Abstract
Nuclear receptors (NRs) are a superfamily of ligand-regulated transcription factors that interact with coregulators and other transcription factors to direct tissue-specific programs of gene expression. Recent years have witnessed a rapid acceleration of the output of high content data platforms in this field, generating discovery-driven datasets that have collectively described: the organization of the NR superfamily (phylogenomics); the expression patterns of NRs, coregulators and their target genes (transcriptomics); ligand- and tissue-specific functional NR and coregulator sites in DNA (cistromics); the organization of nuclear receptors and coregulators into higher order complexes (proteomics); and their downstream effects on homeostasis and metabolism (metabolomics). Significant bioinformatics challenges lie ahead both in the integration of this information into meaningful models of NR and coregulator biology, as well as in the archiving and communication of datasets to the global nuclear receptor signaling community. While holding great promise for the field, the ascendancy of discovery-driven research in this field brings with it a collective responsibility for researchers, publishers and funding agencies alike to ensure the effective archiving and management of these data. This review will discuss factors lying behind the increasing impact of discovery-driven research, examples of high content datasets and their bioinformatic analysis, as well as a summary of currently curated web resources in this field.
Keywords: Nuclear receptors, coregulators, transcriptomics, proteomics, bioinformatics, database
1. Introduction
Nuclear receptors (NRs) comprise a superfamily of conserved transcription factors (encoded by 48 genes in humans and 49 genes in the mouse) that are regulated by small lipophilic ligands and cellular signaling pathways to play essential roles in diverse biological processes [1]. For example, the estrogen, progesterone and androgen receptors are important in reproduction, glucocorticoid receptors in glucose metabolism and stress, the thyroid hormone receptor in oxidative metabolism, and PPARsa in lipid and energy metabolism. NRs also encompass one of the most successful group of targets for drugs currently available, or being developed, to treat a multitude of therapeutic indices, including hypertension, cancer, diabetes, cardiovascular disease, cholesterol gallstone disease and metabolic syndrome [2–5]
Since the cloning of the first NR coactivator, SRC-1/NCOA1, in 1995 [6], intensive characterization has cast coregulators as molecules required by NRs (and other DNA-binding transcription factors) for efficient regulation of gene expression [7, 8]. In contrast to NRs, which are structurally conserved, their coregulators are diverse, both structurally and in the way they contribute to the transcriptional process, namely through a diverse array of reversible enzymatic activities such as acetylation, methylation, ubiquitination and phosphorylation, or as chromatin remodelers. Given their intercession in multiple aspects of NR function, including transcription, translation, splicing and a variety of functional endpoints such as cell motility, coregulators are essential effectors of the biological activities of NRs and their ligands.
A recent phenomenon in the NR signaling and other transcriptional fields is the growing volume of high content or discovery-driven datasets whose data points are generated not in pursuit of a specific research hypothesis, but rather with the intent of affording broad, unbiased perspectives on the myriad processes that accompany the regulation of gene networks in vivo by NRs and coregulators. This review will begin by discussing the political, cultural and technological factors that are shaping the increasing footprint of discovery-driven research. It will go on to describe the various flavors of high content data and the biological processes they inform upon, illustrating this with examples of insights they have afforded that would not have readily accrued from focused hypothesis-directed research. The review will go onto describe how bioinformatic approaches are being applied to integrate these diverse datasets into working systemic models of NR and coregulator biology. Finally it will describe actively-curated Internet web resources developed by NR scientists which complement traditional publication models and help ensure the unrestricted access of the global NR signaling community to the results of discovery-driven research.
2. Evolution of discovery-driven research in nuclear receptor signaling
While its genomic datasets have been in existence for over a decade, a number of forces have combined in recent years to drive the acceleration of discovery-driven research in the nuclear receptor signaling field. Firstly, funding agencies face greater demands from government for accountability and an increased focus on tangible benefits of publicly-funded research in the form of novel therapeutic approaches to disease [9]. Accordingly, they are increasingly turning to collaborative, discovery-driven scientific consortia to complement and enhance the research strategies pursued by individual investigators. A second factor fueling the progress of high content datasets is the robust evolution in instrumentation and the increased affordability of machines that greatly improve on previous generations in capacity, speed, sensitivity and applicability. Mutliplexed reagents allow for the interrogation of large numbers of molecular species in space and time and advanced labeling and detection systems can rapidly and sensitively discern specific assay molecules in complex biological mixtures. In many cases software analysis platforms are integrated into these instruments, providing for immediate end user analysis of data points. The end result is that genome-wide, transcriptome-wide and proteome-wide analyses are increasingly cost effective, providing a wealth of data that, if properly annotated and archived, will be an invaluable environment for hypothesis generation and testing for the entire field. Indeed such is their perceived potential to revolutionize biomedical research that high throughput analysis platforms are candidates for aggressive investment on the part of the National Institiutes of Health [9]. A final factor fueling the evolution of high content datasets is the growing appreciation by scientific publishers of the need to recognize the merits of these studies as valid and meaningful contributions to scientific research and as enduring resources for the wider research community. This will be discussed in more detail later in this review, but the fact that scientists can now publish discovery-driven datasets in peer-reviewed journals, and have these achievements recognized by institutional promotional committees, augurs well for the future growth and importance of such resources in the field.
3. Categories of discovery-driven research in nuclear receptor and coregulator signaling
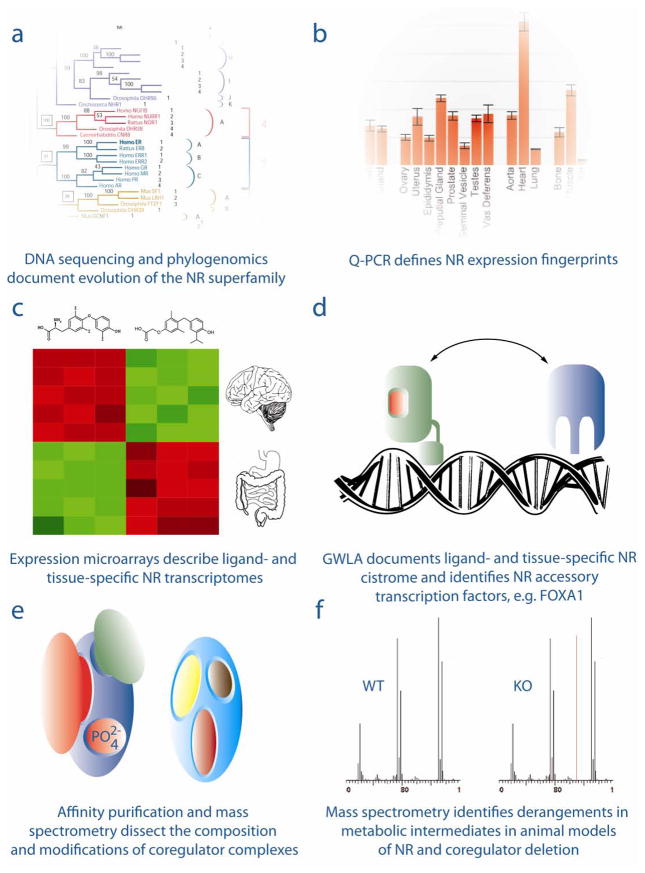
The last decade has seen the evolution of a variety of high content platforms, each of which provides information on distinct but inter-related processes in NR signaling (Figure 1 and Table 1). These include: phylogenomic and genomic analyses which defined the evolution of the NR superfamily in distinct species (Figure 1a); quantitative analysis of the expression patterns of NRs and coregulators (Figure 1b); the definition of ligand- and tissue-specific transcriptomes for NRs and their ligands using expression microarrays (Figure 1c); cistromic analysis of functional genomic sites of occupancy of NRs and related transcription factors (Figure 1d); proteomics-based spatial organization of NRs and coregulators into functional higher order complexes, as well as post-translational modification of these proteins (Figure 1e); and the application of metabolomics to linking disruption of NR or coregulator genes in animal models to imbalances in metabolic intermediates (Figure 1f).
Figure 1.
