Abstract
The molecular mechanisms regulating leukemia-initiating cell (LIC) function are of important clinical significance. We use chronic myelogenous leukemia (CML), as a model of LIC-dependent malignancy and identify the interaction between the ubiquitin ligase Fbw7 and its substrate c-Myc as a regulator of LIC homeostasis. Deletion of Fbw7 leads to c-Myc overexpression, p53-dependent LIC-specific apoptosis and the eventual inhibition of tumor progression. Decrease of either c-Myc protein levels or attenuation of the p53 response rescues LIC activity and disease progression. Further experiments showed that Fbw7 expression is required for survival and maintenance of human CML LIC. These studies identify a ubiquitin ligase:substrate pair regulating LIC activity, suggesting that targeting of the Fbw7:c-Myc axis is an attractive therapy target in refractory CML.
INTRODUCTION
Chronic myeloid leukemia (CML) was the first type of cancer for which a specific chromosomal abnormality was identified, the Philadelphia chromosome (Nowell and Hungerford, 1960). Subsequent studies identified that the translocation event occurred between t(9;22)(q34;q11) fused the breakpoint cluster region gene (BCR) with the Abelson kinase gene (ABL1) to produce the BCR-ABL oncogene (Bartram et al., 1983; Rowley, 1973). This Bcr-Abl fusion protein possesses constitutive tyrosine kinase activity resulting in development of myeloid leukemia through aberrant differentiation of hematopoietic stem cells (HSC) towards the myeloid lineage. CML is dependent on Bcr-Abl induced c-Myc expression (Sawyers et al., 1992). Clinically, CML progresses through at least three different phases: chronic phase (CP), late chronic/accelerated phase (AP) and blast crisis (BC).
Patients diagnosed with CML in the early chronic phase have been successfully treated with tyrosine kinase inhibitors (TKIs), such as imatinib, that inhibit the tyrosine kinase activity of Bcr-Abl and have a 5-year progression free survival rate of 89% (Druker et al., 2006). However, only a fraction of TKI-treated patients achieve long-term remission, suggesting that the compound is unable to target CML initiating cells (de Lavallade et al., 2008; Hochhaus et al., 2009). Indeed, the majority of the patients relapse upon cessation of treatment (Michor et al., 2005). Moreover, some patients, particularly the ones that present with advanced disease can develop resistance to Imatinib treatment (O'Hare et al., 2006). The mechanisms thought to drive resistance and disease relapse include the acquisition of mutations in the kinase domain of Bcr-Abl, amplification of BCR-ABL, and clonal evolution (Gorre et al., 2001; le Coutre et al., 2000; Shah et al., 2002).
An increasing body of work has suggested that disease relapse upon cessation of TKI therapy could be due to a rare population of leukemia initiating cells (LICs) that are resistant or refractory to treatment (Bhatia et al., 2003; Corbin et al., 2011; Jankowska et al., 2009). LICs are thought to possess properties similar to normal hematopoietic stem cells (HSC) such as self-renewal, quiescence and resistance to traditional chemotherapy (Bonnet and Dick, 1997; Huntly and Gilliland, 2005). Thus, the LIC subset might act as a reservoir contributing to relapse by passing BCR-ABL, on to its progeny. In different types of leukemia, evidence in support of the LIC determined that only a small fraction of acute myeloid leukemia cells from patients were able to recapitulate the disease when transplanted into immuno-compromised animals (Bonnet and Dick, 1997; Lapidot et al., 1994). Using similar assays, putative LIC populations were also identified in patients diagnosed with CP and BC CML (Jamieson et al., 2004; Sirard et al., 1996; Wang et al., 1998)
The development of disease animal models, which proved that expression of Bcr-Abl is indeed leukemogenic, provided an important tool to investigate the mechanisms involved in LIC maintenance (Daley et al., 1990; Heisterkamp et al., 1990; Pear et al., 1998). Over the years, Bcr-Abl has been shown to contribute to tumorigenesis through deregulation of molecular pathways that control HSC self-renewal and differentiation (Heidel et al., 2012; Zhao et al., 2007; Zhao et al., 2009). Moreover, transplantation studies in mouse models of Bcr-Abl-induced CP CML suggested that LIC activity is confined to Bcr-Abl-expressing Lineage (Lin)−Sca1+c-Kit+ (LSK) cells, which contains the HSC population (Neering et al., 2007).
Fbw7 is an E3 ubiquitin ligase and is a substrate recognition component of the Cullin-1/SCF complex that targets specific substrate proteins for poly-ubiquitination and degradation by the 26S proteasome. Fbw7 has been shown to regulate a number of oncoproteins such as c-Myc, Notch, and cyclinE (Gupta-Rossi et al., 2001; Koepp et al., 2001; Welcker et al., 2004; Yada et al., 2004). Moreover, we and others have shown that Fbw7 is essential for the maintenance of adult HSC quiescence (Matsuoka et al., 2008; Thompson et al., 2008). Indeed, deletion of Fbw7 in HSCs leads to c-Myc accumulation, aberrant cell cycle entry and eventual HSC exhaustion (Reavie et al., 2010). Here we explore the role of the Fbw7:c-Myc axis and relative abundance of c-Myc protein in the maintenance of CML LIC.
RESULTS
Fbw7 deletion is able to suppress initiation of Bcr-Abl-induced CML
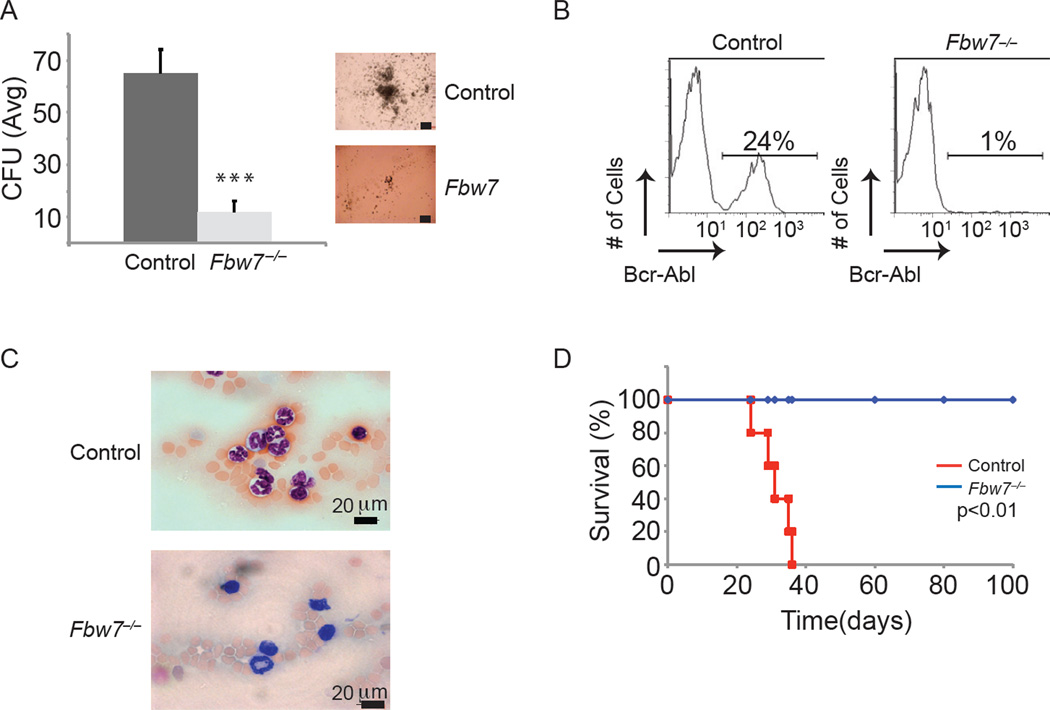
To address the role of Fbw7 in the self-renewal and differentiation of LIC we used a well-established animal model of Bcr-Abl-induced CP CML (Pear et al., 1998). In this model, Bcr-Abl expressing retroviruses are used to infect highly-purified hematopoietic stem and progenitor cells, LSK. Transduced LSKs are then transplanted into lethally irradiated recipients, which develop a CML-like disease that is characterized by the accumulation of Bcr-Abl+CD11b+Gr1+ cells in the peripheral blood (PB), splenomegaly and tissue infiltration. The pathology of the disease replicates chronic phase of CML due to less than 2% of the mononuclear population in the PB has blast morphology, a hallmark of disease progression, whereas the majority displays morphology consistent with mature myeloid cells. Mice succumb to disease starting at ~25–30 days post transplantation (Figure 1 and data not shown). To initially test the role of Fbw7 function in CML in vitro, we used a conditional Fbw7 allele (Vav1cre+;Fbw7f/f) that specifically targets deletion in hematopoietic cells, starting from the HSC subset during development, and purified LSK cells from 2 to 4 week old mice (before any significant alterations of the LSK compartment are evident) (Matsuoka et al., 2008; Stadtfeld and Graf, 2005; Thompson et al., 2007; Thompson et al., 2008). Fbw7−/− and Fbw7+/+ LSK cells were infected with Bcr-Abl retroviruses, sorted by flow cytometry for Bcr-Abl+, and plated on colony-forming unit (CFU) assays. Control (Bcr-Abl+;Fbw7+/+) LSK cells generated colonies at the first plating and were able to serially replate demonstrating extensive proliferation potential (Figure 1A and data not shown). Interestingly, Bcr-Abl+;Fbw7−/− LSK cells generated only a few small colonies at the first plating and were unable to re-plate, suggesting direct effects of Fbw7 deletion on survival of the cells. This was an unexpected finding, as WT (Bcr-Abl−) Fbw7−/− LSK cells are able to efficiently generate colonies at the first plating, and their colony forming ability is only progressively lost (Figure S1) suggesting distinct responses to Fbw7 deletion between physiological and leukemic LSK cells (Thompson et al., 2008). To address Fbw7 function in CML in vivo, LSKs were infected with Bcr-Abl-expressing retroviruses and transplanted into lethally irradiated recipient mice. Control Bcr-Abl expressing cells were able to initiate disease and progress to lethal CML (Figure 1B–D). On the other hand, Bcr-Abl-expressing Fbw7−/− LSK cells were unable to initiate disease and recipient animals did not developed CML (Figure 1B–D). These effects on the initiation of CML were not a consequence of Fbw7 deletion on HSC homing and engraftment suggesting direct role on maintenance of Bcr-Abl+ cells (Figure S1, data not shown). This data suggests that Fbw7 deletion inhibits Bcr-Abl induction of CML due to direct effects on Bcr-Abl+ LSK cell maintenance.
Figure 1. Fbw7 deletion suppresses initiation of Bcr-Abl-induced CML.
