Abstract
Aim: To explore the potential genetic association of CTLA-4 Exon1 +49A/G and 3′UTR (AT)n to susceptibility to systemic lupus erythematosus (SLE), rheumatoid arthritis (RA), and overlapping (OP) autoimmunity; affected with more than one autoimmune disease. Expression of two major CTLA-4 isoforms; full length (mCTLA-4) and soluble (sCTLA-4) were explored in all subjects. A total of 680-age/gender/ethnically matched Kuwaitis were recruited and polymerase chain reaction (PCR)-fragment analysis was employed for genotyping both markers. mCTLA-4 and sCTLA-4 mRNA expression were analyzed using quantitative real time-PCR. The enzyme-linked immunosorbent assay (ELISA) was used to screen sCTLA-4 in all subjects' sera. Results: Only two CTLA-4 3′UTR (AT)n allelotypes; (AT)15 and (AT)6 were detected. The heterozygous (AT)15/6 genotype confers protectivity rather than susceptibility to SLE (p=0.01, odds ratio=0.43, and confidence interval=0.21–0.86). No significant association was observed between Exon 1 +49A/G and any of the tested diseases. A consistently high serum sCTLA-4 level was observed in RA (6.8 ng/mL, p=0.005), SLE (6.34 ng/mL, p=0.007), and OP (8.75 ng/mL, p=0.012) compared to healthy control. A significant increase in the expression of sCTLA-4 mRNA was observed in OP (p=0.05) and SLE (p=0.047), while a significant increase in the expression of mCTLA-4 (p=0.01) was observed only in OP. Conclusion: The present study is the first to report a statistically significant association between OP and serum sCTLA-4. The novelty of our study is the significance of CTLA-4 in the pathogenesis of OP besides SLE and RA.
Introduction
Autoimmune diseases are presumed to be the result of a malfunctioning immune system that fails to maintain nonresponsiveness or tolerance to self (Lesage and Goodnow, 2001). Most frequent autoimmune diseases, such as systemic lupus erythematosus (SLE) (MIM#152700) and rheumatoid arthritis (RA) (MIM# 180300), cluster within the same families, and their inheritance pattern in general is very complex. For the development of efficient disease-preventative and modifying therapies, a deep insight into the genetic and environmental causes that lead to the diseases is imperative.
SLE is a complex prototype of systemic autoimmune disease characterized by sustained abnormal immune activation and autoantibody production. A high concordance rate in monozygotic twins and an increased risk of SLE in first degree relatives compared to the general population demonstrates the underlying genetic basis of this disease (Vyse and Kotzin, 1996; Harley et al., 1998; Bengtsson et al., 2002). RA is a polygenic autoimmune disease that primarily affects the synovial tissue in diathroidal joints causing inflammation disrupting joint integrity (Lee et al., 2003). Both SLE and RA are believed to be T-cell-mediated autoimmune diseases and thus may share a similar pathway of etiology or even steps in a branched pathway. About 25% of patients diagnosed with autoimmune disorders may have a tendency to develop one or more additional autoimmune diseases leading to overlapping autoimmune disease. Such cases share a common causative gene or, more probably, polygenes leading to the assumption that a common pathway of etiology leads to the development of autoimmune diseases in genetically susceptible individuals.
Our previous study on genome scan meta-analysis for non-HLA type candidate loci has confined the risk factor for these autoimmune diseases to locus 2q33, which contains the T lymphocyte regulatory genes, such as CTLA-4, CD28, and ICOS (AlFadhli et al., 2010). Cytotoxic T-lymphocyte-associated antigen-4 (CTLA-4) is found to be a negative regulator of an immune response, while the other two genes appear to positively regulate immune responses. Hence, we primarily decided to select CTLA-4 as a candidate gene for the present study.
CTLA-4 plays a significant role in the immune response, by regulating the adaptation of T cells to a state of proliferative responsiveness and tolerance. It belongs to the immunoglobulin super family and exits in multiple molecular forms and membrane fractions. It generates two major isoforms—membrane bound (mCTLA-4) and soluble form (s-CTLA-4) (Magistrelli et al., 1999). The full length CTLA-4 mRNA contains four exons encoding the CTLA-4 molecule expressed at the membrane surface. The soluble form, lacking exon three encoding the transmembrane region, is the most widely studied alternate splice variant of the CTLA-4 gene (Oaks and Hallet, 2000; Wan et al., 2006; Saverino et al., 2007; Daroszewski et al., 2009; Chan et al., 2010). Several studies have previously implicated the elevated levels of sCTLA-4 proteins in sera of patients with autoimmune thyroiditis (Saverino and Brizzolara, 2007), SLE (Wong et al., 2005), Myasthenia gravis (Wang et al., 2002), systemic sclerosis (Sato et al., 2004), celiac disease (Simone et al., 2009), and autoimmune pancreatitis (Umemura et al., 2008). Similarly, CTLA-4 polymorphisms are found to be associated with several autoimmune disorders; namely, type I diabetes, autoimmune thyroid disease, celiac disease, grave's disease, RA, and multiple sclerosis.