Six generations of discovery-driven research in nuclear receptor signaling, 1999-2011: a) Phylogenomics, b) NR & NR ligand target gene transcriptomics, c) NR and coregulator gene transcriptomics, d) cistromics, e) proteomics and f) metabolomics. See text for details.
Table 1. Examples of high content datasets and associated publications in the nuclear receptor signaling field.
Examples of high content datasets and associated publications in the nuclear receptor signaling field. Literature references for transcriptomics and cistromics of NR & NR ligand target genes can be found in the Transcriptomics and Cistromics section of specific Molecule Pages on the NURSA website (www.nursa.org).
| System | Subsystem | Technique | Species | Tissue/context | References |
|---|---|---|---|---|---|
| Transcriptomic | NRs | Q-PCR | M. musculus | Multiple adult | [19] |
| M. musculus | Metabolic circadian | [20] | |||
| H. sapiens | Activated macrophages | [29] | |||
| M. musculus | Adipogenesis in 3T3L1 cells | [34] | |||
| M. musculus | Adult brain | [24] | |||
| M. musculus | Osteoblasts, osteogenesis v adipogenesis | [23] | |||
| M. musculus | Development | [21] | |||
| M. musculus | Adult endocrine pancreas | [22] | |||
| H. sapiens | Cancer cell lines | [30] | |||
| H. sapiens | HeLa and HepG2 cells | [25] | |||
| M. musculus & H. sapiens | Embryonic stem cells | [59] | |||
| Coregulators | Q-PCR | M. musculus | Multiple adult | [60] | |
| NR & NR ligand target genes | Expression microarray | Multiple | Multiple | See Legend | |
| Coregulator target genes | Expression microarray | M. musculus | Livers of SRC null mutants | [61] | |
| GWLA (Cistromics) | NR & NR ligand target genes | ChIP-chip and ChIP-seq | Multiple | Multiple | See Legend |
| Coregulator target genes | ChIP-chip and ChIP-seq | H. sapiens | SRC-3 in 17β-estradiol stimulated MCF-7 cells | [45] | |
| Proteomic | Coregulator-coregulator | Affinity purification-mass spectrometry | H. sapiens | HeLa cells | In Press |
| YTH/in silico | Multiple | Multiple | [31] | ||
| NR-coregulator | Affinity purification-mass spectrometry | H. sapiens | ERα-SRC-3 in 17β-estradiol stimulated MCF-7 cells | [45] | |
| Metabolomic | NR | Mass spectrometry | M. musculus | NR models of diabetes and fatty liver disease | |
| Coregulator | Mass spectrometry | M. musculus | Livers of SRC-2 null mutants | [44] |
3.1 Genomics and Phylogenomics
The protein sequences of NRs deduced from their cloned cDNAs had by the late 1980s established the existence of a NR superfamily. It was not until the late 1990s however that the pioneering bioinformatic milestone in the field was reached, namely the drafting of a systematic phylogeny and nomenclature for the superfamily, based on the comparative evolution of the conserved DNA- and ligand-binding domains of its members [10]. The phylogeny described 6 subfamilies and 26 groups of receptors; gene names comprised the prefix "NR" followed by an Arabic numeral for the subfamily, a capital letter for the group and another arabic numeral for individual genes. The largest family, family 1, includes the TR, PPAR, RAR, RevERB, ROR and LXR subfamilies, in addition to FXR, CAR, VDR, PXR. Family 2 includes the COUP-TF, HNF4 and RXR subfamilies, as well as PNR, TR2 and TR4. Additional families include: family 3, which includes the steroid receptors (ERs, GR, MR, PR and AR) and the ERR subfamily; family 4 (NURR1, NOR1, NUR77); family 5 (SF-1 and LRH-1); and family 6 (GCNF). In addition, family 0 included receptors containing only one of the conserved domains, including SHP and DAX1.
Assisted by an almost religious commitment to data archiving, the intensive, heavily-funded genome sequencing efforts of the late 1990s and early 21st century documented the distribution of genes encoding NRs in a variety of species, including 48, 49 and 47 in the human [11], mouse [12] and rat [13] genomes respectively, 21 in the Drosophila melanogaster genome [14] and a surprisingly large number (270) in Caenorhabditis elegans [15]. A comparative genomic analysis of the human, mouse and rat NR superfamilies found that although a high degree of sequence conservation existed across species, the variation was sufficient between some members, such as PXR and CAR to suggest ongoing species-specific adaptations to environmental factors [13]. These initial discovery-driven surveys of NR genes in the context of their cognate genomes laid the groundwork for the rapid accumulation of data that described their transcriptional and functional biologies.
3.2 Transcriptomics
3.2.1 NR and coregulator target gene transcriptomics
The NR signaling field has been extremely active in the production of expression array datasets –a recent study by NURSA archived nearly 1200 individual datasets across over 300 unique tissues and cell lines, 91 ligands and 35 NRs [16] which are accessible in the Molecule Pages for individual NRs and ligands on the NURSA website (see later in this review for a fuller discussion). Expression arrays, when analyzed using appropriate statistical methods [17] have shown themselves in this and other fields to be powerful tools for linking genome-wide fluctuations in gene expression to specific signaling inputs and in turn to broader cellular function. These datasets have emerged largely from hypothesis-driven studies focused on individual genes and while they have been useful as far as those studies have gone, they have been largely under-utilized as a basis for a database of target genes of NRs and their ligands. In a field which is, at its essence, transcriptional in nature, there is a glaring need for such a resource and it is hoped that funding agencies eventually recognize and redress the wide gap in knowledge that exists in its absence.
3.2.2 NR and coregulator gene transcriptomics
3.2.2.1 Broad strokes: anatomical and circadian profiling of nuclear receptors
Until the mid 2000s, NRs as a superfamily had been classified solely based on the phylogenomic approach described above [10], but a pioneering discovery-driven initiative to classify them according to expression pattern, thereby garnering some insight into their functional relationships, was undertaken under the auspices of the Nuclear Receptor Signaling Atlas (NURSA). The NURSA Consortium was the first example in the field of a collaborative consortium model organized to generate and distribute to the community a series of inter-related discovery-driven datasets [18]. The Q-PCR-based study established anatomical expression profiles for the 49 members of the NR superfamily in 39 tissues from two different strains of the most widely used mouse models, C57Bl/6 and 129SvJ [19]. While piecemeal approaches to characterizing NR expression patterns were available in a variety of published reports, they lacked common methodological platforms and protocols, making meaningful comparisons between them difficult. The Q-PCR NR expression profiling depicted anatomical NR expression depicting as a circular dendrogram through bioinformatic clustering of the NR tissue expression profile [19]. The dataset highlighted groups of NRs whose patterns of expression hinted at their potential to coordinate the transcriptional programs necessary to affect physiological pathways along two major axes: 1) reproduction, development, and growth and (2) nutrient uptake, metabolism, and excretion. [19]. In a parallel study to the anatomical profiling, NURSA established for the community a comprehensive, unbiased and quantitative cartography of the expression of all NRs over a 24 hr cycle in white and brown adipose tissue, liver, and skeletal muscle [20]. While these studies are tempered with caveats such as cell-specific expression of receptors, as well as the relationship between mRNA and protein, they were nevertheless an important step towards establishing a framework for hypothesis generation and testing in the field.
3.2.2.2 Filling in the gaps: NR expression in normal physiology and disease
The two initial NURSA Q-PCR NR profiling studies were the first attempt to define, on a single technological platform and using a highly sensitive quantitative method, the steady-state expression levels of NRs. These initial low-resolution passes set the stage for subsequent discovery-driven efforts pursuing more detailed analyses of NR expression during development [21] and in the adult endocrine pancreas [22], bone [23] and brain [24], as well as in two workhorse cell lines, HepG2 and HeLa [25], providing a valuable resource for the many researchers using these cell lines. As with the original NURSA study, these expression profiles identified potential functional relationships between receptors that had not been previously considered.
