutation plays a significant role in the progression of the chronic HBV infection to HCC [23].
It has been demonstrated that HBV genotype C is associated frequently with mutation in the core promoter region and these particular mutations have been shown to be independent risk factors for HCC development [24, 25]. Also, in the present study, the BCP double mutation, A1762T/G1764A, tended to occur more frequently in the HCC group compared with the CHB group although it was not statistically significant. The exact mechanism of this double mutation on hepatocellular carcinogenesis remains unknown. However, it has been speculated that the BCP mutations may enhance viral replication by increased transcription of the pregenomic RNA through the removal of the nuclear receptor–binding motif while creating a binding site for hepatocyte nuclear factor [26, 27]. The combination of these changes may lead to the suppression of the precore mRNA and an increase in pgRNA transcription resulting in an overall increase in viral replication [24].
In the HBx region, the presence of the C1653T mutation was significantly associated with the development of HCC in earlier studies [17, 28–30]. This mutation is located in box α, which is a strong activation element of both enhancer II and the core promoter; the α box element (nucleotides 1646–1668) individually stimulates promoter activity more than 100-fold [31]. The C1653T mutation converts the box α binding site for CCAAT/enhancer-binding protein EBP and related factors into the perfect palindromic sequence 1648-TCTTATATAAGA, which might enhance binding affinity and enhancer II/core promoter activity [31, 32]. Hence, the C1653T could influence HBeAg production and viral replication through BCP activity [17, 32]. In view of this synergistic effect, our study results showed that the combined BCP A1762T/G1764A and C1653T mutations occurred more frequently in the HCC than in the CHB group and suggest that the C1653T in addition to the BCP double mutation may be a promoter of HCC development in patients with chronic HBV infection.
In conclusion, the BCP A1762T/G1764A double mutation may act in synergy with the C1653T mutation to increase the risk of HCC in patients with genotype C2 HBV infection. Also, the C1653T and G1896A PC mutations are both independent risk factors for the development of HCC.
Abbreviations
- A1762T/G1764A
nucleotide substitution of A to T at nucleotide 1762 and of G to A at nucleotide 1764
- AFP
alpha-fetoprotein
- BCP
basal core promoter
- C1653T
C to T at nucleotide 1653
- CHB
chronic hepatitis B
- G1896A
G to A at nucleotide 1896
- HBeAg
hepatitis B e antigen
- HBV
hepatitis B virus
- HCC
hepatocellular carcinoma
- PC
precore
- T1753V
T to V (C/A/G) at nucleotide 1753
Footnotes
A part of this work was presented at the 46th annual meeting of the European Association for the Study of the Liver (EASL) in 2011 (The International Liver Congress 2011).
Financial Disclosure
None.
Conflicts of Interest
None.
REFERENCES
- 1.van Zonneveld M, Honkoop P, Hansen BE, et al. Long-term follow-up of alpha-interferon treatment of patients with chronic hepatitis B. Hepatology. 2004;39:804–810. doi: 10.1002/hep.20128. [DOI] [PubMed] [Google Scholar]
- 2.Liu S, Xie J, Yin J, et al. A matched case-control study of hepatitis B virus mutations in the preS and core promoter regions associated independently with hepatocellular carcinoma. J Med Virol. 2011;83:45–53. doi: 10.1002/jmv.21829. [DOI] [PubMed] [Google Scholar]
- 3.Nguyen VT, Law MG, Dore GJ. Hepatitis B-related hepatocellular carcinoma: epidemiological characteristics and disease burden. J Viral Hepat. 2009;16:453–463. doi: 10.1111/j.1365-2893.2009.01117.x. [DOI] [PubMed] [Google Scholar]