In the present study, we aimed to evaluate the genetic association and contribution of two individual markers from the CTLA-4 gene known to be involved in susceptibility to various autoimmune diseases, the CTLA-4 Exon 1+49 A/G SNP (rs231775) resulting in a threonine to alanine substitution in the signal peptide of the molecule (Anjos et al., 2002) and the CTLA-4 3′UTR (AT)n repeat proposed to influence CTLA4 mRNA stability and turnover (Conne et al., 2000; Lowe et al., 2000; Kuersten and Goodwin, 2003), in patients with individual autoimmune diseases, such as SLE, RA and in a group of patients with overlap autoimmune diseases. We further aim to explore the functional role of the CTLA-4 gene at the mRNA and protein level.
Materials and Methods
A total of 680 subjects were recruited in this study; 236 SLE, 124 RA, 38 overlap syndrome, and 282 healthy controls. Blood samples were collected from outpatient clinics of the Mubarak Al-Kabeer Hospital, Kuwait, during 2005–2010. The SLE and RA patients were selected when they had at least four of the Systemic Lupus International Collaboration Clinics/American College of Rheumatology (SLICC/ACR) criteria. Patients with overlapping (OP) autoimmune disease had coexistence of at least two of the following autoimmune diseases, such as SLE, RA, Hashimoto's thyroiditis (HT), diabetes mellitus, and alopecia areata (AA) (Table 1). Healthy volunteers within the age range 19 to 68 years (median 46 years) were randomly selected from five provinces of Kuwait. Inclusion criteria were general good health and no first degree relatives with autoimmune diseases. Exclusion criteria were any recent history of acute or chronic debilitating illness. Ethnic bias within the population studied was minimized by excluding patients who were not of Arab origin. The conduct of the study was approved by the institution's ethics committee and all the participants provided written informed consent.
Table 1.
Stratification of Overlapping Autoimmune Diseases
| Overlapping autoimmune diseases | Number of cases |
|---|---|
| SLE+RA | 10 |
| SLE+AA | 2 |
| SLE+HT | 8 |
| RA+HT | 10 |
| RA+HT+Diabetes Mellitus type 1 | 3 |
| HT+Diabetes Mellitus type 1 | 5 |
| Total | 38 |
SLE, systemic lupus erythematosus; RA, rheumatoid arthritis; AA, alopecia areata; HT, hashimoto's thyroiditis.
Each participant donated peripheral blood for DNA and RNA extraction and serum isolation. Patient and control sera were stored at −20°C for up to 1 month or for long-term storage at −80°C. DNA was isolated from peripheral nucleated blood cells using the Gentra kit and RNA using QIAamp RNA blood kits (QIAGEN). cDNA was synthesized using the Hi Capacity Reverse Transcription kit purchased from Applied Biosystems. Reactions were performed according to the manufacturer's instructions.
DNA analysis/genotyping of CTLA-4 3′UTR (AT)n
CTLA-4 3′UTR (AT)n repeat was genotyped using polymerase chain reaction (PCR) followed by fragment analysis. A 10 μL reaction mix containing 5 pmol of each labeled forward primer: FAM 5′-GCCAGTGAT GCTTAAAGGTTG-3′and reverse primer: 5′-AACATACGTGGCTCTATGC A-3′ was prepared. The thermal amplification program consisted of an initial denaturation at 95°C (5 min), followed by thirty cycles at 94°C (30 s), 55°C (45 s), and 72°C (1 min), and a final extension at 72°C (8 min). One μL of the PCR amplicon was mixed with 10 μL of Hi Di formamide and 0.3 μL genetic marker ROX400HD and subjected to fragment analysis using the ABI3100 (Applied Biosystem). Genotyping results were further confirmed by sequencing randomly selected samples.