(A) Average CFU from VavCre+;Fbw7+/+ and VavCre+;Fbw7−/− cells infected with Bcr-Abl expressing retrovirus at the first plating. Image on right are representative colonies from control or Fbw7−/− Bcr-Abl+ LSKs. Scale bar 100 µm. (B–C) FACS analysis (B) and blood smears (C) of PB taken from host mice transplanted with VavCre+;Fbw7+/+ (Control) or VavCre+;Fbw7−/− Bcr-Abl+ LSK cells. D) Kaplan Meier survival curves of irradiated animals that were transplanted with VavCre+;Fbw7+/+ or VavCre+;Fbw7−/− Bcr-Abl+ LSKs. (n=5 for each genotype). Error bars indicate +/− SD. p<0.0001. See also Figure S1.
Fbw7 deletion is able to suppress Bcr-Abl-induced disease progression
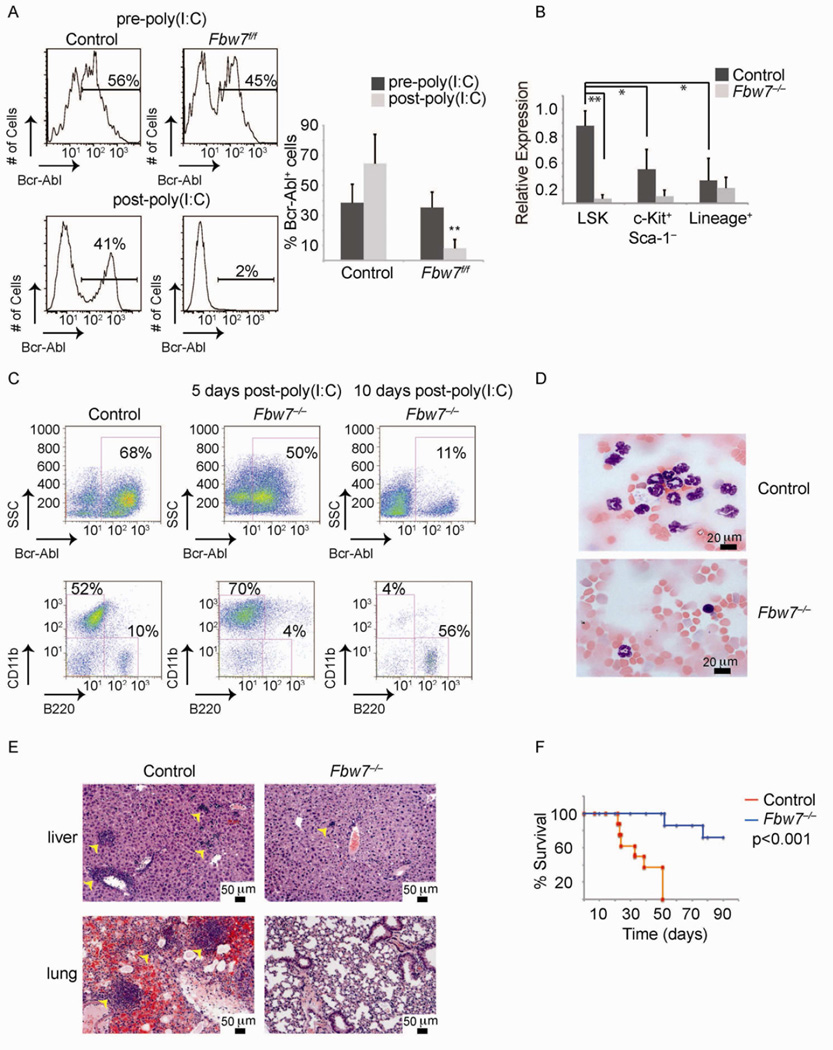
The above experiments address effects of Fbw7 on transformation but do not study its role during progression of CML in vivo. To experimentally address this question we took advantage of an inducible Fbw7 allele and crossed these mice to the Mx1cre strain, of which Crerecombinase is expressed as early as the HSC stage upon administration of poly(I:C) (Mx1cre+;Fbw7f/f) and allows for gene deletion after the onset of the disease. In these experiments CML was established by Bcr-Abl-expressing Mx1cre+;Fbw7f/f and littermate control LSK cells. Disease onset was verified by flow cytometry 7 days post-transplantation (Figure 2A). Fbw7 deletion was achieved by three poly(I:C) injections and confirmed by quantitative reverse transcriptase PCR (qRT-PCR) analysis (Figure 2B). Notably, tumor LSKs expressed the highest levels of Fbw7 when compared to more differentiated subsets (Figure 2B). Control recipients developed CML as characterized by the accumulation of Bcr-Abl+CD11b+Gr1+ cells in the bone marrow (BM) (Figure 2C), PB (Figure 2A and D) and peripheral organs such as the spleen, liver and lung (Figure 2E and not shown). In contrast, poly(I:C) -mediated deletion of Fbw7 led to a rapid reversal of CML progression (as judged by both Bcr-Abl+ and CD11b+ absolute cell numbers, Figure 2A and C) resulting in almost no infiltration of secondary tissues by leukemic cells (Figure 2E). More importantly, while all control mice succumbed to the disease by day 50 post-transplantation, the majority of recipient animals transplanted with Bcr-Abl+;Mx1cre+;Fbw7f/f cells and injected with poly(I:C) survived (Figure 2F). These studies demonstrated that Fbw7 deletion is also able to suppress further development of CML and lead to disease remission, suggesting effects on putative leukemia-initiating cells.
Figure 2. Fbw7 is essential for progression of established CML in vivo.
(A) PB from mice transplanted with MxCre+;Fbw7+/+ and MxCre+;Fbw7−/− Bcr-Abl infected LSK cells. The bar graph on right is a quantification of Bcr-Abl+ cells in the PB. (B) qRT-PCR analysis of Fbw7 expression in sorted populations from WT and Fbw7−/− CML 5 days after the post-poly(I:C) injection. (C) FACS analysis of the BM of animals transplanted with Bcr-Abl+ LSK cells. (D) Blood smears ~10 days post poly(I:C) injections. (E) Hematoxylin and eosin (H&E) staining of liver and lung. (F) Kaplan Meier survival curves of irradiated animals that were transplanted with Fbw7+/+ or Fbw7−/− Bcr-Abl+ LSKs. (n=9 for each genotype). Error bars indicate +/−SD. * p<0.01, ** p<0.001. See also Figure S2
Fbw7 deficient Bcr-Abl cells have no leukemia-initiating activity in vivo
To directly test the self-renewal capacity of the LIC fraction, we performed secondary transplantation experiments using whole spleen cells isolated ~10 days after poly(I:C)-treatment from Mx1cre+;Fbw7−/− and littermate controls. To ensure that identical numbers of Bcr-Abl+ LSKs were transplanted in both cohorts, we normalized the total number of spleen cells based on the frequency of Bcr-Abl+ LSK cells. In agreement with our previous findings, recipients of Fbw7−/− tumor cells did not develop CML (Figure S2). In contrast, control Bcr-Abl+ cells harbored LIC activity when transplanted into secondary recipients and transferred disease exhibiting the same hallmarks as the primary CML (Figure S2). These results strongly suggested that Fbw7 deletion specifically inhibits CML LIC activity.
Fbw7 deletion affects survival of CML-initiating cell populations
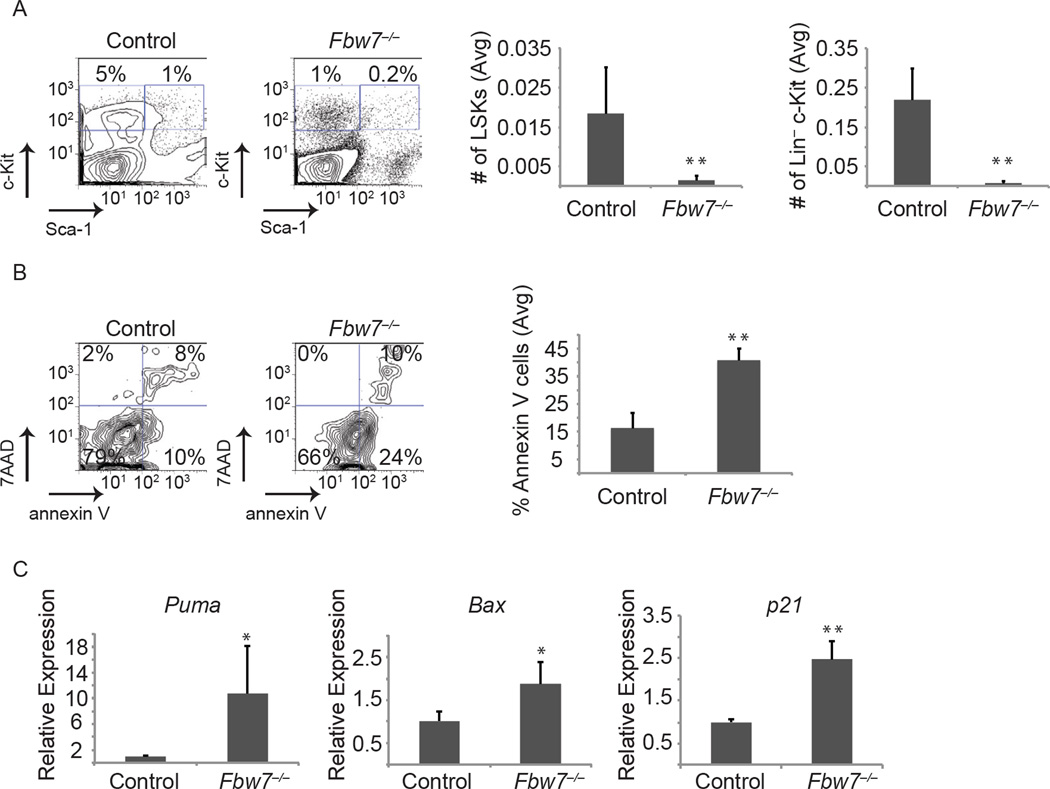
It was previously shown that the LIC activity in Bcr-Abl-induced CML is confined to the Lin−c-Kit+ and specifically the LSK subset of the Bcr-Abl-expressing tumor (Neering et al., 2007). To directly study putative effects of Fbw7 deletion in these subsets we studied both their relative representation and their absolute numbers in response to Fbw7 deletion (using the inducible Mx1cre+;Fbw7f/f in vivo model). Fbw7 deletion in established CML led to the rapid and significant loss of Bcr-Abl+ Lin−c-Kit+ and more specifically the Bcr-Abl+ LSK population (Figure 3A). Interestingly, at the same time points, we were able to detect more differentiated Bcr-Abl-expressing tumor cells suggesting that deletion of Fbw7 specifically targets immature LIC subsets (Figure 2C). The acute loss of Bcr-Abl+ LSK cells following Fbw7 deletion was significantly more rapid than what has been reported for WT LSK cells, which takes 3–4 months (Matsuoka et al., 2008; Thompson et al., 2008). QRT-PCR studies showed that CML LSK cells express slightly higher levels of Fbw7 mRNA than WT LSK but is not statistically significant (Figure S1D). To identify a putative mechanism to explain the impact of Fbw7 deletion on the Bcr-Abl+ LSK population we evaluated apoptosis and cell death using Annexin-V and 7AAD. As shown in Figure 3B, Fbw7 deletion led to a rapid and significant increase (5–8 fold) in the fraction of the Bcr-Abl+ LSK cells undergoing apoptosis suggesting direct induction of cell death in this stem and progenitor subset. We further evaluated p53 pathway target genes associated with cell survival by qRT-PCR and found Puma, Bax, p21 and Noxa up-regulated in Fbw7−/− tumor LSKs (Figure 3C and data not shown), suggesting that p53 pathway activation mediates the induced death of Fbw7 deficient Bcr-Abl+ LSK cells. These studies provide the biological mechanism explaining the loss of Bcr-Abl+ LICs and the suppression of disease progression in response to Fbw7 deletion.