One of the first key follow-up studies [21] to the NURSA study was one that profiled NR expression during development of a higher organism, namely the zebrafish Danio rerio, which was chosen since its transparency during embryogenesis lent itself to systematic whole mount in situ hybridization. While less amenable to quantification than quantitative PCR, this approach offers the advantage of affording a perspective on the cellular distribution of NRs within a given organ or tissue. Charting the spatiotemporal expression 101 NR and coregulator genes during zebrafish development, this study offered a whole-organism view of their sites of action, and identified their potential functions in central nervous system development. As with a previous NURSA dataset, tissue-to-tissue fluctuations in the expression of coregulators were less dramatic relative to NRs. Based on this the authors asserted that the tissue-specificity of NR action was due to NRs rather than coregulators, but this is likely an oversimplification given the widely-acknowledged potential of coregulator complex composition and post-translational modification to influence the regulation of NR target genes [26, 27]. Extending the embryonic theme into a mammalian setting, a NURSA study profiled NR expression in human and mouse embryonic stem cells and embryoid bodies [28]. The surprisingly large discrepancy in the expression patterns between the two species of certain receptors, ERRβ, DAX-1 and LRH-1 in particular, was taken to indicate distinct species-specific functions in these cases.
Follow-up studies have been extended to several tissues types, including the pancreas, in which substantial divergence in expression of members of the NR superfamily was demonstrated across distinct cell types [22]. Moreover, this study was extended to evaluate the relative expression of NRs during hyperglycemia and showed that16 NRs were significantly altered mRNA levels compared to normal mouse islets. A study in bone provided interesting insights into the possible role of NRs in determining the fate of multipotent cell types, one of which is the calvarial osteoblast, which can follow either an osteogenic or adipogenic differentiation pathway. The study binned NRs into one of four expression clusters, namely: those upregulated during osteogenic, but not adipogenic, differentiation; upregulated in both conditions, with greater upregulation during adipogenesis; upregulated equally in both conditions; and downregulated during adipogenic, but not osteogenic, differentiation [23]. Finally, a Q-PCR-based brain study demonstrated unambiguous grouping of some NRs with no previously known functional implication, such as Coup-TFI and Rev-erbα in a subgroup of brain regions involved in learning and memory [24]. This type of quantum leap in our understanding of the role of NRs normal physiology can only be afforded by large-scale profiling projects of this type, and in concert they establish a solid rationale for future hypothesis-based research.
A substantial amount of evidence implicates individual NRs as causative or preventative agents in a variety of disease states, and so a clear rationale exists to investigate expression patterns across the superfamily in established models of human disease. Macrophage activation plays a central role in atherogenesis, autoimmunity, and a variety of other inflammatory diseases. Barish et al. [29] demonstrated that 28 NRs were expressed in macrophages activated by bacterial lipopolysaccharide or by interferon-γ, with specific temporal induction patterns unique to each stimulus. Extending the analysis to the NCI60 panel of cancer cell lines, a subsequent NURSA study [30] showed that specific NR expression patterns were predictive of the drug responses of individual cell lines, suggesting that multiplex profiling of NR expression patterns in tumors might afford predictability of the sensitivity of tumors to specific NR-based therapeutics.
3.3 Proteomics
NR signaling is driven by protein-protein interactions between NRs and coregulators, as well as between coregulators which associate as components of large modular complexes [8]. Understanding the composition of functional NR-coregulator complexes in specific signaling contexts could provide a basis for the development of novel NR- and coregulator-targeted therapeutics. In comparison to expression arrays and, to a lesser extent Q-PCR, high-content proteomics in the NR signaling field is at a relatively early stage of development. An initial informatics study [31] combined literature mining with yeast two-hybrid screening to assemble a database of NR-coregulator interactions.
More recently the NURSA Consortium has devoted considerable effort documenting the composition of native HeLa cell coregulator complexes using a combination of affinity purification and mass spectrometry [32]. The immunoprecipitation (IP) / mass spectrometry (MS) protocol involves cellular fractionation to generate high concentrations of protein extracts; (ii) a rapid two-step IP protocol with reducing stringency of washes; (iii) SDS-PAGE of the immuno-complex and division of each IP gel lane into 6 regions for sequencing in separate chromatographic runs [33]. The group has developed a software-based approach to filtering out non-specifically binding proteins, which has been a perennial problem in coimmunoprecipitation. These efforts have culminated in the recent release of a large dataset documenting 5000 individual coregulator-coregulator interactions (Malovannaya et al., In Press). Although at an early point its development, high-content analysis of the many post-translational modifications in NR and coregulator signal transduction [8] will be an important source of information in building models for the role of cellular signaling pathways in regulating the activity of NRs and coregulators.
The NURSA proteomics datasets demonstrate how a discovery-driven dataset can provide an experimental framework for drawing parallels between distinct biological systems that were not previously inferred. Kittler et al. [34] carried out a genome wide screen for RNA interference (RNAi) screen for genes important for cell division. Among other findings, they identified a set of transcriptional regulators whose knockdown resulted in defects in cytokinesis, one of which was SMRT/NCOR2. Bioinformatic analysis implied a functional interaction between SMRT/NCOR2, TBL1X and MLL5, an inference the authors were able to confirm with reference to the presence of TBL1X and MLL5 in a HeLa cell SMRT complex from Mitch Lazar’s project (10.1621/datasets.01002). Another study [35] described the cloning and characterization of a novel coregulator of ERα, CCAR1. Again, the authors leveraged the observation of the presence of CCAR1 proteolytic fragments in a TRAP230/MED12 complex characterized by the NURSA proteomics effort [32] to confirm their own data suggesting that CCAR1 was associated with Mediator complexes.
3.4 Cistromics and Epigenomics
While expression microarrays provide a valuable global perspective on the collective transcriptional response to a given stimulus, they are insufficient, in the context of NR-mediated signaling, to implicate the specific receptors, coregulators or ancillary transcription factors that mediate those responses. Moreover they are not designed to distinguish between directly and indirectly-regulated genes, nor the contribution, if any, of pre-genomic ligand functions to the transcriptional output. The relatively new discipline of genome-wide location analysis (GWLA) encompasses a group of techniques which provide data on the specific genomic locations of NRs and coregulators in specific signaling contexts. The term “cistromics’ was first coined by Myles Brown’s laboratory –recognized as the pioneer in the field - in the latter part of the last decade to describe “the complete set of cis-acting targets of a transacting factor across the genome [36]. Early adopters of GWLA used affinity purification of protein-DNA mixtures (chromatin IP, or ChIP) coupled to solid phase tiled arrays (ChIP-chip) but this technique has been largely supplanted by the advent of massively-parallel sequencing platforms in the form of ChIP-seq. The GWLA waters are muddied to an extent by the fact that DNA-bound NRs can influence transcriptional events at promoters far distant from their sites of occupancy, both proximal and distal, such that tying transcription of a specific gene to a specific DNA-binding event can be done only tentatively. Brown’s groundbreaking papers demonstrated unequivocally that crosstalk on DNA of ERα with transcription factors such as FoxA1 was an early event in the regulation of 17β-estradiol/ERα target genes in MCF-7 cells [37, 38] (Figure 1d). A more recent variant on the GWLA theme is GRO-Seq (Global Run-On-seq), a deep sequencing approach that acts as a measure of direct transcriptional output by mapping the location and orientation of RNA Polymerase II [39]. Lee Kraus’ laboratory has applied the technique to 17β-estradiol stimulation of MCF-7 cells, opening up the possibility of defining the time course of regulation of 17βE2 directly-regulated transcripts [40]. In addition to cistromics, a nascent discipline is epigenomics, namely the genome-wide study of covalent modifications of DNA by NRs and associated coregulators, and the impact of these interactions on tissue-specific NR-regulated transcriptomes. Initial studies in this area have cast histones in the role of fine-tuners of NR binding and suggesting a role in tissue-specific patterns of NR recruitment to target genes. For example, PPARγ binds preferentially to genomic regions divested of repressive histone H3 lysine 9 dimethylation (H3K9me2) and H3K27me3 marks in 3T3-L1 cells undergoing adipogenesis [41]. Moreover, the H3K4me2 mark is a key determinant of tissue-specific binding by FOXA1 and ERα [42] and enrichment of the histone variant H2A.z is observed in hormone-induced GR-accessible sites [43]. Future studies will undoubtedly shed further light on the role of chromatin-NR-coregulator interactions in regulating cell-type specific expression of NR cistromes.