Exon1+49 A/G genotyping
Allelic determinations of the CTLA-4 Exon1+49A/G SNP (rs231775) were studied using a modified PCR- restriction digestion, followed by fragment analysis. The PCR reaction was performed in a total volume of 10 μL, including 5 pmol primers (NED) labeled F-5′-GCTCTACTTCCTGAAGA CCT-3′ and R-5′-AGTCTCACTCACCTTTGCAG-3′). Five μL of the amplified product was subjected to restriction digestion in a total volume of 10 μL having 5% of Bbv1. Two μL of the restricted fragment was then mixed with 10 μL of Hi Di formamide and 0.3 μL of the genetic marker ROX 400HD, and subjected to ABI3100 (Applied Biosystem). Allele A results in an undigested PCR product of 162 bp, and allele G results in a digested PCR product of 90 and 72 bp; the 72-bp fragment contains the fluorescence-labeled forward primer and could be visualized on the ABI-3100. Genotyping results were further confirmed by sequencing randomly selected samples.
Real-time PCR
Quantitative Real-time PCR (TaqMan probe-based detection) was performed to study the expression of the human sCTLA-4 and mCTLA-4 gene, relative to human GAPDH as the endogenous control. Gene-specific primers and probes for CTLA-4 mRNA were designed using the NCBI server and purchased from Applied Biosystem, USA. The quantitative real time PCR reactions were carried out according to the manufacturer's instructions.
Detection of serum sCTLA-4 by enzyme-linked immunosorbent assay
Specific enzyme-linked immunosorbent assay (ELISA) kits (Cayman Chemical Company) were used for measuring serum sCTLA-4 levels in patients with RA, SLE, OP, and control subjects, according to the manufacturer's protocol. The concentration of serum sCTLA-4 in a total of 134 patients and 68 healthy subjects were evaluated. The results are expressed in ng/mL.
Statistical analysis
The fit to the Hardy–Weinberg equilibrium was verified. The distribution of allelic and genotype frequencies were analyzed using the Pearson's χ2 test. Statistical significance was defined as p<0.05. The p-value, odds ratio (OR), and 95% confidence interval (CI) were calculated using online statistical programs (http://faculty.vassar.edu/lowry/odds2x2.html). Relative expression of mCTLA-4 and sCTLA-4 mRNA were analyzed using the Student's t-test (Graph Pad Prism). The Mann–Whitney test was used to analyze the differences in the serum sCTLA-4 concentration between patient groups and healthy subjects (www.socr.ucla.edu).
Results
Allelic and genotypic frequencies of the Exon 1+49A/G polymorphism in 680 RA, SLE, and OP autoimmune patients and healthy subjects were detected (Table 2). The 1+49A/G allele G (20%) was less available compared to allele A (80%), which dominated the population. The frequencies of CTLA-4 1+49A/G genotypes AA, AG, and GG in autoimmune patients were not significantly different from the healthy control. The GG genotype was less frequent in all three disease groups and healthy subjects (<10%). Lack of association of the 1+49A/G SNP with the risk of having any of the tested autoimmune diseases might indicate that, there is no effect of this SNP on disease susceptibility in the Kuwaiti population.
Table 2.
Distribution of Allelic and Genotypic Frequencies of CTLA 4 Exon 1+49 A/G SNP in Four Tested Cohorts
| Case (n) | Allele | Case no. (f) | Control no. (f) | Chi square | p-Value |
|---|---|---|---|---|---|
| SLE (472) | A | 382 (0.81) | 0.21 | 0.65 | |
| G | 90 (0.19) | ||||
| RA (228) | A | 178 (0.78) | A-450 (0.8) | 0.29 | 0.59 |
| G | 50 (0.22) | G-114 (0.2) | |||
| OP (76) | A | 60 (0.79) | 0.03 | 0.86 | |
| G | 16 (0.21) |
| Case (n) | Genotype | Case no. (f) | Control no. (f) | Chi square | p-Value |
|---|---|---|---|---|---|
| SLE (236) | AA | 152 (0.64) | 0 | 1 | |
| RA (114) | 74 (0.65) | 182 (0.65) | 0 | 1 | |
| OP (38) | 24 (0.63) | 0.03 | 0.86 | ||
| SLE (236) | 78 (0.33) | 0.39 | 0.53 | ||
| RA (114) | AG | 30 (0.26) | 86 (0.31) | 0.69 | 0.4 |
| OP (38) | 12 (0.32) | 0.02 | 0.89 | ||
| SLE (236) | 6 (0.03) | 2.03 | 0.15 | ||
| RA (114) | GG | 10 (0.09) | 14 (0.05) | 2.07 | 0.15 |
| OP (38) | 2 (0.05) | 0.006 | 0.94 |
OP, overlapping.