Figure 3. Fbw7 deletion affects CML-initiating cell survival through the activation of the p53 pathway.
(A) FACS plots depicting the relative percentage of Bcr-Abl expressing stem and progenitor (LSK), and progenitors (Lin−c-Kit+Sca−) cells in the BM of MxCre+;Fbw7+/+ or MxCre+;Fbw7−/−mice. Bar graphs depict number of tumor stem and progenitor cells based on frequency of LSK in total number of BM cells. (B) FACS plots showing relative annexin V and 7-AAD positive cells in the Bcr-Abl+ LSK subset in the BM. Graph on right represents percent of annexin V+ cells in the Bcr-Abl+ LSK. (C) qRT-PCR analysis showing expression of the p53 target genes, Puma, Bax, and p21, in sorted control or Fbw7−/− LSKs from the tumor. Error bars indicate +/−SD. (n=4 for each genotype). * p<0.05, ** p<0.01.
c-Myc is the key substrate targeted by Fbw7 in CML-initiating cell populations
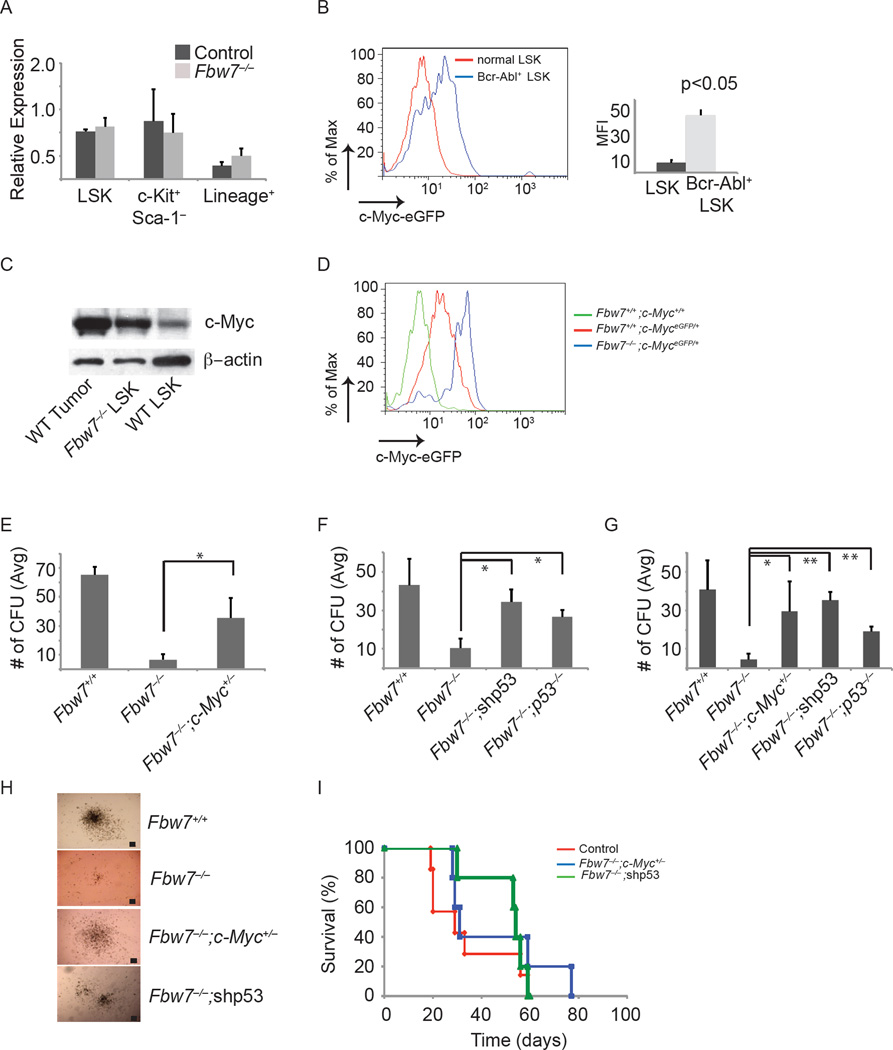
As we have previously shown that c-Myc is an Fbw7 substrate during early hematopoiesis (Reavie et al., 2010) and CML is dependent on c-Myc induced by Bcr-Abl (Sawyers et al., 1992),we investigated the possibility that “unphysiologically” high levels of the oncogenic c-Myc protein could cause the cell death observed upon loss of Fbw7 expression in CML. We further hypothesized that this was most likely through activation of the p53 pathway since p53 target genes were upregulated in Fbw7−/− Bcr-Abl+ LSKs (Figure 3C). As we have shown differential effects of Fbw7 deletion on wild-type and Bcr-Abl+ LSK function in vitro (Figures 1 and S1), we directly compared levels of c-Myc protein in these two subsets using a targeted c-Myc allele that expresses a c-Myc-eGFP fusion protein (c-MyceGFP), which has been shown to be a functional protein fusion and a faithful indicator of endogenous c-Myc protein levels (Huang et al., 2008). Despite the fact that c-Myc mRNA levels were unchanged between control and Fbw7 deficient Bcr-Abl+ LSKs (Figure 4A), using this allele (Mx1cre+;Fbw7f/f;c-MyceGFP/+), we showed that Bcr-Abl+ LSK cells expressed significantly higher c-Myc protein levels than WT LSK cells (Figure 4B). This data was further corroborated by western blot (Figure 4C). Notably, c-Myc protein levels were higher in leukemic LSKs when compared to both WT LSK and Fbw7−/− LSKs, explaining the different physiological responses observed between non-leukemic and leukemic LSKs in response to Fbw7 deletion (Figure 4C). In Bcr-Abl+ LSK cells, loss of Fbw7 expression further induced the levels of c-Myc protein beyond that observed in WT leukemic LSK cells (Figure 4D). These observations suggested that slight changes in c-Myc protein abundance could result in distinct phenotypic responses.
Figure 4. Decrease of c-Myc protein levels and inhibition of p53 activation are able to rescue CML-initiating activity.
(A) qRT-PCR analysis of c-Myc expression in sorted tumor subsets (LSK, c-Kit+ and Lin+) (B) c-Myc protein expression in normal and Bcr-Abl+ LSK cells in the BM. Graph on right shows mean fluorescence intensity (MFI) for eGFP (c-Myc protein). (C) Western blot analysis of c-Myc protein expression in LSKs sorted from WT tumor, Fbw7−/− and WT mice. (D) c-Myc protein expression in MxCre+;Fbw7+/+ or MxCre+;Fbw7−/− Bcr-Abl+LSK. E) Average CFU from sorted Bcr-Abl+ LSK cells from MxCre+;Fbw7+/+, MxCre+;Fbw7−/−, or MxCre+;Fbw7−/−;Myc+/− mice. (F, G) Average CFU from sorted Bcr-Abl+ LSK cells from MxCre+;Fbw7+/+, MxCre+;Fbw7−/−, MxCre+;Fbw7−/−;shp53, or MxCre+;Fbw7−/−;p53−/− mice on the first (F) and secondary (G) platings. (H) Images of Bcr-Abl+ colonies generated from the indicated genotypes (n=3 for each genotype). Scale bar 100 µm. (I) Kaplan Meier survival curves of animals transplanted with control (red), Fbw7−/−;Myc+/− (blue) LSKs transduced with a retrovirus expressing Bcr-Abl, or Fbw7−/− LSKs transduced with a retrovirus expressing Bcr-Abl and a shRNA targeting p53 (green). Error bars indicate +/−SD. *p<0.01, **p<0.001. See also Figure S3.
Additional Fbw7 substrates, particularly Notch, have been previously implicated in CML progression (Ito et al., 2010) and could influence the observed LIC defects upon Fbw7 deletion in Bcr-Abl+ LSKs. To address this question, we initially evaluated the expression level of cleaved Notch1 in WT and Fbw7−/− Bcr-Abl+ c-Kit+ cells. Expression of Notch1 was not detected in either population (Figure S3A). Of note, Notch1 and Notch2 do not appear to be important Fbw7 substrates in WT HSCs as generation of triple knockout mice (MxCre+;Fbw7f/f;Notch1f/f;Notch2f/f) could not rescue the HSC defects observed in Fbw7−/− LSKs (non-leukemic) (Figure S3B–E). More specifically, the frequency (total cell number) of CD150+ CD48− LSKs and aberrant cell cycle status were unaffected by reducing Notch levels in Fbw7 deficient mice (Figure S3B–D) ((Thompson et al., 2008) and unpublished). These in vivo studies have defined the effects of Fbw7 on leukemia initiating cell populations. They have also demonstrated that c-Myc (and not Notch1/2) is the major Fbw7 substrate in CML. Although we cannot exclude that other Fbw7 substrates may play a part in the regulation of CML LIC.