3.5 Metabolomics
The research output of laboratories such as Bruce Spiegelman, Ronald Evans, Bert O’Malley and others over the past 20 years has drawn the field inexorably to the realization that both NRs and coregulators are key players in carbohydrate, fat and lipid metabolism and, as such, are eminently positioned as therapeutic leverage points in a broad spectrum of diseases of Western society including diabetes, obesity and the metabolic syndrome. Metabolomics is the systematic profiling of metabolic intermediates involved in biochemical processes in cellular systems, and two studies from the NURSA Consortium illustrate the power of an unbiased metabolomic approach to identify subtle but critical fluctuations in metabolic intermediates that clarify the phenotypes of animal models with altered expression of NRs or coregulators. The first demonstrated that knockout of SRC-2/Ncoa2 has a very specific metabolic phenotype of hypoglycemia and hepatic overstorage of glycogen, similar to the human genetic disorder type 1 glycogen storage disease or Von Gierke's disease [44]. More recently, work from the NURSA Consortium has demonstrated that knockout of SRC-1/Ncoa1 has a more complex metabolic phenotype, involving impaired gluconeogenesis, decreased glycolytic flux, and compensatory increases in fatty acid and amino acid oxidation (Louet et al. Cell, in revision, 2009).
3.6 Small Molecule Modulators
As druggable proteins, NRs and their associated coregulators are eminently positioned as therapeutic targets in a wide variety of diseases and disorders. While NR modulator-based research platforms are well established in private pharmaceutical companies, notably GlaxoSmithKline, Johnson & Johnson, Wyeth Pharmaceuticals and others, the field to date has lacked a public discovery-driven effort to generate small molecule modulators and ligands for the use of the research community. Similarly, while for-fee access databases such as that of the American Chemical Society are comprehensive, public resources such as NCBI PubChem remain in a developmental stage– ligands for specific NRs are not yet linked to records for those receptors in NCBI Entrez. A recent effort to redress this imbalance and provide the broader research community with a high throughput screening service is the Molecular Libraries Program (MLP). The goal of the MLP is to provide researchers with what would otherwise be the prohibitively expensive chemistry, screening and informatics support required to probe the role of small molecule modulators in cellular pathways.
4. The role of bioinformatics
At its essence, bioinformatics complements discovery-driven research efforts by (i) developing computational and mathematical approaches to detect statistically significant patterns and trends across complex and diverse discovery-driven datasets; and (ii) developing web applications designed to facilitate the annotation, widespread accessibility and enduring availability of these datasets to a userbase of technologically diverse biologists. In this section I will provide a recent example of an integrative analysis of high content datasets, as well as discussing currently-curated web resources of relevance to this field.
4.1 Bioinformatics challenge I: integrative analysis of discovery-driven datasets
An important bioinformatic goal is the successful integration of the myriad types of high content data depicted in Figure 1 with the goal of developing reliable paradigms for NR signaling, within which the posing and testing of hypotheses might take place. In a landmark study Lanz et al. established a paradigm for how bench transcriptomics, cistromics and proteomics could be married by integrative bioinformatics in a well-established model for NR action, namely 17β-estradiol stimulation of MCF-7 cells [45]. In doing so, they framed a working model for how functional interactions between activated NRs, coregulators and DNA give rise to efficient regulation of gene networks in vivo. The first component of the analysis was an affinity purification/mass spectrometric analysis of ERα and SRC-3-associated proteins in MCF-7 cells treated with 17βE2. The second component was a previous study from NURSA, namely the GEMS 17βE2/MCF-7 expression microarray meta-analysis dataset [46], which drew upon the combined statistical power of multiple aligned microarray experiments to establish a consensus transcriptional response in this system. The final components were an SRC-3/NCOA3 GWLA analysis in 17βE2-treated MCF-7 cells, in addition to previously published GWLA experiments in the same model system [38]. The increased confidence resulting from integrating multiple experimental approaches allowed the group to paint ontology annotations onto the 17βE2 transcriptome and common ERα/SRC-3 ChIP binding signatures, effectively correlating the system’s endpoint biology with promoter occupancy. While confirming previously characterized functions of SRC-3 on 17βE2 target genes (TFF1, XBP1), this analysis also identified SRC-3 and ERα binding on previously unrecognized ERα targets such as ERBB2. Moreover, the unbiased approach identified an anticipated high degree of convergence between ERα and SRC-3 binding sites in addition to significant overlap between SRC-3 and FoxA1 which was taken to represent an unexpected convergence in the biology of these proteins [45].
4.2 Bioinformatics challenge II: archiving and communicating discovery-driven research
Since the mid 19th century, the peer-reviewed primary research article that establishes a hypothesis, describes the methods, reports the results of the experiments and interprets their significance, has been the mainstay of scientific communication. While printed journals were adequately scoped to accommodate the discrete, circumscribed content these articles contained, they were never designed to communicate the high content datasets this review has discussed. Expression microarrays, for example, demonstrate the glaring lack of infrastructure in place in journals, even those on advanced electronic publishing platforms, to effectively accommodate high content datasets. The expedient in this case has been that due to printed page constraints, the authors select a handful of genes that show significant regulation - the so-called gene list - and communicate those in the paper. The actual array data, if available at all, appears often as a flat PDF in supplemental materials, and even rarer still, is submitted to a publically-accessible database. This situation is unsatisfactory at best, leaving the vast majority of expression array data not only unreported, but refractory to further analysis by other investigators [47].
The opacity of the traditional research article to data mining, and the rapid evolution in Internet technologies that provide for rich, intuitive user experiences in websites, have driven the evolution of a new generation of biological databases in which a firm emphasis is placed on usability, that is, the interface between the human researcher and the biological data. Many of these sites – NCBI (National Center for Biotechnology Information) PubMedb, NCBI PubChem, NCBI GEO (Gene Expression Omnibus) and EBI UniProt to name a few – are generic, broad spectrum resources with whole genome coverage. In addition to these, the NR and coregulator field has a number of actively-curated, freely-accessible specialist databases (Table 2).
Table 2. Specialist web resources in nuclear receptor signaling.
Web and bioinformatics resources in the nuclear receptor signaling field. Note that only currently curated, unrestricted-access sites are listed here. Many other databases existed at one time but are either no longer actively curated or available on the Internet. URLs: NURSA, www.nursa.org; Nuclear Receptor Resource, http://nrresource.org/; IUPHAR, http://www.iuphar-db.org/DATABASE/ReceptorFamiliesForward?type=NHR; Androgen Receptor Mutation Database http://androgendb.mcgill.ca/; Androgen-Responsive Gene Database, http://argdb.fudan.edu.cn.