Distribution of the allelic and genotypic frequency of CTLA-4 3′UTR (AT)n is shown in Table 3. Only two alleles of (AT)n were detected in the Kuwaiti population, (AT)15 and (AT)6. The allele with (AT)6 repeats (>87%) was more frequent in cases and controls than the (AT)15 repeat. Allelic frequencies of (AT)n failed to show any significant difference in the tested autoimmune cases compared to healthy control (p>0.05). Among the three genotypes of (AT)n repeat detected in this study (6/6, 15/15, and 15/6), homozygous (AT)15/15 represents the rare genotype (<13%). The heterozygous (AT)15/6 genotype showed a significant association that confers protectivity rather than susceptibility to SLE (p=0.01, OR=0.4283, 95% CI=0.2133–0.8598), whereas the other tested cohorts failed to show any significant association.
Table 3.
Distribution of Allelic and Genotypic Frequencies of CTLA 4 3′UTR (AT)n in Four Tested Cohorts
| Case (n) | Allele | Case no. (f) | Control no. (f) | Chi square | p-Value |
|---|---|---|---|---|---|
| SLE (420) | 6 | 376 (0.90) | 1.18 | 0.28 | |
| 15 | 44 (0.10) | ||||
| RA (224) | 6 | 200 (0.89) | 6–422 (0.87) | 0.63 | 0.43 |
| 15 | 24 (0.11) | 15–62 (0.13) | |||
| OP (76) | 6 | 66 (0.87) | 0.01 | 0.92 | |
| 15 | 10 (0.13) |
| Case (n) | Genotype | Case no. (f) | Control no. (f) | Chi square | p-Value |
|---|---|---|---|---|---|
| SLE (210) | 182 (0.87) | 2.64 | 0.1 | ||
| RA (112) | 6/6 | 94 (0.84) | 196 (0.81) | 0.45 | 0.5 |
| OP (38) | 30 (0.79) | 0.09 | 0.76 | ||
| SLE (210) | 16 (0.08) | 0.17 | 0.68 | ||
| RA (112) | 15/15 | 6 (0.05) | 16 (0.07) | 0.21 | 0.65 |
| OP (38) | 2 (0.13) | 0.09 | 0.75 | ||
| SLE (210) | 12 (0.06) | 5.9 | 0.01 | ||
| RA (112) | 15/6 | 12 (0.11) | 30 (0.12) | 0.52 | 0.47 |
| OP (38) | 6 (0.16) | 0.34 | 0.56 |
Bold value indicates significance.
Expression of CTLA-4 isoforms
A total of 94 autoimmune patients (32 SLE, 42 RA, and 20 OP) and 28 healthy donors were tested for the expression of mCTLA-4 and sCTLA-4 isoforms of CTLA-4 mRNA. A significant increase in the expression of sCTLA-4 mRNA was observed in SLE (p=0.047) and a suggestive significance in OP patients (p=0.05) compared to healthy controls. A fold difference of 0.84–2.8 and 0.69–4.6 were observed in the expression of target mRNA in SLE and OP patients, respectively, relative to healthy control. Similarly, analysis of mCTLA-4 mRNA revealed a significant difference (p=0.01) only between OP patients and healthy subjects, where a 0.01–5.7-fold increase in the expression of mCTLA-4 mRNA was observed. No significant difference in the expression of sCTLA-4 or mCTLA-4 mRNA was observed between RA patients and healthy control (p>0.05). Analysis of (delta threshold cycle) the ΔCT sCTLA-4/mCTLA-4 ratio revealed no significant differences between patients and control groups (p>0.05). Furthermore, differences concerning mCTLA-4 and sCTLA-4 expressions were not statistically significant between any of the tested cohorts (RA vs. SLE, SLE vs. OP, and OP vs. RA, p>0.05).