Decrease of c-Myc protein levels and p53 silencing rescues Fbw7−/− LIC function
To directly address the mechanisms of action of Fbw7 in CML-initiating cells we attempted to genetically rescue Fbw7 deletion effects on the survival of the Bcr-Abl+ LSK population. We generated Mx1cre+;Fbw7f/f;c-Mycf/+ mice and silenced p53 expression in Mx1cre+;Fbw7f/f cells by using a p53-specific shRNA or by deleting p53, Mx1cre+;Fbw7f/f;p53−/− mice (Bric et al., 2009). We hypothesized that a decrease in c-Myc protein levels or the inhibition of p53 response could rescue the ability of Fbw7−/− Bcr-Abl+LSK cells to maintain CML disease progression in vivo and serially replate in vitro. As shown in Figures 4E–I both genetic modifications led to a significant rescue of the ability of the Fbw7 deficient LSK cells to generate colonies in vitro and to induce disease in-vivo. The Fbw7−/−;c-Myc−/+, Fbw7−/−;shp53 and Fbw7−/−;p53−/− colonies were almost indistinguishable in numbers from colonies generated by WT cells and no lineage differences were noted (Figure 4E–H). Fbw7−/−;c-Myc−/+ and Fbw7−/−;p53−/− colonies were able to serially replate in a fashion identical to WT counterparts (Figure 4G). To directly assess the self-renewal ability of LICs, we transplanted transduced LSKs from Fbw7−/−;c-Myc−/+ or Fbw7−/−;shp53 into lethally irradiated recipients. Importantly, mice receiving Bcr-Abl+ LSKs from Fbw7−/−;c-Myc−/+ developed a CML-like disease with similar kinetics to Bcr-Abl+ control cells restoring Fbw7−/− LIC self-renewal capacity (Figure 4I). However, as previously shown by Lowe and collegues loss of p53 in CML lead to disease progression, and Bcr-Abl+ shp53+ LSKs from Fbw7−/− progressed to an accelerated phase based on pathology and ~5% blasts in the periphery (data not shown) (Wendel et al., 2006). These experiments demonstrate that c-Myc over-expression and p53-mediated cell death are responsible for the apoptotic phenotype of the Fbw7 deficient LIC.
In vivo visualization of c-Myc protein expression in CML
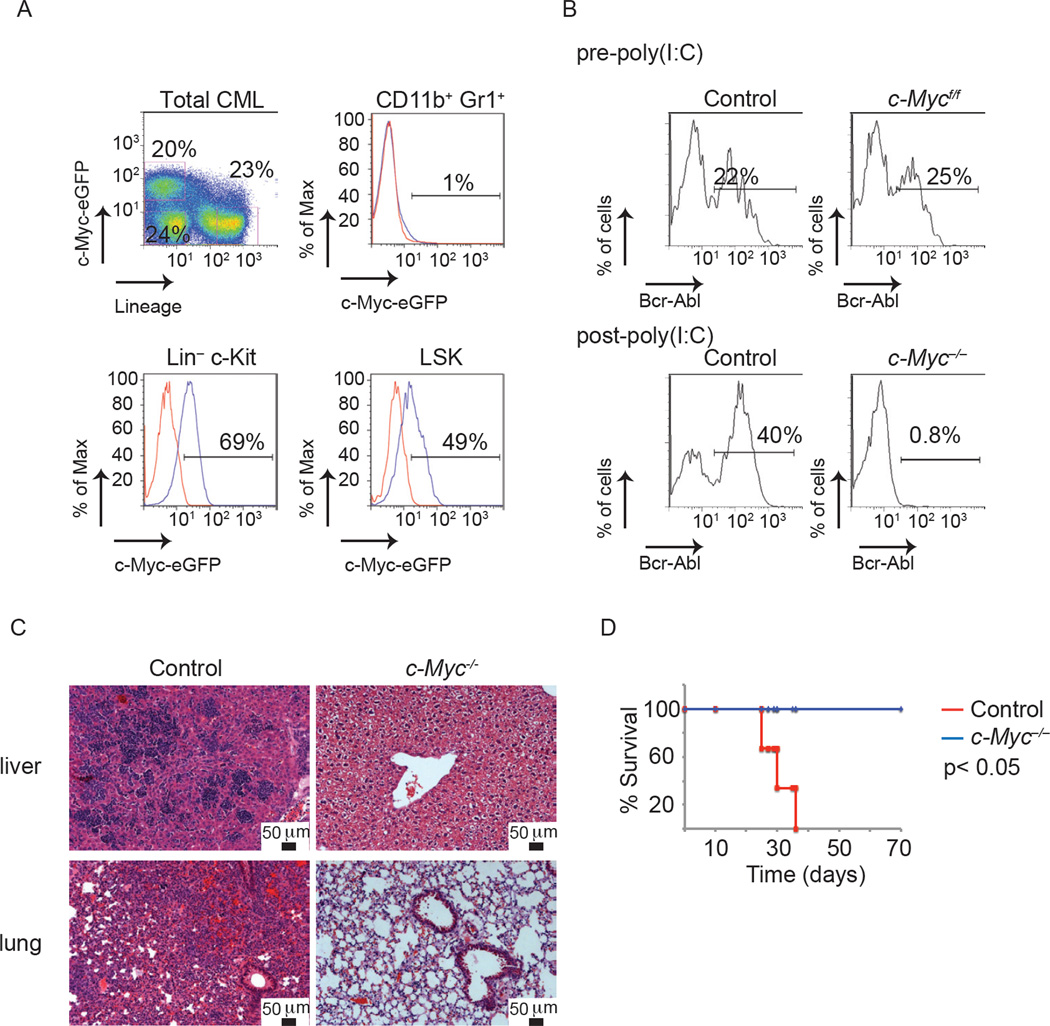
These studies suggested that CML-initiating cells express c-Myc protein and depend on its activity. Although it was previously shown that Bcr-Abl is able to induce the transcription of c-Myc (Nakamura et al., 2011; Xie et al., 2002), it is unclear whether c-Myc function is essential for the initiation or/and the progression of the disease in vivo. To address this question we utilized the c-MyceGFP genetic model and visualized c-Myc expression in established CML. As shown in Figure 5A, only a minority of CML (Bcr-Abl+) cells express detectable levels of c-Myc protein. All c-Myc protein expression is confined within the Lin− fraction and comprises approximately 10–20% of the bulk of the tumor. Myc protein expression was detected in both Bcr-Abl+ Lin− c-Kit+ and Bcr-Abl+ LSK populations. In contrast, Bcr-Abl+CD11b+Gr1+ cells are negative for GFP expression (Figure 5A). To test whether there is a correlation between leukemia-initiating activity and c-Myc protein expression, identical numbers of purified different tumor subsets were transplanted into secondary recipients. Neither the Lin+ c-Myc/eGFP− nor the Lin− c-Myc/eGFP− fractions were able to transfer disease (Figure S4 and data not shown). On the other hand, all leukemia-initiating activity was confined to the c-MyceGFP leukemic cell fraction (Figure S4B and C). However, flowcytometry based separation of Bcr-Abl+Lin−c-Kit+Sca1− and Bcr-Abl+ Lin−c-Kit+Sca1+ fractions, coupled to subsequent transplantation experiments demonstrated that only the LSK fraction could transfer disease in secondary hosts (Figure S4B and C). This finding contrasts with c-Myc protein expression and function in normal LSK cells, where two distinct populations exist. The c-Myc− population contains HSC activity and a c-Myc+ population contains multipotential progenitors (Reavie et al., 2010). These studies demonstrate in vivo c-Myc visualization in leukemia and suggest that although LIC activity lies within the c-Myc-expressing fraction, c-Myc protein expression is not sufficient to guarantee leukemia initiation.
Figure 5. CML-initiating cell activity and disease progression depends on c-Myc expression and activity.
(A) c-Myc protein expression in Bcr-Abl+ CML tumor subsets. (B–D) Mice transplanted with LSKs expressing Bcr-Abl from MxCre+;c-Myc+/+ and MxCre+;c-Mycf/f mice and treated with poly(I:C) following disease initiation. (B) PB analysis of mice, (C) H&E staining of liver and lung, and (D) Kaplan Meier survival curves of animals transplanted with MxCre+;Myc+/+ (red), MxCre+;Myc−/− (blue) (n=5 for each genotype). See also Figure S4.
Bcr-Abl-induced CML is addicted to c-Myc expression and function
To test the importance of c-Myc protein expression in CML initiation and progression we used a conditional c-Myc allele (Mx1cre+;c-Mycf/f). All genotypes prior to deletion were able to initiate disease as verified by PB analysis (Figure 5B). Once disease onset was verified, c-Myc was deleted using poly(I:C) administration. Deletion of c-Myc led to an almost complete absence of Bcr-Abl+ cells from PB and infiltration in secondary tissues such as liver and lung within three weeks (Figure 5B and C). Mice carrying c-Myc−/− Bcr-Abl+ cells were followed up to 6–8 months post cell transplantation and never developed any signs of a CML-like disease. On the other hand, control mice carrying Mx1cre+;c-Myc+/+ Bcr-Abl+ cells succumbed to a lethal CML-like disease within five weeks post transplantation (Figure 5D).
To further quantify c-Myc protein levels, we utilized mice carrying only one allele of c-Myc (Mx1cre+;c-Mycf/w). We had previously shown that these LSK cells express lower levels of c-Myc protein (Reavie et al., 2010). Bcr-Abl+c-Myc+/− LSK cells were able to generate colonies in vitro, at similiar efficiency to their Bcr-Abl+c-Myc+/+ counterparts in both primary and secondary platings (Figure S4D). We then initiated disease by transplanting Bcr-Abl-expressing LSK (Bcr-Abl+c-Myc+/−) cells and upon verification of CML initiation deleted one c-Myc allele by poly(I:C) administration. Interestingly, a single allele of c-Myc was sufficient to maintain disease progression (Figure S4E–G). These studies suggest that there are well-defined thresholds of c-Myc protein expression, which is controlled by Fbw7-mediated ubiquitination, essential for CML induction and progression. Indeed, both lack and non-physiologically increased levels of c-Myc severely affect CML progression.