| Database | Curated Information and Data1 | Origin |
|---|---|---|
|
NURSA
www.nursa.org |
Q-PCR expression of nuclear receptors | NURSA |
| Affinity purification/Mass spectrometry of coregulator complexes | NURSA | |
| Expression microarrays of NR target genes | NURSA | |
| ChIP-seq of NR cistromes | NURSA | |
| NR and CoR interactions | NURSA, Entrez Gene | |
| Ortholog information | Various | |
| NR ligands | NCBI PubChem | |
| NR and NR ligand transcriptomes and cistromes | NCBI PubMed, GEO | |
| NR & CoR pathology | NCBI PubMed OMIM Morbid Map HPRD, GAD |
|
| NR & CoR animal model phenotypes | Jackson Laboratory | |
| Literature | NCBI PubMed, Molecular Endocrinology | |
| Educational Resources | NURSA | |
|
Nuclear Receptor Resource http://nrresource.org |
Pathways | NRR |
| Educational Resources | NRR | |
| Expression | NURSA | |
|
IUPHAR
http://www.iuphar-db.org/DATABASE/ReceptorFamiliesForward?type=NHR |
Ortholog information | Various |
| Ligands | IUPHAR | |
| NR and CoR Interactions | NCBI PubMed | |
| Target Genes | NCBI PubMed | |
| Animal models | NCBI PubMed | |
|
Androgen Receptor Mutation Database
http://androgendb.mcgill.ca/ |
Androgen receptor gene mutations | NCBI PubMed |
| Androgen receptor-interacting proteins | NCBI PubMed | |
|
Androgen-Responsive Gene Database http://argdb.fudan.edu.cn |
Androgen & androgen receptor target genes | NCBI PubMed |
4.2.1 NURSA
The Molecule Pages of the NURSA website have evolved since their launch in 2003 into a portal which encompasses a broad variety of biological resources of relevance to NRs, their ligands, coregulators and target genes, both from datasets generated as part of the NURSA Consortium as well as from wider community sources (Table 2). These pages include links to cDNA and genomic DNA sequence repositories such as the NCBI database collection (including NCBI RefSeq, GenBank and UniGene); protein sequence repositories such as the UniProt/EBI-databases SwissProt, Pfam and the protein structural and crystallographic database PDB; portals for species-specific gene collections such as HUGO (human), MGI (for mouse data), RGD (rat); gene expression resources (NCBI GEO, MousePAT, Allen Brain Atlas); reciprocal links with specialist resources such as the FAST-DB transcript & splicing resource and the Phosphosite Plus post-translational protein modification resource; and NCBI's literature resource, NCBI PubMed. NURSA has engaged members of both the academic and industrial communities in developing its NR ligand database, which integrates information from resources such as NCBI PubChem and PDB on nearly 250 NR ligands. Other content includes a Diseases and Phenotypes module which integrates curated, literature-based information on the disease involvement and animal model phenotypes of NRs and coregulators from generic resources such as OMIM, HPRD, GAD (Table 2). An Interactions module combines literature-based information on the protein-protein interactions of NRs and coregulators from NCBI, as well as coregulator proteomics data generated by NURSA (Figure 1e). The most recent addition is a Transcriptomics and Cistromics module, which catalogs published expression microarray and GWLA studies related to NRs and their ligands and annotates them for tissue or cell line and species of study [16].
The website also has an electronic publishing alliance with the premier specialist research journal in the field, Molecular Endocrinology, through which reciprocal links are established between journal articles and related Molecule Pages on the NURSA website. These annotations provide article readers with one-click access to contextual information, and NURSA website users with targeted content from a respected journal in the NR signaling field.
4.2.2 IUPHAR
The International Union of Basic and Clinical Pharmacology (IUPHAR), formerly the International Union of Pharmacology, was established in 1959 as a forum for scientists in a variety of pharmacology-related fields, including NRs, G-protein coupled receptors and others. IUPHAR has taken a leading role in establishing nomenclatures for gene families and regularly solicits reviews from leading investigators in specific fields to summarize recent research [48, 49]. IUPHAR has recently undertaken an initiative to migrate expert-curated content from its reviews into relational databases, and to establish links with other generic and specialist resources to provide contextual information for its userbase. The IUPHAR database on NRs (Table 2) provides information on NR orthologs, ligands, interactions, target genes and animal models that was originally published in a series of reviews on the NR field. As a specialist pharmacology resource, IUPHAR is uniquely placed to provide expert curation in the area of NR ligands, and it is hoped that future versions of the database will continue to add to its content in this area for the benefit of users of this and other databases in the field.
4.2.3 Nuclear Receptor Resource
The Nuclear Receptor Resource was the first presence for the NR community on the Internet and included several interconnected but distinct web resources focussed on difference receptors or receptor subfamilies [50], see also below. Several of the sites are unfortunately no longer actively curated and those that remain have been consolidated into a single resource that provides educational resources, information on NR-regulated pathways, NR interactions and NR expression patterns, reproduced from the original NURSA expression studies [19, 20] (see Table 2 for URL). The site also links to a tool for searching for PPAR response elements in any given genomic DNA sequence.
4.2.4 Androgen Receptor Mutations Database
This well-established resource, launched in 1998 and updated monthly, documents published and unpublished somatic mutations and polymorphisms in the human AR gene in a variety of diseases, including prostate cancer and diseases associated with CAG tract length variations. It also includes a list of AR-interacting proteins curated from the published literature (see Table 2 for URL).
4.2.5 Androgen-Responsive Gene Database
This web resource is based on intensive curation of the published literature to assemble a database of a total of 3300 human, mouse and rat androgen-regulated genes, along with essential hand-curated metadata such as expression fold change, androgen-responsive sequence (where available), response time, tissue/cell type, experimental method, and ligand identity and concentration, The database is integrated with multiple external resources, including NCBI, Gene Ontology, and Kyoto Encyclopedia of Genes and Genomes pathway, to afford the user convenient access to information on the biological characteristics and context of androgen-regulated genes (see Table 2 for URL).
In addition to these active bioinformatic resources, many useful databases existed at one time but have since gone offline or are no longer actively curated, presumably due to lack of financial support. These include NUREBASE [51], NucleaRDB [52], NRSAS [53], NRMD [54], ERGDB [55], ERTargetDB [56] and KBERG [57]. Some of the earliest efforts to establish a presence on the Internet for the NR signaling community occurred under the auspices of the Nuclear Receptor Resource [50], and included Mark Danielsen’s Glucocorticoid Receptor Resource and David Moore’s Thyroid Receptor Resource, both of which are now defunct. The loss of the time and effort that were in invested in collecting, curating and maintaining these and other databases is highly regrettable, and there is a glaring need for funding agencies worldwide to recognize the value to the research community of continuing investing in web-based resources in this field in order to ensure their ongoing curation. Conceivably, funding could be made available, at least for American resources, through the US National Library of Medicine to maintain and ensure the continued curation of some of the larger resources for posterity.
5. Concluding remarks
Supported by an increasingly sophisticated data gathering analysis and distribution infrastructure, discovery-driven research has made tangible progress over the last decade or so in deconstructing the baroque biology of NRs and their coregulators. The next challenge at the bench level is one of resolution: to date, most large scale data acquisitions have been at the level of the tissue or organ, glossing over issues such as the cell- and promoter-specificity of NR and coregulator function. Single cell- and organelle- level analyses will provide valuable information on the subcellular dynamics of NRs and coregulators, the function of splice forms of these factors, the role of specific post-translational modifications in fine-tuning their modes of action. Extrapolating these observations to fluctuations in single-cell metabolomes will conceivably provide for more accurate information on the relationship between ligands, NRs, coregulators and homeostatic control at the cellular level. Finally, defining the relationship at the single cell level between these factors and the regulation of cell division and motility will undoubtedly expose leverage points for therapeutic intervention in the myriad neoplastic diseases in which they are implicated. Gathering, archiving and analyzing these data in anintegrated fashion will be a tremendous experimental and bioinformatic challenge, but the payoff in terms of more focused, efficient and cost-effective therapies, is likely to be commensurate.
Despite its size and demonstrable relevance to health and disease however, the field of NR signaling has a relatively small number of specialist, publically-available web and bioinformatic resources. Given the number of established journals in the field this has until recent years not been a pressing concern, but the growing number and diversity of high content technological platforms indicates that dataset archiving, analysis and distribution will become critical elements of future scientific progress in this field. The daunting volume of data points that will be generated by these and other initiatives over the next decade warrants a committed effort on the part of researchers, funding agencies and publishers to ensure that the bioinformatic infrastructure is in place to ensure the effective management of these data resources, and that data mining does not develop into a bottleneck to restrict progress in the field.