- 4.Kao JH, Chen PJ, Lai MY, Chen DS. Basal core promoter mutations of hepatitis B virus increase the risk of hepatocellular carcinoma in hepatitis B carriers. Gastroenterology. 2003;124:327–334. doi: 10.1053/gast.2003.50053. [DOI] [PubMed] [Google Scholar]
- 5.Miyakawa Y, Mizokami M. Classifying hepatitis B virus genotypes. Intervirology. 2003;46:329–338. doi: 10.1159/000074988. [DOI] [PubMed] [Google Scholar]
- 6.Asim M, Malik A, Sarma MP, et al. Hepatitis B virus BCP, Precore/core, X gene mutations/genotypes and the risk of hepatocellular carcinoma in India. J Med Virol. 2010;82:1115–1125. doi: 10.1002/jmv.21774. [DOI] [PubMed] [Google Scholar]
- 7.Arauz-Ruiz P, Norder H, Robertson BH, Magnius LO. Genotype H: a new Amerindian genotype of hepatitis B virus revealed in Central America. J Gen Virol. 2002;83:2059–2073. doi: 10.1099/0022-1317-83-8-2059. [DOI] [PubMed] [Google Scholar]
- 8.Norder H, Courouce AM, Magnius LO. Complete genomes, phylogenetic relatedness, and structural proteins of six strains of the hepatitis B virus, four of which represent two new genotypes. Virology. 1994;198:489–503. doi: 10.1006/viro.1994.1060. [DOI] [PubMed] [Google Scholar]
- 9.Stuyver L, De Gendt S, Van Geyt C, et al. A new genotype of hepatitis B virus: complete genome and phylogenetic relatedness. J Gen Virol. 2000;81:67–74. doi: 10.1099/0022-1317-81-1-67. [DOI] [PubMed] [Google Scholar]
- 10.Yuen MF, Tanaka Y, Shinkai N, et al. Risk for hepatocellular carcinoma with respect to hepatitis B virus genotypes B/C, specific mutations of enhancer II/core promoter/ precore regions and HBV DNA levels. Gut. 2008;57:98–102. doi: 10.1136/gut.2007.119859. [DOI] [PubMed] [Google Scholar]
- 11.Chan HL, Hui AY, Wong ML, et al. Genotype C hepatitis B virus infection is associated with an increased risk of hepatocellular carcinoma. Gut. 2004;53:1494–1498. doi: 10.1136/gut.2003.033324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song BC, Cui XJ, Kim H. Hepatitis B virus genotypes in Korea: an endemic area of hepatitis B virus infection. Intervirology. 2005;48:133–137. doi: 10.1159/000081740. [DOI] [PubMed] [Google Scholar]
- 13.Kim H, Jee YM, Song BC, et al. Analysis of hepatitis B virus quasispecies distribution in a Korean chronic patient based on the full genome sequences. J Med Virol. 2007;79:212–219. doi: 10.1002/jmv.20789. [DOI] [PubMed] [Google Scholar]
- 14.Liu CJ, Chen BF, Chen PJ, et al. Role of hepatitis B virus precore/core promoter mutations and serum viral load on noncirrhotic hepatocellular carcinoma: a case-control study. J Infect Dis. 2006;194:594–599. doi: 10.1086/505883. [DOI] [PubMed] [Google Scholar]
- 15.Okamoto H, Yotsumoto S, Akahane Y, et al. Hepatitis B viruses with precore region defects prevail in persistently infected hosts along with seroconversion to the antibody against e antigen. J Virol. 1990;64:1298–1303. doi: 10.1128/jvi.64.3.1298-1303.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Omata M, Ehata T, Yokosuka O, Hosoda K, Ohto M. Mutations in the precore region of hepatitis B virus DNA in patients with fulminant and severe hepatitis. N Engl J Med. 1991;324:1699–1704. doi: 10.1056/NEJM199106133242404. [DOI] [PubMed] [Google Scholar]