Fbw7 deletion inhibits progression of established, Bcr-Abl-induced B-ALL
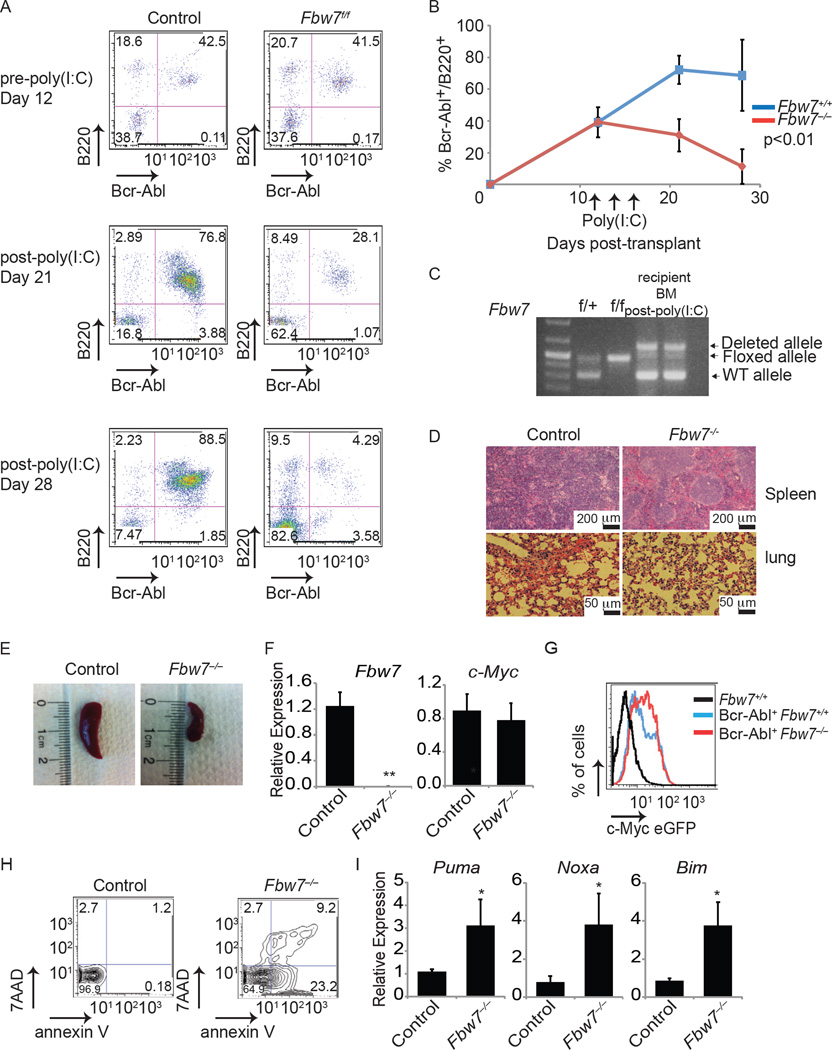
The BCR-ABL translocation is also found in B-cell acute lymphoblastic leukemia (B-ALL). We thus determined whether Fbw7 plays a role in progression of B-ALL. To establish B-ALL, we transduced MxCre+;Fbw7+/+ or MxCre+;Fbw7f/f whole BM with retrovirus expressing Bcr-Abl-GFP followed by transplantation into lethally irradiated recipient mice. PB was analyzed 12 days post transplantation to determine initiation of disease by Bcr-Abl+B220+. Both cohorts of mice showed approximately 40% Bcr-Abl+B220+ cells. At that point, deletion of Fbw7 was initiated by administration of poly(I:C) (Figure 6A) and disease progression was monitored. As expected, mice transplanted with Bcr-Abl+MxCre+;Fbw7+/+ BM had an increase in the percentage of Bcr-Abl+B220+ cells in the PB. However, mice transplanted with Bcr-Abl+MxCre+;Fbw7f/f BM showed a significant reduction in Bcr-Abl+B220+ cells (Figure 6A) and these cells were virtually undetectable three weeks after the initiation of Fbw7 deletion (Figure 6B). MxCre+;Fbw7+/+ mice showed signs of B-ALL including infiltration of secondary tissues and splenomegaly, whereas mice transplanted with MxCre+;Fbw7f/f showed no sign of disease following treatment with poly(I:C) (Figure 6C–E). Once more, utilizing the c-MyceGFP mouse model, we evaluated c-Myc protein expression in the tumor to determine whether loss of B-ALL was due to stabilization of c-Myc as seen in the CML model. Unlike the CML model, approximately 100% of Bcr-Abl+ cells were B220+ and Bcr-Abl+ LSKs were not observed. Although a significantly greater percentage of the tumor in Fbw7−/− expressed c-MyceGFP, no overall increase in expression was observed suggesting that Fbw7 could have additional substrates in B-ALL (Figure 6F and G). Analysis of Annexin V and 7-AAD in the Bcr-Abl+ B220+ BM cells showed a significant increase in cell apoptosis and cell death along with induction of apoptosis associated p53 targets in MxCre+;Fbw7−/− BM (Figure 6H and I). This is an exciting finding as it suggests that Fbw7 could be an attractive therapeutic target also in Bcr-Abl+ B-ALL. In agreement with this notion, sequencing of FBW7 in human B-ALL patient cDNAs failed to identify any inactivating mutations (0/50 samples), suggesting that FBW7 function is required for B-ALL disease progression (data not shown).
Figure 6. Depletion of Fbw7 inhibits progression of B-ALL.
(A) FACS analysis of PB from mice transplanted with MxCre+;Fbw7+/+ and MxCre+;Fbw7−/− total BM cells expressing Bcr-Abl. Upper panel: 12 days post-transplant but prior to poly(I:C) treatment. Middle and lower panels: Day 21 and 28 days post-poly(I:C) treatment, respectively. (B) Graph depicting the percent of Bcr-Abl+ B220+ cells in the PB of both cohorts. (C) Genotyping PCR from recipient BM. (D) H&E staining of spleen and lung. (E) Representative spleen at day 28. (F) qRT-PCR analysis of Fbw7 and c-Myc expression in sorted tumor. (G) c-Myc protein expression in spleen of recipient animals gated on Bcr-Abl+ B220+. (H) FACS plots showing annexin V+ and 7-AAD+ cells in the Bcr-Abl+ B220+ cells in the BM. (I) qRT-PCR analysis showing the expression of p53 target genes, Puma, Bim, and Noxa, in sorted control or Fbw7−/− tumors. Error bars indicate +/−SD. *p<0.01, **p<0.001
Human CML leukemia-initiating cells require FBW7 function
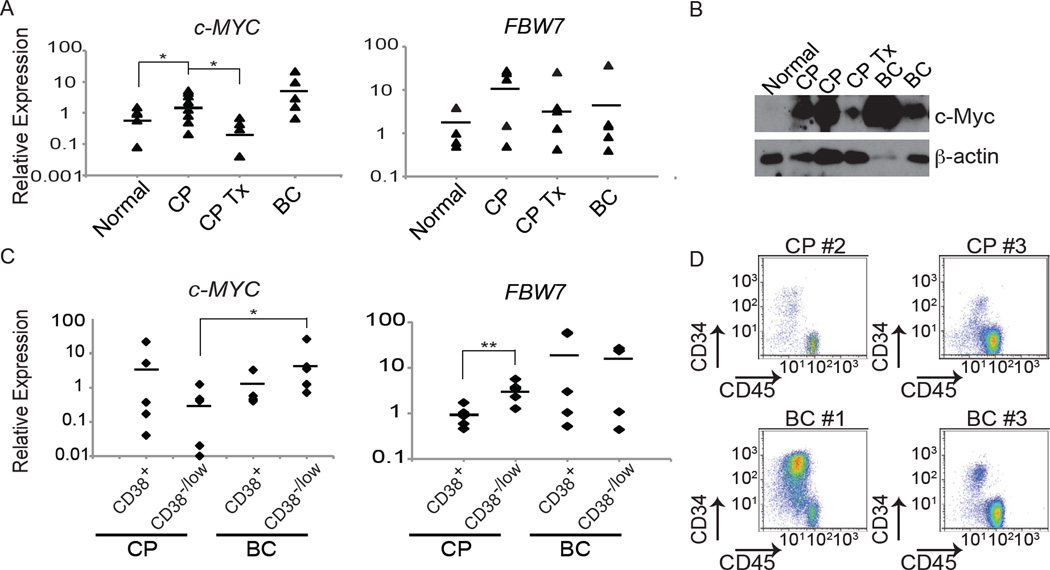
Depletion of Fbw7 in the mouse model eradicates the CML LIC, we next asked whether these findings are relevant to the human disease. Consistent with the findings that Bcr-Abl induces c-MYC expression (Xie et al., 2002), we observed that c-MYC expression level in PB mononuclear cells (PBMNCs) of patients is higher in newly diagnosed or untreated CP CML patients than in normal PBMNC. TKI treated CML chronic phase patients displayed almost physiological levels of c-MYC (Figure 7A). c-Myc protein levels followed similar patterns of expression (Figure 7B). On the other hand no significant differences in the levels of FBW7 expression were noted (Figure 7A), consistent with the idea that FBW7 is also controlled at the activity level and not merely transcriptionally.
Figure 7. FBW7 and c-MYC expression patterns in human CML.
(A and B) FBW7 and c-MYC mRNA levels (A, determined by qRT-PCR, bar indicates average) and c-Myc protein levels (B, determined by Western blot) from total PBMNCs in healthy individuals (normal), CML patients in CP without treatment (CP) or being treated with Imatinib (CP Tx), or in BC (BC). (C) qRT-PCR for c-MYC and FBW7 mRNA levels from CD34+CD38+ and CD34+CD38low populations from BM of patients in CP, and BC normalized to normal UCB derived CD34+CD38low. Bar indicates average. (D) FACS plots showing CD45 and CD34 expression in human CML patients used to sort CD34+CD38+ and CD34+CD38low cell populations. *p<0.01, **p<0.001
We further determined the levels of c-MYC and FBW7 expression in stem and progenitor populations in CML patients by sorting CD34+CD38+ and CD34+CD38low populations, respectively, from CP, and BC patient BM samples (Figure 7C and D). c-MYC mRNA expression was detected in all subsets, its highest level was however in BC CD34+CD38low cells. Interestingly, although FBW7 was also expressed in all samples, CP CD34+CD38low cells expressed significantly higher levels than CD34+CD38+ cells from the same samples, in agreement with our animal modeling data that detected the highest Fbw7 expression in the LSK LIC population.