While this review has to date focused on NR and coregulator signaling-specific resources, their multifaceted roles in physiology and disease argues strongly for existing and future bioinformatics efforts to engage resources in parallel disciplines with a view to sharing database resources for the mutual benefit of their respective user bases. Consider, for example, a clinician searching a diabetes-focused database who is empowered to view expression of NRs and coregulators in the diabetic endocrine pancreas, or a NR scientist who is able to tap into cancer gene expression data from another database, filtered by NR or coregulator Gene IDs. The synergy that arises from the intersection of the data in distinct databases in these and otherinstances can be readily appreciated, and the incipient efforts of the National Institute of Diabetes, Digestive and Kidney Diseases in establishing dkCOIN [58] bodes well in this regard. This network of databases is being developed to establish interconnectivity between molecule-, disease- and organ- and mouse model-centric databases to provide clinicians and scientists with access to data resources beyond their own sphere of research or interest.
Their full value of high-content datasets will be realized only with their full unrestricted availability, and it is incumbent on the community to ensure they are archived and annotated with the same alacrity as were DNA sequences in the 1980s and 1990s. Unlike sequence data, these datasets are highly contextual in nature and must be associated with detailed critical metadata that articulate these contexts. Without a commitment on the part of funding agencies to funding and maintaining databases curated by experts in the field, there can be little doubt that much of it will be lost to posterity. Equally, if publishers do not apply rigorous standard to the deposition of high content datasets in appropriate public repositories, much of it will never reach the public domain, as has regrettably been the case for expression array datasets [47]. If these goals can be achieved however, discovery-driven research and bioinformatics will undoubtedly form the foundation of significant strides in our understanding of NR and coregulator biology in the next decade and beyond.
Acknowledgments
NJM is supported by NIDDK U19-DK62434. The author regrets the omission of many important contributions to the field on the basis of space constraints. The comments of Dr Austin Cooneyare gratefully acknowledged.
Footnotes
Familiar and official symbols for all molecules described in this review can be found on the Nuclear Receptor Signaling Atlas (NURSA) home page.
URLs for generic bioinformatics resources are as follows: NCBI PubMed: www.ncbi.nlm.nih.gov/pubmed; NCBI PubChem: http://pubchem.ncbi.nlm.nih.gov; NCBI GEO www.ncbi.nlm.nih.gov/geo; NCBI OMIM: www.ncbi.nlm.nih.gov/omim; EBI UniProt www.uniprot.org; NCBI RefSeq: www.ncbi.nlm.nih.gov/refseq; NCBI GenBank: www.ncbi.nlm.nih.gov/genbank; NCBI UniGene www.ncbi.nlm.nih.gov/unigene; EBI UniProt http://www.uniprot.org; Pfam: http://pfam.sanger.ac.uk; PDB: www.pdb.org; HUGO: www.genenames.org; MGI: www.informatics.jax.org; RGD: http://rgd.mcw.edu; HPRD: www.hprd.org; GAD: http://geneticassociationdb.nih.gov; Phosphosite Plus: www.phosphosite.org; Molecular Libraries Program: http://mli.nih.gov.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tsai MJ, O'Malley BW. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu Rev Biochem. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- 2.Huang W, Glass CK. Nuclear receptors and inflammation control: molecular mechanisms and pathophysiological relevance. Arterioscler Thromb Vasc Biol. 30:1542–1549. doi: 10.1161/ATVBAHA.109.191189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berlin M. Recent advances in the development of novel glucocorticoid receptor modulators. Expert Opin Ther Pat. 20:855–873. doi: 10.1517/13543776.2010.493876. [DOI] [PubMed] [Google Scholar]
- 4.Sharifi N. New agents and strategies for the hormonal treatment of castration-resistant prostate cancer. Expert Opin Investig Drugs. 19:837–846. doi: 10.1517/13543784.2010.494178. [DOI] [PubMed] [Google Scholar]
- 5.Warner M, Gustafsson JA. The role of estrogen receptor beta (ERbeta) in malignant diseases--a new potential target for antiproliferative drugs in prevention and treatment of cancer. Biochem Biophys Res Commun. 396:63–66. doi: 10.1016/j.bbrc.2010.02.144. [DOI] [PubMed] [Google Scholar]
- 6.Onate SA, Tsai SY, Tsai MJ, O'Malley BW. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 7.Lonard DM, O'Malley BW. The expanding cosmos of nuclear receptor coactivators. Cell. 2006;125:411–414. doi: 10.1016/j.cell.2006.04.021. [DOI] [PubMed] [Google Scholar]
- 8.McKenna NJ, O'Malley BW. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108:465–474. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- 9.Collins FS. Research agenda. Opportunities for research and NIH. Science. 327:36–37. doi: 10.1126/science.1185055. [DOI] [PubMed] [Google Scholar]
- 10.NRNC. A unified nomenclature system for the nuclear receptor superfamily. Cell. 1999;97:161–163. doi: 10.1016/s0092-8674(00)80726-6. [DOI] [PubMed] [Google Scholar]
- 11.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowski J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 12.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, Antonarakis SE, Attwood J, Baertsch R, Bailey J, Barlow K, Beck S, Berry E, Birren B, Bloom T, Bork P, Botcherby M, Bray N, Brent MR, Brown DG, Brown SD, Bult C, Burton J, Butler J, Campbell RD, Carninci P, Cawley S, Chiaromonte F, Chinwalla AT, Church DM, Clamp M, Clee C, Collins FS, Cook LL, Copley RR, Coulson A, Couronne O, Cuff J, Curwen V, Cutts T, Daly M, David R, Davies J, Delehaunty KD, Deri J, Dermitzakis ET, Dewey C, Dickens NJ, Diekhans M, Dodge S, Dubchak I, Dunn DM, Eddy SR, Elnitski L, Emes RD, Eswara P, Eyras E, Felsenfeld A, Fewell GA, Flicek P, Foley K, Frankel WN, Fulton LA, Fulton RS, Furey TS, Gage D, Gibbs RA, Glusman G, Gnerre S, Goldman N, Goodstadt L, Grafham D, Graves TA, Green ED, Gregory S, Guigo R, Guyer M, Hardison RC, Haussler D, Hayashizaki Y, Hillier LW, Hinrichs A, Hlavina W, Holzer T, Hsu F, Hua A, Hubbard T, Hunt A, Jackson I, Jaffe DB, Johnson LS, Jones M, Jones TA, Joy A, Kamal