- 17.Shinkai N, Tanaka Y, Ito K, et al. Influence of hepatitis B virus X and core promoter mutations on hepatocellular carcinoma among patients infected with subgenotype C2. J Clin Microbiol. 2007;45:3191–3197. doi: 10.1128/JCM.00411-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen BF, Liu CJ, Jow GM, Chen PJ, Kao JH, Chen DS. High prevalence and mapping of pre-S deletion in hepatitis B virus carriers with progressive liver diseases. Gastroenterology. 2006;130:1153–1168. doi: 10.1053/j.gastro.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 19.Fang ZL, Sabin CA, Dong BQ, et al. Hepatitis B virus pre-S deletion mutations are a risk factor for hepatocellular carcinoma: a matched nested case-control study. J Gen Virol. 2008;89:2882–2890. doi: 10.1099/vir.0.2008/002824-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurbanov F, Tanaka Y, Mizokami M. Geographical and genetic diversity of the human hepatitis B virus. Hepatol Res. 2010;40:14–30. doi: 10.1111/j.1872-034X.2009.00601.x. [DOI] [PubMed] [Google Scholar]
- 21.Carman WF, Jacyna MR, Hadziyannis S, et al. Mutation preventing formation of hepatitis B e antigen in patients with chronic hepatitis B infection. Lancet. 1989;2:588–591. doi: 10.1016/s0140-6736(89)90713-7. [DOI] [PubMed] [Google Scholar]
- 22.Hunt CM, McGill JM, Allen MI, Condreay LD. Clinical relevance of hepatitis B viral mutations. Hepatology. 2000;31:1037–1044. doi: 10.1053/he.2000.6709. [DOI] [PubMed] [Google Scholar]
- 23.Tong MJ, Blatt LM, Kao JH, Cheng JT, Corey WG. Basal core promoter T1762/A1764 and precore A1896 gene mutations in hepatitis B surface antigen-positive hepatocellular carcinoma: a comparison with chronic carriers. Liver Int. 2007;27:1356–1363. doi: 10.1111/j.1478-3231.2007.01585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuen MF, Sablon E, Tanaka Y, et al. Epidemiological study of hepatitis B virus genotypes, core promoter and precore mutations of chronic hepatitis B infection in Hong Kong. J Hepatol. 2004;41:119–125. doi: 10.1016/j.jhep.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 25.Chu CM, Lin CC, Lin SM, Lin DY, Liaw YF. Viral load, genotypes, and mutants in hepatitis B virus-related hepatocellular carcinoma: special emphasis on patients with early hepatocellular carcinoma. Dig Dis Sci. 2012;57:232–238. doi: 10.1007/s10620-011-1844-2. [DOI] [PubMed] [Google Scholar]
- 26.Kidd AH, Kidd-Ljunggren K. A revised secondary structure model for the 3′-end of hepatitis B virus pregenomic RNA. Nucleic Acids Res. 1996;24:3295–3301. doi: 10.1093/nar/24.17.3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J, Buckwold VE, Hon MW, Ou JH. Mechanism of suppression of hepatitis B virus precore RNA transcription by a frequent double mutation. J Virol. 1999;73:1239–1244. doi: 10.1128/jvi.73.2.1239-1244.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim JK, Chang HY, Lee JM, et al. Specific mutations in the enhancer II/core promoter/precore regions of hepatitis B virus subgenotype C2 in Korean patients with hepatocellular carcinoma. J Med Virol. 2009;81:1002–1008. doi: 10.1002/jmv.21501. [DOI] [PubMed] [Google Scholar]
- 29.Kaneko M, Uchida T, Moriyama M, et al. Probable implication of mutations of the X open reading frame in the onset of fulminant hepatitis B. J Med Virol. 1995;47:204–208. doi: 10.1002/jmv.1890470304. [DOI] [PubMed] [Google Scholar]
- 30.Uchida T, Saitoh T, Shinzawa H. Mutations of the X region of hepatitis B virus and their clinical implications. Pathol Int. 1997;47:183–193. doi: 10.1111/j.1440-1827.1997.tb04479.x. [DOI] [PubMed] [Google Scholar]
- 31.Yuh CH, Chang YL, Ting LP. Transcriptional regulation of precore and pregenomic RNAs of hepatitis B virus. J Virol. 1992;66:4073–4084. doi: 10.1128/jvi.66.7.4073-4084.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ito K, Tanaka Y, Orito E, et al. T1653 mutation in the box alpha increases the risk of hepatocellular carcinoma in patients with chronic hepatitis B virus genotype C infection. Clin Infect Dis. 2006;42:1–7. doi: 10.1086/498522. [DOI] [PubMed] [Google Scholar]