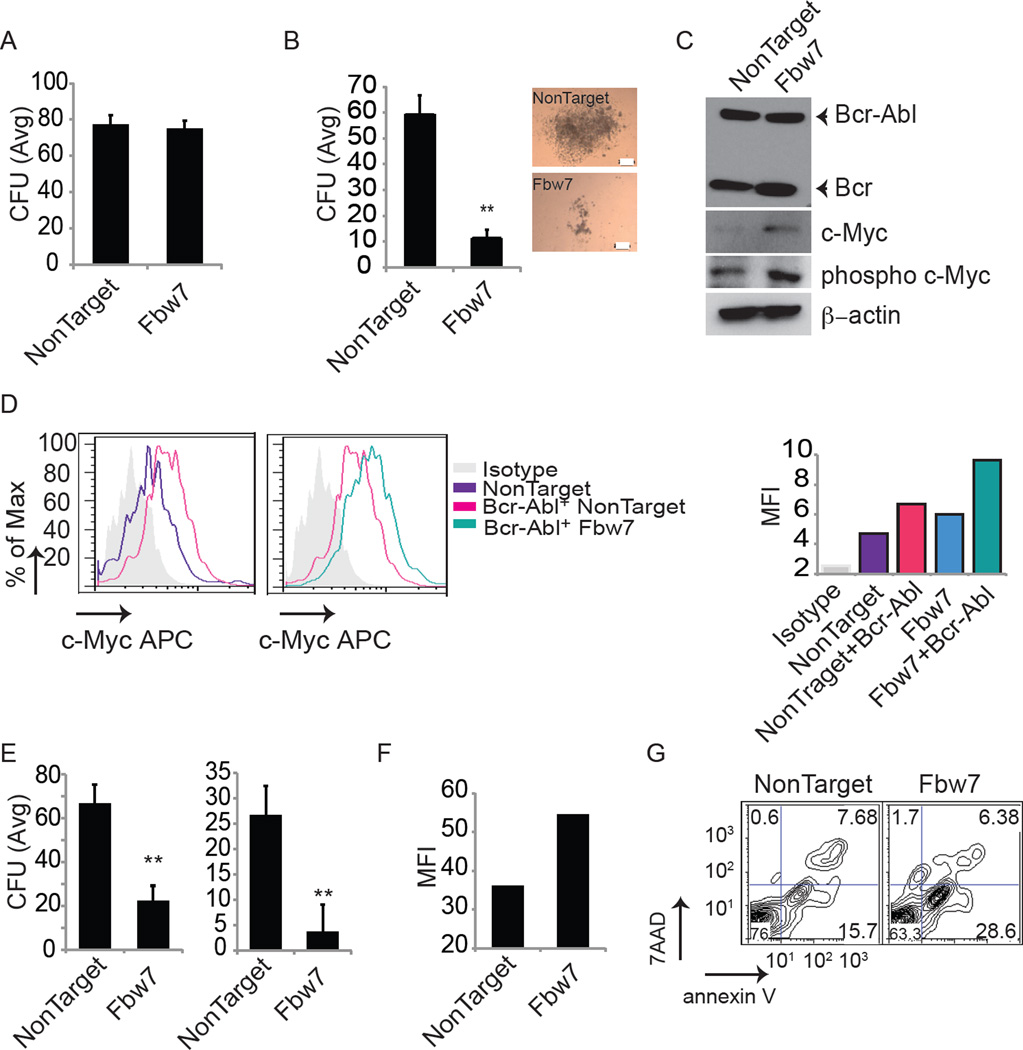
To address whether Fbw7 possesses a similar functional role in human CML, we transduced the Bcr-Abl+ human CML cell line KU812with lentiviruses expressing shRNAs against Fbw7. Efficient knockdown was confirmed by qRT-PCR (Figure S5A). Loss of Fbw7 induced apoptosis and lead to the accumulation of c-Myc (Figure S5B and C). Degradation of c-Myc requires a priming phosphorylation event on T58 by GSK3β (Gregory et al., 2003), and in agreement with this Fbw7 silencing lead to the specific enrichment of phosphorylated c-Myc (Figure S5C). To further study potential Fbw7 functions in human cells, we first silenced Fbw7 using lentiviruses expressing shRNAs against Fbw7 in normal umbilical cord blood (UCB) derived CD34+ with or without co-infection with Bcr-Abl retrovirus and subsequently plated the cells in colony forming assays. Fbw7 silencing showed no alterations in the colony forming ability of normal CD34+ cells, but significantly decreased plating capacity of Bcr-Abl+ CD34+ cells (Figure 8A and B). Fbw7 silencing led to accumulation of both c-Myc and phospo-c-Myc protein in total progeny derived from Bcr-Abl+ infected CD34+ cells (Figure 8C). Overexpression of c-Myc protein was further verified in the Bcr-Abl+ CD34+ population compared to normal CD34+ population by intracellular FACS for c-Myc expression. Consistent with the finding in the mouse model, Bcr-Abl expression results in increased c-Myc expression, which is further increased by silencing of Fbw7 (Figure 8D). Finally, FBW7 silencing in purified primary human CP (from either newly diagnosed or untreated patients) CML CD34+ cells led to a significant loss of plating ability in CFU assays (Figure 8E and Figure S5D and E), most likely due to the elevated levels of c-Myc, and significant induction of cell death (Figures 8F and G, and Figure S5F). These combined human data are in agreement with our animal experiments.
Figure 8. Fbw7 silencing in human CML-initiating cells leads to induction of apoptosis and loss of differentiation potential.
(A) Average CFU from normal UCB derived CD34+ cells expressing shRNA against either NonTarget or Fbw7. (B–D) Normal UCB derived CD34+ expressing Bcr-Abl together with shRNAs against either NonTarget or Fbw7. Progeny of cells were plated in CFU assay (B, Scale bar 200µm) and western blot for c-Myc, Phospho-c-Myc, and Bcr-Abl in total progeny of CD34+ cells (C). (D) c-Myc expression determined by intracellular FACS gated on CD34+ population. Graph on right shows MFI for c-Myc protein. (E–G) CP patient BM derived CD34+ cells were transduced with lentivirus expressing either control or shRNAs against Fbw7, puromycin selected for 48 hours. (E) Average CFU assay from CD34+ progeny in two different patients. (F) MFI for c-Myc protein for representative CML patient derived CD34+ cells following Fbw7 silencing. (G) Representative annexin V staining of patient derived CD34+ cells following lentiviral transduction. Error bars indicate +/− SD. *p<0.01, **p<0.001. See also Figure S5.
DISCUSSION
We demonstrate here the essential function of the Fbw7 E3 ligase for the initiation and the progression of CML. Fbw7 deletion leads to LIC apoptosis due to aberrantly high levels of c-Myc protein expression and activation of the p53 pathway. Interestingly, p53 mutations can accompany disease progression in human CML and p53 loss in some cases impedes the anti-leukemic response to Bcr-Abl inhibition (Kelman et al., 1989; Wendel et al., 2006), suggesting that loss of p53 in some tumors could constitute an adaptive response to the increase in the levels of c-Myc during CML progression. Overall, our experiments suggest that Fbw7 expression is absolutely essential for the maintenance of non-toxic levels of c-Myc protein within CML LIC cells. Interestingly, although Fbw7 is a ubiquitin ligase capable of targeting a large number of substrates, our results suggest that c-Myc is its key substrate in CML, in contrast with human T-ALL, where Notch1 appears to be a main targets (O'Neil et al., 2007; Thompson et al., 2008). However, we cannot exclude that additional Fbw7 substrates, except for Notch1, play roles in its function. Further studies are required to address the identity and function of such substrates.
We were able to visualize, using flow cytometry, c-Myc protein abundance in vivo, and show that c-Myc expression is restricted within the LIC population, with the bulk of the tumor being c-Myc−. These results proved that Fbw7 function is specifically required by cells with leukemia-initiating activity but is dispensable for the maintenance of the more differentiated CML subset. Moreover, genetic deletion of c-Myc during disease progression showed that Bcr-Abl-driven CML is addicted to physiological c-Myc function, suggesting that the disease requires well-defined and Fbw7-regulated thresholds of c-Myc abundance and activity. This is an intriguing idea with potential important clinical ramifications in the field of cancer biology, as it would suggest that both depletion and over-abundance of c-Myc protein levels in tumors could lead to similar clinical outcomes, albeit with distinct mechanisms of action. Recent development of small molecules targeting c-Myc co-activator bromodomain inhibitors (Delmore et al., 2011), opens the way for therapeutic protocols that include c-Myc activity inhibition in established CML.
Classic experiments have shown that introduction of v-abl in a myeloid cell line can specifically induce c-Myc expression in a tyrosine kinase-dependent manner (Cleveland et al., 1989). Subsequent seminal studies by Sawyers and colleagues demonstrated that Bcr-Abl-induced transformation could be suppressed by dominant negative Myc mutants in vitro (Sawyers et al., 1992). In agreement with these studies we were able to visualize c-Myc protein levels in vivo in progressing CML and identify the populations that retain c-Myc protein expression. Interestingly, only a minority of the established leukemia cells express c-MyceGFP. These cells are characterized by the expression of c-Kit and include the CML LSK population, previously suggested to possess all LIC activity (Neering et al., 2007; Reynaud et al., 2011). When we subdivided the c-Myc-expressing population using the c-MyceGFP reporter we were able to transplant disease only using the c-MyceGFP LSK fraction suggesting that although LIC activity lies within the c-Myc-expressing fraction, c-Myc protein expression is not sufficient to guarantee leukemia-initiating properties. This is an intriguing distinction between normal and leukemic hematopoiesis as we previously showed that normal c-MyceGFP LSK cells are multi-potential progenitors but not bona fide HSC (Reavie et al., 2010).
We have previously shown that Fbw7 is required for normal hematopoiesis, as its deletion leads to progressive adult stem cell exhaustion. This would suggest that inhibition of Fbw7 activity is not an ideal therapeutic target in leukemia. However, the response of normal and malignant stem and progenitor cells to the deletion of Fbw7 is vastly distinct. Putative LIC respond acutely, as Fbw7 deletion leads to rapid (<2weeks) loss of cell numbers and activity, whereas the response of normal HSC is delayed and there are no significant changes in the number of HSC cells till 10–12 weeks post Fbw7 deletion (Matsuoka et al., 2008; Thompson et al., 2008) (Data not shown). We believe that this differential response to Fbw7 deletion can be explained by the significantly higher levels of c-Myc protein in the Bcr-Abl-expressing LIC. This data suggest that it should be possible to define a “therapeutic window” altering either the concentration of the inhibitor or the length of the treatment. Drug combination could be another therapeutic avenue, especially as TKI fail to target CML LIC. Indeed, Nakayama and colleagues have demonstrated that Fbw7 inhibition can be used in combination with other established CML treatments, including Imatinib to achieve efficient targeting of CML-initiating cells (Takeishi, 2013), both in animal models and primary human disease. At this point no Fbw7 inhibitors have been identified. However, recent studies suggested that small molecule targeting Fbw7 is a feasible approach. Tyers and collegues have recently identified a biplanar dicarboxylic acid compound as an inhibitor of substrate recognition by the yeast Fbw7 ortholog (Cdc4) (Aghajan et al., 2010; Orlicky et al., 2010). Moreover, as Fbw7-mediated c-Myc recognition is induced by the priming phosphorylation of c-MycThr58 by GSK3, GSK3 inhibitors could also be used to target Fbw7 function and c-Myc stability. Such inhibitors have been developed and their efficacy in vivo was tested using MLL-induced models of AML (Wang et al., 2008). These GSK3 inhibitors are currently in Phase II clinical trials for the treatment of Alzheimer’s disease (Martinez et al., 2011), opening the way for their future use for the treatment of CML in combination with tyrosine kinase inhibitors. Finally, as c-Myc activity is a driver of distinct tumor types, it is conceivable that Fbw7 inhibitors could be promising therapeutic tools in a wide range to blood and solid tumors.
EXPERIMENTAL PROCEDURES
Animals
Fbw7flox mice were previously published (Thompson et al., 2008). Mycflox mice were a kind gift from F. Alt (de Alboran et al., 2001), and MyceGFP mice were a kind gift from Dr. Barry Sleckman (Huang et al., 2008). IFNα-inducible Mx1cre, p53 germline knockout, and C57Bl6 recipient mice were purchased from Jackson Laboratories. All mice were housed in a pathogen-free animal facility at the NYU School of Medicine. All animal procedures were approved by Institutional Animal Care and Use Committee of the NYU School of Medicine and carried out in compliance with NIH guidelines.