M, Karlsson EK, Karolchik D, Kasprzyk A, Kawai J, Keibler E, Kells C, Kent WJ, Kirby A, Kolbe DL, Korf I, Kucherlapati RS, Kulbokas EJ, Kulp D, Landers T, Leger JP, Leonard S, Letunic I, Levine R, Li J, Li M, Lloyd C, Lucas S, Ma B, Maglott DR, Mardis ER, Matthews L, Mauceli E, Mayer JH, McCarthy M, McCombie WR, McLaren S, McLay K, McPherson JD, Meldrim J, Meredith B, Mesirov JP, Miller W, Miner TL, Mongin E, Montgomery KT, Morgan M, Mott R, Mullikin JC, Muzny DM, Nash WE, Nelson JO, Nhan MN, Nicol R, Ning Z, Nusbaum C, O'Connor MJ, Okazaki Y, Oliver K, Overton-Larty E, Pachter L, Parra G, Pepin KH, Peterson J, Pevzner P, Plumb R, Pohl CS, Poliakov A, Ponce TC, Ponting CP, Potter S, Quail M, Reymond A, Roe BA, Roskin KM, Rubin EM, Rust AG, Santos R, Sapojnikov V, Schultz B, Schultz J, Schwartz MS, Schwartz S, Scott C, Seaman S, Searle S, Sharpe T, Sheridan A, Shownkeen R, Sims S, Singer JB, Slater G, Smit A, Smith DR, Spencer B, Stabenau A, Stange-Thomann N, Sugnet C, Suyama M, Tesler G, Thompson J, Torrents D, Trevaskis E, Tromp J, Ucla C, Ureta-Vidal A, Vinson JP, Von Niederhausern AC, Wade CM, Wall M, Weber RJ, Weiss RB, Wendl MC, West AP, Wetterstrand K, Wheeler R, Whelan S, Wierzbowski J, Willey D, Williams S, Wilson RK, Winter E, Worley KC, Wyman D, Yang S, Yang SP, Zdobnov EM, Zody MC, Lander ES. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Z, Burch PE, Cooney AJ, Lanz RB, Pereira FA, Wu J, Gibbs RA, Weinstock G, Wheeler DA. Genomic analysis of the nuclear receptor family: new insights into structure, regulation, and evolution from the rat genome. Genome Res. 2004;14:580–590. doi: 10.1101/gr.2160004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, George RA, Lewis SE, Richards S, Ashburner M, Henderson SN, Sutton GG, Wortman JR, Yandell MD, Zhang Q, Chen LX, Brandon RC, Rogers YH, Blazej RG, Champe M, Pfeiffer BD, Wan KH, Doyle C, Baxter EG, Helt G, Nelson CR, Gabor GL, Abril JF, Agbayani A, An HJ, Andrews-Pfannkoch C, Baldwin D, Ballew RM, Basu A, Baxendale J, Bayraktaroglu L, Beasley EM, Beeson KY, Benos PV, Berman BP, Bhandari D, Bolshakov S, Borkova D, Botchan MR, Bouck J, Brokstein P, Brottier P, Burtis KC, Busam DA, Butler H, Cadieu E, Center A, Chandra I, Cherry JM, Cawley S, Dahlke C, Davenport LB, Davies P, de Pablos B, Delcher A, Deng Z, Mays AD, Dew I, Dietz SM, Dodson K, Doup LE, Downes M, Dugan-Rocha S, Dunkov BC, Dunn P, Durbin KJ, Evangelista CC, Ferraz C, Ferriera S, Fleischmann W, Fosler C, Gabrielian AE, Garg NS, Gelbart WM, Glasser K, Glodek A, Gong F, Gorrell JH, Gu Z, Guan P, Harris M, Harris NL, Harvey D, Heiman TJ, Hernandez JR, Houck J, Hostin D, Houston KA, Howland TJ, Wei MH, Ibegwam C, Jalali M, Kalush F, Karpen GH, Ke Z, Kennison JA, Ketchum KA, Kimmel BE, Kodira CD, Kraft C, Kravitz S, Kulp D, Lai Z, Lasko P, Lei Y, Levitsky AA, Li J, Li Z, Liang Y, Lin X, Liu X, Mattei B, McIntosh TC, McLeod MP, McPherson D, Merkulov G, Milshina NV, Mobarry C, Morris J, Moshrefi A, Mount SM, Moy M, Murphy B, Murphy L, Muzny DM, Nelson DL, Nelson DR, Nelson KA, Nixon K, Nusskern DR, Pacleb JM, Palazzolo M, Pittman GS, Pan S, Pollard J, Puri V, Reese MG, Reinert K, Remington K, Saunders RD, Scheeler F, Shen H, Shue BC, Siden-Kiamos I, Simpson M, Skupski MP, Smith T, Spier E, Spradling AC, Stapleton M, Strong R, Sun E, Svirskas R, Tector C, Turner R, Venter E, Wang AH, Wang X, Wang ZY, Wassarman DA, Weinstock GM, Weissenbach J, Williams SM, Woodage T, Worley KC, Wu D, Yang S, Yao QA, Ye J, Yeh RF, Zaveri JS, Zhan M, Zhang G, Zhao Q, Zheng L, Zheng XH, Zhong FN, Zhong W, Zhou X, Zhu S, Zhu X, Smith HO, Gibbs RA, Myers EW, Rubin GM, Venter JC. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 15.Sluder AE, Maina CV. Nuclear receptors in nematodes: themes and variations. Trends Genet. 2001;17:206–213. doi: 10.1016/s0168-9525(01)02242-9. [DOI] [PubMed] [Google Scholar]
- 16.Ochsner SA, Watkins CM, Lagrone BS, Steffen DL, McKenna NJ. Tissue-Specific Transcriptomics and Cistromics of Nuclear Receptor Signaling: A Web Research Resource. Mol Endocrinol. doi: 10.1210/me.2010-0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Solomon PJ. Some statistics in bioinformatics: the fifth Armitage Lecture. Stat Med. 2009;28:2833–2856. doi: 10.1002/sim.3668. [DOI] [PubMed] [Google Scholar]
- 18.McKenna NJ, Cooney AJ, DeMayo FJ, Downes M, Glass CK, Lanz RB, Lazar MA, Mangelsdorf DJ, Moore DD, Qin J, Steffen DL, Tsai MJ, Tsai SY, Yu R, Margolis RN, Evans RM, O'Malley BW. Minireview: Evolution of NURSA, the Nuclear Receptor Signaling Atlas. Mol Endocrinol. 2009;23:740–746. doi: 10.1210/me.2009-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bookout AL, Jeong Y, Downes M, Yu RT, Evans RM, Mangelsdorf DJ. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell. 2006;126:789–799. doi: 10.1016/j.cell.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, Mangelsdorf DJ, Evans RM. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 21.Bertrand S, Thisse B, Tavares R, Sachs L, Chaumot A, Bardet PL, Escriva H, Duffraisse M, Marchand O, Safi R, Thisse C, Laudet V. Unexpected novel relational links uncovered by extensive developmental profiling of nuclear receptor expression. PLoS Genet. 2007;3:2085–2100. doi: 10.1371/journal.pgen.0030188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chuang JC, Cha JY, Garmey JC, Mirmira RG, Repa JJ. Nuclear hormone receptor expression in the endocrine pancreas. Mol Endocrinol. 2008;22:2353–2363. doi: 10.1210/me.2007-0568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pirih FQ, Abayahoudian R, Elashoff D, Parhami F, Nervina JM, Tetradis S. Nuclear Receptor Profile in Calvarial Bone Cells Undergoing Osteogenic Versus Adipogenic Differentiation. J Cell Biochem. 2008;105:1316–1326. doi: 10.1002/jcb.21931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gofflot F, Chartoire N, Vasseur L, Heikkinen S, Dembele D, Le Merrer J, Auwerx J. Systematic gene expression mapping clusters nuclear receptors according to their function in the brain. Cell. 2007;131:405–418. doi: 10.1016/j.cell.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 25.Jeong Y, Bookout AL, Mangelsdorf DJ. Quantitative mRNA Expression Profile of Nuclear Receptor Superfamily in HepG2 and HeLa Cells (Nuclear Receptor Signaling Atlas) 2007. http://www.nursa.org/datasets.cfm?doi=10.1621/datasets.04011.
- 26.Korzus E, Torchia J, Rose DW, Xu L, Kurokawa R, McInerney EM, Mullen TM, Glass CK, Rosenfeld MG. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 27.Wu RC, Smith CL, O'Malley BW. Transcriptional regulation by steroid receptor coactivator phosphorylation. Endocr Rev. 2005;26:393–399. doi: 10.1210/er.2004-0018. [DOI] [PubMed] [Google Scholar]
- 28.Xie CQ, Jeong Y, Fu M, Bookout AL, Garcia-Barrio MT, Sun T, Kim BH, Xie Y, Root S, Zhang J, Xu RH, Chen YE, Mangelsdorf DJ. Expression profiling of nuclear receptors in human and mouse embryonic stem cells. Mol Endocrinol. 2009;23:724–733. doi: 10.1210/me.2008-0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barish GD, Downes M, Alaynick WA, Yu RT, Ocampo CB, Bookout AL, Mangelsdorf DJ, Evans RM. A Nuclear Receptor Atlas: macrophage activation. Mol Endocrinol. 2005;19:2466–2477. doi: 10.1210/me.2004-0529. [DOI] [PubMed] [Google Scholar]
- 30.Holbeck S, Chang J, Best AM, Bookout AL, Mangelsdorf DJ, Martinez ED. Expression profiling of nuclear receptors in the NCI60 cancer cell panel reveals receptor-drug and receptor-gene interactions. Mol Endocrinol. 24:1287–1296. doi: 10.1210/me.2010-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Albert S, Gaudan S, Knigge H, Raetsch A, Delgado A, Huhse B, Kirsch H, Albers M, Rebholz-Schuhmann D, Koegl M. Computer-assisted generation of a protein-interaction database for nuclear receptors. Mol Endocrinol. 2003;17:1555–1567. doi: 10.1210/me.2002-0424. [DOI] [PubMed] [Google Scholar]
- 32.Jung SY, Malovannaya A, Wei J, O'Malley BW, Qin J. Proteomic analysis of steady-state nuclear hormone receptor coactivator complexes. Mol Endocrinol. 2005;19:2451–2465. doi: 10.1210/me.2004-0476. [DOI] [PubMed] [Google Scholar]
- 33.Malovannaya A, Li Y, Bulynko Y, Jung SY, Wang Y, Lanz RB, O'Malley BW, Qin J. Streamlined analysis schema for high-throughput identification of endogenous protein complexes. Proc Natl Acad Sci U S A. 107:2431–2436. doi: 10.1073/pnas.0912599106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fu M, Sun T, Bookout AL, Downes M, Yu RT, Evans RM, Mangelsdorf DJ. A Nuclear Receptor Atlas: 3T3-L1 adipogenesis. Mol Endocrinol. 2005;19:2437–2450. doi: 10.1210/me.2004-0539. [DOI] [PubMed] [Google Scholar]
- 35.Kim JH, Yang CK, Heo K, Roeder RG, An W, Stallcup MR. CCAR1, a key regulator of mediator complex recruitment to nuclear receptor transcription complexes. Mol Cell. 2008;31:510–519. doi: 10.1016/j.molcel.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lupien M, Brown M. Cistromics of hormone-dependent cancer. Endocr Relat Cancer. 2009;16:381–389. doi: 10.1677/ERC-09-0038. [DOI] [PubMed] [Google Scholar]
- 37.Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, Fox EA, Silver PA, Brown M. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 38.Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, Wang Q, Bekiranov S, Sementchenko V, Fox EA, Silver PA, Gingeras TR, Liu XS, Brown M. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 39.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hah N, Danko CG, Core LJ, Siepel AC, Lis JT, Kraus WL. Exploring the direct estrogen-regulated transcriptome in breast cancer cells using GRO-Seq. The 2010 Endocrine Society Meeting; 2010. [Google Scholar]
- 41.Lefterova MI, Steger DJ, Zhuo D, Qatanani M, Mullican SE, Tuteja G, Manduchi E, Grant GR, Lazar MA. Cell-specific determinants of peroxisome proliferator-activated receptor gamma function in adipocytes and macrophages. Mol Cell Biol. 30:2078–2089. doi: 10.1128/MCB.01651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.John S, Sabo PJ, Johnson TA, Sung MH, Biddie SC, Lightman SL, Voss TC, Davis SR, Meltzer PS, Stamatoyannopoulos JA, Hager GL. Interaction of the glucocorticoid receptor with the chromatin landscape. Mol Cell. 2008;29:611–624. doi: 10.1016/j.molcel.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 44.Chopra AR, Louet JF, Saha P, An J, Demayo F, Xu J, York B, Karpen S, Finegold M, Moore D, Chan L, Newgard CB, O'Malley BW. Absence of the SRC-2 coactivator results in a glycogenopathy resembling Von Gierke's disease. Science. 2008;322:1395–1399. doi: 10.1126/science.1164847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lanz RB, Bulynko Y, Malovannaya A, Labhart P, Wang L, Li W, Qin J, Harper M, O'Malley BW. Global Characterization of Transcriptional Impact of the SRC-3 Coregulator. Mol Endocrinol. doi: 10.1210/me.2009-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ochsner SA, Steffen DL, Hilsenbeck SG, Chen ES, Watkins C, McKenna NJ. GEMS (Gene Expression MetaSignatures), a Web resource for querying meta-analysis of expression microarray datasets: 17beta-estradiol in MCF-7 cells. Cancer Res. 2009;69:23–26. doi: 10.1158/0008-5472.CAN-08-3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ochsner SA, Steffen DL, Stoeckert CJ, Jr, McKenna NJ. Much room for improvement in deposition rates of expression microarray datasets. Nat Methods. 2008;5:991. doi: 10.1038/nmeth1208-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Germain P, Chambon P, Eichele G, Evans RM, Lazar MA, Leid M, De Lera AR, Lotan R, Mangelsdorf DJ, Gronemeyer H. International Union of Pharmacology. LXIII. Retinoid X receptors. Pharmacol Rev. 2006;58:760–772. doi: 10.1124/pr.58.4.7. [DOI] [PubMed] [Google Scholar]
- 49.Dahlman-Wright K, Cavailles V, Fuqua SA, Jordan VC, Katzenellenbogen JA, Korach KS, Maggi A, Muramatsu M, Parker MG, Gustafsson JA. International Union of Pharmacology. LXIV. Estrogen receptors. Pharmacol Rev. 2006;58:773–781. doi: 10.1124/pr.58.4.8. [DOI] [PubMed] [Google Scholar]
- 50.Martinez E, Moore DD, Keller E, Pearce D, Vanden Heuvel JP, Robinson V, Gottlieb B, MacDonald P, Simons S, Jr, Sanchez E, Danielsen M. The Nuclear Receptor Resource: a growing family. Nucleic Acids Res. 1998;26:239–241. doi: 10.1093/nar/26.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruau D, Duarte J, Ourjdal T, Perriere G, Laudet V, Robinson-Rechavi M. Update of NUREBASE: nuclear hormone receptor functional genomics. Nucleic Acids Res. 2004;32:D165–167. doi: 10.1093/nar/gkh062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Horn F, Vriend G, Cohen FE. Collecting and harvesting biological data: the GPCRDB and NucleaRDB information systems. Nucleic Acids Res. 2001;29:346–349. doi: 10.1093/nar/29.1.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bettler E, Krause R, Horn F, Vriend G. NRSAS: Nuclear Receptor Structure Analysis Servers. Nucleic Acids Res. 2003;31:3400–3403. doi: 10.1093/nar/gkg505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Durme JJ, Bettler E, Folkertsma S, Horn F, Vriend G. NRMD: Nuclear Receptor Mutation Database. Nucleic Acids Res. 2003;31:331–333. doi: 10.1093/nar/gkg122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tang S, Han H, Bajic VB. ERGDB: Estrogen Responsive Genes Database. Nucleic Acids Res. 2004;32:D533–536. doi: 10.1093/nar/gkh083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jin VX, Sun H, Pohar TT, Liyanarachchi S, Palaniswamy SK, Huang TH, Davuluri RV. ERTargetDB: an integral information resource of transcription regulation of estrogen receptor target genes. J Mol Endocrinol. 2005;35:225–230. doi: 10.1677/jme.1.01839. [DOI] [PubMed] [Google Scholar]
- 57.Tang S, Zhang Z, Tan SL, Tang MH, Kumar AP, Ramadoss SK, Bajic VB. KBERG: KnowledgeBase for Estrogen Responsive Genes. Nucleic Acids Res. 2007;35:D732–736. doi: 10.1093/nar/gkl816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.NIDDK. DKCOIN (National Institute of Diabetes, Digestive and Kidney Diseases); 2010. www.dkcoin.org. [Google Scholar]
- 59.Jeong Y, Mangelsdorf DJ. Expression Profiling of Nuclear Receptors in Human and Mouse Embryonic Stem Cells (Nuclear Receptor Signaling Atlas) 2009. http://www.nursa.org/datasets.cfm?doi=10.1621/datasets.05005. [DOI] [PMC free article] [PubMed]
- 60.Bookout AL, Lanz RB, McKenna NJ, Mangelsdorf DJ. Tissue-specific expression patterns of nuclear receptor coregulators (Nuclear Receptor Signaling Atlas) 2007. http://www.nursa.org/datasets.cfm?doi=10.1621/datasets.04002.
- 61.Jeong Y, Kwak I, Lee KY, White LD, Wang XP, Brunicardi FC, O'Malley BW, DeMayo FJ. Genomic analysis of the impact of steroid receptor coactivators (SRCs) ablation on hepatic metabolism (Nuclear Receptor Signaling Atlas) 2007. http://www.nursa.org/datasets.cfm?doi=10.1621/datasets.04010. [DOI] [PubMed]