Generation and analysis of CML animals
LSK were isolated from 4–6 week old mice using the mouse hematopoietic progenitor enrichment kit (Stem Cell Technologies) per manufacturer’s protocol and stained with a lineage cocktail, c-Kit, and Sca-1 antibodies followed by FACS purification. LSK were cells were infected with Bcr-Abl-NGFR or Bcr-Abl-GFP retrovirus (Wertheim et al., 2002) and spun at 2500rpm for 90 minutes at 30° C. 48 to 72 hrs post transfection ~20,000–40,000 Lin− Bcr-Abl+ LSKs were transplanted intravenously into lethally irradiated recipient mice with 2–5×105 support BM cells. For donor cells delted post-transplantation, deletion was initiated on day 7 post transplantation 3 injections with poly(I:C) (Amsersham) at a concentration of 5 µg/g of body weight, and disease was monitored by flowcytometry. Further details of culture and analysis are provided in the Supplemental Experimental Procedures.
Analysis and culture of human CML samples
Primary patient samples were obtained with informed consent from all donors in accordance with the Declaration of Helsinki and studies were approved by the Institutional Review Boards at NYU Medical Center, and Memorial Sloan-Kettering Cancer Center. UCB or patient BM CD34+ cells were isolated using CD34+ selection kit following manufacturer’s instructions (Stem Cell Technologies). Cells were cultured in Stemspan (Stem Cell Technologies), supplemented with 50 ng/ml SCF, 50 ng/ml Flt3L, and 100 ng/ml Tpo for 24 hours followed lentiviral transduction. Further details of culture and analysis are provided in the Supplemental Experimental Procedures.
FACS Analysis
All antibodies used for FACS analysis were procured from e-Bioscience. Specifically, the antibodies we used were as follows: c-Kit (2B8), Sca-1 (D7), Mac-1 (M1/70), Gr-1 (RB6–8C5), NK1.1 (PK136), TER-119, CD3 (145-2C11), CD19 (1D3), IL7R (RAM34), CD4 (RM4–5), CD8 (53 – 6.7), CD271 (NGFR) (ME20.4). BM lineage antibody cocktail includes the following: Mac-1, Gr-1, NK1.1, TER-119, CD4, CD8, IL7R and CD19. Apoptosis was detected using Annexiv-V PE-conjugated detection kit (BD Pharmingen) along with 7-AAD following manufacturers protocol. For intracellular c-Myc staining, cells were stained with anti-CD34 (BD Pharmingen), washed, fixed, and permeabilized using BD cytofix/cytoperm kit following manufactuors protocol. Stainings were performed with rabbit anti-c-Myc (Cell Signaling) followed by goat anti-rabbit Alexa Fluor 647 (Invitrogen).
Quantitative real-time PCR
Total RNA was harvested from cells using the Qiagen RNeasy Kit (Qiagen, Germany). RNA was quantified by absorbance at A260 nm and 2 µg of total RNA used for cDNA synthesis using Superscript III first strand synthesis kit (Invitrogen) qRT-PCR was carried out using SYBR green universal mix PCR reaction buffer (Roche) using an Roche lightcycler 480 II (Roche). All signals are normalized to levels of Gapdh.
Statistical analysis
All the statistical analyses were performed using un-paired two-tailed Student's t-test, unless otherwise specified.
Supplementary Material
Highlights.
Fbw7 function is essential for the initiation and progression of CML.
Fbw7 deletion leads to c-Myc overexpression and activation of the p53 pathway.
c-Myc is an Fbw7 substrate in CML initiating cells.
FBW7 is required for maintenance of human CML initiating cells.
SIGNIFICANCE.
CML is initiated by the BCR-ABL translocation and maintained by LIC. Although current therapies can suppress disease, they are insufficient to target LIC. Utilizing a Bcr-Abl model of CML and human CML samples, we demonstrate that the E3 ligase Fbw7 is required for the initiation and progression of CML as well as maintenance of the LIC. We demonstrate that interaction between Fbw7 and its protein substrate c-Myc is required for CML progression, and Fbw7 deletion leads to p53-mediated apoptosis of LIC. In agreement with these findings, silencing of FBW7 leads to loss of human CML LIC self-renewal. These studies identify Fbw7 ligase as an essential regulator of CML LIC maintenance and open the way for targeting Fbw7 activity in CML.
ACKNOWLEDGEMENTS
We would like to thank Dr. B. Sleckman for providing the c-MyceGFP knock in mice, J. Silva for the anti-p53 shRNA vector and W. Pear for the Bcr-Abl vectors. We would also like to thank the members of the Aifantis lab for valuable advice and discussions. Also, the NYU Flow Cytometry facility for expert cell sorting. IA is supported by the National Institutes of Health (RO1CA133379, RO1CA105129, 1RO1CA173636, RO1CA149655, RO1GM088847), the Leukemia and Lymphoma Society (TRP program grants), the Chemotherapy Foundation, the William Lawrence Blanche Hughes Foundation and the V Foundation for Cancer Research. L.R. is supported by a NIH Ruth L. Kirchstein Award, S.M.B. is supported by the NYU Hematology/Oncology NIH training grant (5T32HL007151-33) and the NIH institutional training grant (1T32CA160002-01). B.A–O. is supported by the Alexander von Humboldt Foundation. I.A is a Howard Hughes Medical Institute Early Career Scientist. We would like to dedicate this paper to our mice lost by the fury of hurricane Sandy.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aghajan M, Jonai N, Flick K, Fu F, Luo M, Cai X, Ouni I, Pierce N, Tang X, Lomenick B, et al. Chemical genetics screen for enhancers of rapamycin identifies a specific inhibitor of an SCF family E3 ubiquitin ligase. Nat Biotechnol. 2010;28:738–742. doi: 10.1038/nbt.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartram CR, de Klein A, Hagemeijer A, van Agthoven T, Geurts van Kessel A, Bootsma D, Grosveld G, Ferguson-Smith MA, Davies T, Stone M, et al. Translocation of c-ab1 oncogene correlates with the presence of a Philadelphia chromosome in chronic myelocytic leukaemia. Nature. 1983;306:277–280. doi: 10.1038/306277a0. [DOI] [PubMed] [Google Scholar]
- Bhatia R, Holtz M, Niu N, Gray R, Snyder DS, Sawyers CL, Arber DA, Slovak ML, Forman SJ. Persistence of malignant hematopoietic progenitors in chronic myelogenous leukemia patients in complete cytogenetic remission following imatinib mesylate treatment. Blood. 2003;101:4701–4707. doi: 10.1182/blood-2002-09-2780. [DOI] [PubMed] [Google Scholar]
- Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- Bric A, Miething C, Bialucha CU, Scuoppo C, Zender L, Krasnitz A, Xuan Z, Zuber J, Wigler M, Hicks J, et al. Functional identification of tumor-suppressor genes through an in vivo RNA interference screen in a mouse lymphoma model. Cancer Cell. 2009;16:324–335. doi: 10.1016/j.ccr.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland JL, Dean M, Rosenberg N, Wang JY, Rapp UR. Tyrosine kinase oncogenes abrogate interleukin-3 dependence of murine myeloid cells through signaling pathways involving c-myc: conditional regulation of c-myc transcription by temperature-sensitive v-abl. Mol Cell Biol. 1989;9:5685–5695. doi: 10.1128/mcb.9.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbin AS, Agarwal A, Loriaux M, Cortes J, Deininger MW, Druker BJ. Human chronic myeloid leukemia stem cells are insensitive to imatinib despite inhibition of BCR-ABL activity. J Clin Invest. 2011;121:396–409. doi: 10.1172/JCI35721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley GQ, Van Etten RA, Baltimore D. Induction of chronic myelogenous leukemia in mice by the P210bcr/abl gene of the Philadelphia chromosome. Science. 1990;247:824–830. doi: 10.1126/science.2406902. [DOI] [PubMed] [Google Scholar]
- de Alboran IM, O'Hagan RC, Gartner F, Malynn B, Davidson L, Rickert R, Rajewsky K, DePinho RA, Alt FW. Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity. 2001;14:45–55. doi: 10.1016/s1074-7613(01)00088-7. [DOI] [PubMed] [Google Scholar]
- de Lavallade H, Apperley JF, Khorashad JS, Milojkovic D, Reid AG, Bua M, Szydlo R, Olavarria E, Kaeda J, Goldman JM, Marin D. Imatinib for newly diagnosed patients with chronic myeloid leukemia: incidence of sustained responses in an intention-to-treat analysis. J Clin Oncol. 2008;26:3358–3363. doi: 10.1200/JCO.2007.15.8154. [DOI] [PubMed] [Google Scholar]
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druker BJ, Guilhot F, O'Brien SG, Gathmann I, Kantarjian H, Gattermann N, Deininger MW, Silver RT, Goldman JM, Stone RM, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006;355:2408–2417. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- Gorre ME, Mohammed M, Ellwood K, Hsu N, Paquette R, Rao PN, Sawyers CL. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- Gregory MA, Qi Y, Hann SR. Phosphorylation by glycogen synthase kinase-3 controls c-myc proteolysis and subnuclear localization. The Journal of biological chemistry. 2003;278:51606–51612. doi: 10.1074/jbc.M310722200. [DOI] [PubMed] [Google Scholar]
- Gupta-Rossi N, Le Bail O, Gonen H, Brou C, Logeat F, Six E, Ciechanover A, Israel A. Functional interaction between SEL-10, an F-box protein, and the nuclear form of activated Notch1 receptor. J Biol Chem. 2001;276:34371–34378. doi: 10.1074/jbc.M101343200. [DOI] [PubMed] [Google Scholar]
- Heidel FH, Bullinger L, Feng Z, Wang Z, Neff TA, Stein L, Kalaitzidis D, Lane SW, Armstrong SA. Genetic and Pharmacologic Inhibition of beta-Catenin Targets Imatinib-Resistant Leukemia Stem Cells in CML. Cell Stem Cell. 2012;10:412–424. doi: 10.1016/j.stem.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisterkamp N, Jenster G, ten Hoeve J, Zovich D, Pattengale PK, Groffen J. Acute leukaemia in bcr/abl transgenic mice. Nature. 1990;344:251–253. doi: 10.1038/344251a0. [DOI] [PubMed] [Google Scholar]
- Hochhaus A, O'Brien SG, Guilhot F, Druker BJ, Branford S, Foroni L, Goldman JM, Muller MC, Radich JP, Rudoltz M, et al. Six-year follow-up of patients receiving imatinib for the first-line treatment of chronic myeloid leukemia. Leukemia. 2009;23:1054–1061. doi: 10.1038/leu.2009.38. [DOI] [PubMed] [Google Scholar]
- Huang CY, Bredemeyer AL, Walker LM, Bassing CH, Sleckman BP. Dynamic regulation of c-Myc proto-oncogene expression during lymphocyte development revealed by a GFP-c-Myc knock-in mouse. Eur J Immunol. 2008;38:342–349. doi: 10.1002/eji.200737972. [DOI] [PubMed] [Google Scholar]
- Huntly BJ, Gilliland DG. Leukaemia stem cells and the evolution of cancer-stem-cell research. Nat Rev Cancer. 2005;5:311–321. doi: 10.1038/nrc1592. [DOI] [PubMed] [Google Scholar]
- Ito T, Kwon HY, Zimdahl B, Congdon KL, Blum J, Lento WE, Zhao C, Lagoo A, Gerrard G, Foroni L, et al. Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature. 2010;466:765–768. doi: 10.1038/nature09171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson CH, Ailles LE, Dylla SJ, Muijtjens M, Jones C, Zehnder JL, Gotlib J, Li K, Manz MG, Keating A, et al. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N Engl J Med. 2004;351:657–667. doi: 10.1056/NEJMoa040258. [DOI] [PubMed] [Google Scholar]
- Jankowska AM, Szpurka H, Tiu RV, Makishima H, Afable M, Huh J, O'Keefe CL, Ganetzky R, McDevitt MA, Maciejewski JP. Loss of heterozygosity 4q24 and TET2 mutations associated with myelodysplastic/myeloproliferative neoplasms. Blood. 2009 doi: 10.1182/blood-2009-02-205690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelman Z, Prokocimer M, Peller S, Kahn Y, Rechavi G, Manor Y, Cohen A, Rotter V. Rearrangements in the p53 gene in Philadelphia chromosome positive chronic myelogenous leukemia. Blood. 1989;74:2318–2324. [PubMed] [Google Scholar]
- Koepp DM, Schaefer LK, Ye X, Keyomarsi K, Chu C, Harper JW, Elledge SJ. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science. 2001;294:173–177. doi: 10.1126/science.1065203. [DOI] [PubMed] [Google Scholar]
- Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, Minden M, Paterson B, Caligiuri MA, Dick JE. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- le Coutre P, Tassi E, Varella-Garcia M, Barni R, Mologni L, Cabrita G, Marchesi E, Supino R, Gambacorti-Passerini C. Induction of resistance to the Abelson inhibitor STI571 in human leukemic cells through gene amplification. Blood. 2000;95:1758–1766. [PubMed] [Google Scholar]
- Martinez A, Gil C, Perez DI. Glycogen synthase kinase 3 inhibitors in the next horizon for Alzheimer's disease treatment. Int J Alzheimers Dis. 2011;2011:280502. doi: 10.4061/2011/280502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Oike Y, Onoyama I, Iwama A, Arai F, Takubo K, Mashimo Y, Oguro H, Nitta E, Ito K, et al. Fbxw7 acts as a critical fail-safe against premature loss of hematopoietic stem cells and development of T-ALL. Genes Dev. 2008 doi: 10.1101/gad.1621808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michor F, Hughes TP, Iwasa Y, Branford S, Shah NP, Sawyers CL, Nowak MA. Dynamics of chronic myeloid leukaemia. Nature. 2005;435:1267–1270. doi: 10.1038/nature03669. [DOI] [PubMed] [Google Scholar]
- Nakamura S, Yokota D, Tan L, Nagata Y, Takemura T, Hirano I, Shigeno K, Shibata K, Fujisawa S, Ohnishi K. Down-regulation of Thanatos-associated protein 11 by BCR-ABL promotes CML cell proliferation through c-Myc expression. Int J Cancer. 2011 doi: 10.1002/ijc.26065. [DOI] [PubMed] [Google Scholar]
- Neering SJ, Bushnell T, Sozer S, Ashton J, Rossi RM, Wang PY, Bell DR, Heinrich D, Bottaro A, Jordan CT. Leukemia stem cells in a genetically defined murine model of blast-crisis CML. Blood. 2007;110:2578–2585. doi: 10.1182/blood-2007-02-073031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowell PC, Hungerford DA. Chromosome studies on normal and leukemic human leukocytes. J Natl Cancer Inst. 1960;25:85–109. [PubMed] [Google Scholar]
- O'Hare T, Corbin AS, Druker BJ. Targeted CML therapy: controlling drug resistance, seeking cure. Curr Opin Genet Dev. 2006;16:92–99. doi: 10.1016/j.gde.2005.11.002. [DOI] [PubMed] [Google Scholar]
- O'Neil J, Grim J, Strack P, Rao S, Tibbitts D, Winter C, Hardwick J, Welcker M, Meijerink JP, Pieters R, et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J Exp Med. 2007;204:1813–1824. doi: 10.1084/jem.20070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlicky S, Tang X, Neduva V, Elowe N, Brown ED, Sicheri F, Tyers M. An allosteric inhibitor of substrate recognition by the SCF(Cdc4) ubiquitin ligase. Nat Biotechnol. 2010;28:733–737. doi: 10.1038/nbt.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pear WS, Miller JP, Xu L, Pui JC, Soffer B, Quackenbush RC, Pendergast AM, Bronson R, Aster JC, Scott ML, Baltimore D. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood. 1998;92:3780–3792. [PubMed] [Google Scholar]
- Reavie L, Della Gatta G, Crusio K, Aranda-Orgilles B, Buckley SM, Thompson B, Lee E, Gao J, Bredemeyer AL, Helmink BA, et al. Regulation of hematopoietic stem cell differentiation by a single ubiquitin ligase-substrate complex. Nat Immunol. 2010;11:207–215. doi: 10.1038/ni.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynaud D, Pietras E, Barry-Holson K, Mir A, Binnewies M, Jeanne M, Sala-Torra O, Radich JP, Passegue E. IL-6 controls leukemic multipotent progenitor cell fate and contributes to chronic myelogenous leukemia development. Cancer Cell. 2011;20:661–673. doi: 10.1016/j.ccr.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley JD. Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- Sawyers CL, Callahan W, Witte ON. Dominant negative MYC blocks transformation by ABL oncogenes. Cell. 1992;70:901–910. doi: 10.1016/0092-8674(92)90241-4. [DOI] [PubMed] [Google Scholar]
- Shah NP, Nicoll JM, Nagar B, Gorre ME, Paquette RL, Kuriyan J, Sawyers CL. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell. 2002;2:117–125. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- Sirard C, Lapidot T, Vormoor J, Cashman JD, Doedens M, Murdoch B, Jamal N, Messner H, Addey L, Minden M, et al. Normal and leukemic SCID-repopulating cells (SRC) coexist in the bone marrow and peripheral blood from CML patients in chronic phase, whereas leukemic SRC are detected in blast crisis. Blood. 1996;87:1539–1548. [PubMed] [Google Scholar]
- Stadtfeld M, Graf T. Assessing the role of hematopoietic plasticity for endothelial and hepatocyte development by non-invasive lineage tracing. Development (Cambridge, England) 2005;132:203–213. doi: 10.1242/dev.01558. [DOI] [PubMed] [Google Scholar]
- Takeishi S, Matsumoto A, Onoyama I, Naka K, Hirao A, Nakayama KI. Ablation of Fbw7 eliminates leukemia-intiating cells by preventing quiescence. 2013 doi: 10.1016/j.ccr.2013.01.026. [DOI] [PubMed] [Google Scholar]
- Thompson BJ, Buonamici S, Sulis ML, Palomero T, Vilimas T, Basso G, Ferrando A, Aifantis I. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J Exp Med. 2007;204:1825–1835. doi: 10.1084/jem.20070872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson BJ, Jankovic V, Gao J, Buonamici S, Vest A, Lee JM, Zavadil J, Nimer SD, Aifantis I. Control of hematopoietic stem cell quiescence by the E3 ubiquitin ligase Fbw7. J Exp Med. 2008;205:1395–1408. doi: 10.1084/jem.20080277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC, Lapidot T, Cashman JD, Doedens M, Addy L, Sutherland DR, Nayar R, Laraya P, Minden M, Keating A, et al. High level engraftment of NOD/SCID mice by primitive normal and leukemic hematopoietic cells from patients with chronic myeloid leukemia in chronic phase. Blood. 1998;91:2406–2414. [PubMed] [Google Scholar]
- Wang Z, Smith KS, Murphy M, Piloto O, Somervaille TC, Cleary ML. Glycogen synthase kinase 3 in MLL leukaemia maintenance and targeted therapy. Nature. 2008;455:1205–1209. doi: 10.1038/nature07284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welcker M, Orian A, Jin J, Grim JE, Harper JW, Eisenman RN, Clurman BE. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. P Natl Acad Sci USA. 2004;101:9085–9090. doi: 10.1073/pnas.0402770101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel HG, de Stanchina E, Cepero E, Ray S, Emig M, Fridman JS, Veach DR, Bornmann WG, Clarkson B, McCombie WR, et al. Loss of p53 impedes the antileukemic response to BCR-ABL inhibition. P Natl Acad Sci USA. 2006;103:7444–7449. doi: 10.1073/pnas.0602402103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheim JA, Forsythe K, Druker BJ, Hammer D, Boettiger D, Pear WS. BCR-ABL-induced adhesion defects are tyrosine kinase-independent. Blood. 2002;99:4122–4130. doi: 10.1182/blood.v99.11.4122. [DOI] [PubMed] [Google Scholar]
- Xie S, Lin H, Sun T, Arlinghaus RB. Jak2 is involved in c-Myc induction by Bcr-Abl. Oncogene. 2002;21:7137–7146. doi: 10.1038/sj.onc.1205942. [DOI] [PubMed] [Google Scholar]
- Yada M, Hatakeyama S, Kamura T, Nishiyama M, Tsunematsu R, Imaki H, Ishida N, Okumura F, Nakayama K, Nakayama KI. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. Embo J. 2004;23:2116–2125. doi: 10.1038/sj.emboj.7600217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Blum J, Chen A, Kwon HY, Jung SH, Cook JM, Lagoo A, Reya T. Loss of beta-catenin impairs the renewal of normal and CML stem cells in vivo. Cancer Cell. 2007;12:528–541. doi: 10.1016/j.ccr.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Chen A, Jamieson CH, Fereshteh M, Abrahamsson A, Blum J, Kwon HY, Kim J, Chute JP, Rizzieri D, et al. Hedgehog signalling is essential for maintenance of cancer stem cells in myeloid leukaemia. Nature. 2009;458:776–779. doi: 10.1038/nature07737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.