Serum sCTLA-4
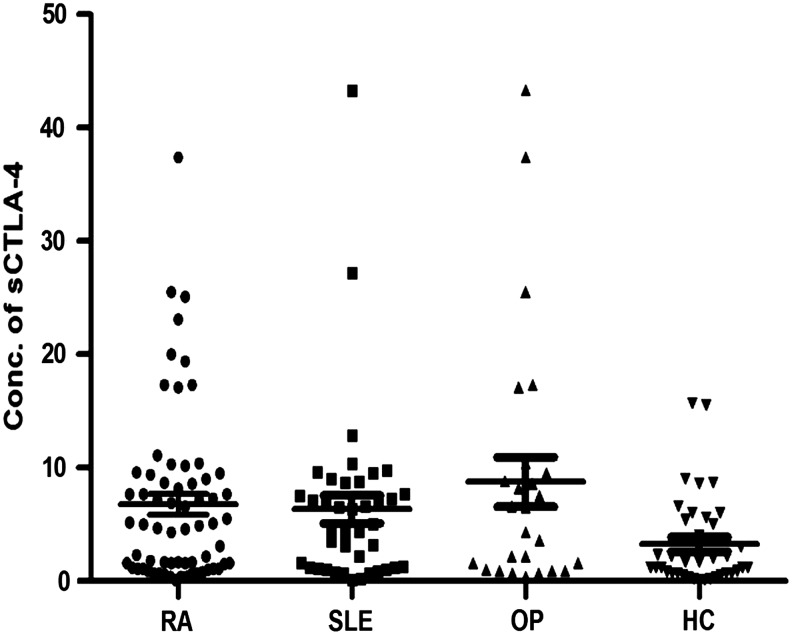
The range of serum sCTLA-4 detected in autoimmune patients and healthy subjects are presented in Figure 1.Only 36/68 healthy subjects (52.9%) had detectable serum sCTLA-4 with a detection limit of 0.1–9 ng/mL. However, 26/28 OP patients (92%) had serum sCTLA-4 in the range of 0.4–43.3 ng/mL. All the patients with SLE (n=41) and RA (n=65) had a detectable level of sCTLA-4 in the range of (0.1–43.3 ng/mL) and (0.1–37.4 ng/mL), respectively. The average serum concentration of sCTLA-4 was found to be higher in OP patients (8.75 ng/mL) compared to RA (6.8 ng/mL) and SLE (6.34 ng/mL). However, no significant differences in serum sCTLA-4 levels were observed between the patient groups (p>0.05). While the serum sCTLA-4 concentration was found to be significantly higher in autoimmune patients, SLE (6.34±7.8 ng/mL, p=0.007), RA (6.8±7.5 ng/mL, p=0.005), and OP (8.75±11.19 ng/mL, p=0.012) compared to healthy control (2.6–2.7 ng/mL).
FIG. 1.
Serum soluble form CTLA-4 (sCTLA-4) found in autoimmune patients, such as rheumatoid arthritis (RA), systemic lupus erythematosus (SLE), overlapping (OP), and healthy control (HC). The concentration of serum sCTLA-4 in a total of 134 autoimmune patients and 68 healthy controls were analyzed using the ELISA test. Results are expressed in ng/mL.
Discussion
The allelic and genotypic frequencies of CTLA-4 Exon1+49A/G in all tested autoimmune patients failed to show any significant association when compared to healthy control. Lack of association of the A-G polymorphism with autoimmune diseases could be due to the predominance of the A allele (80%) in the Kuwaiti population. The G allele suspected for susceptibility in other populations (Ban et al., 2003) was less (20%) available compared to the A allele in the Kuwaiti population. Several studies have previously shown the association of the G allele with the occurrence of endocrine autoimmune diseases, such as the Grave's disease (Donner et al., 1997b; Kouki et al., 2000), Addison's disease (Donner et al., 1997a), and insulin-dependent diabetes mellitus (Donner et al., 1997b). Only 6 out of 22 population studies have so far reported the predominance of the G allele (>65%). However, noninvolvement of the G allele in the function and expression of the CTLA-4 gene have also been reported (Xu et al., 2002). This could be explained by the fact that another polymorphism in the linkage disequilibrium may be responsible for the disease; a possibility as the threonine to alanine substitution does not affect the function of the leader peptide or merely because the CTLA-4 A/G polymorphism alone is not sufficient to influence disease expression.
A comparative analysis of our experimental data with other ethnicities were attempted, 9 out of 18 studies on the exon 1+49A/G polymorphism in SLE patients did not show any significant association with SLE, which includes the Spanish (D'Alfonso et al., 2000; Aguilar et al., 2003), English (Heward et al., 1999), Japanese (Takeuchi et al., 2003), Korean (Hudson et al., 2002), Chinese (Liu et al., 2001), Mexican (Mehrian et al., 1998), African Americans (Parks et al., 2004), and Malaysian (Chua et al., 2010). Recently, Chang et al. (2012) have reported a detailed meta-analysis on the CTLA-4 exon-1+49A/G SNP suggesting the significance of this polymorphism as a risk factor for SLE susceptibility at least in Asians.
Likewise, 5 out of 10 RA studies failed to show any significant association (Seidl et al., 1998; Gonzalez-Escribano et al., 1999; Barton et al., 2000; Yanagawa et al., 2000; Hadj-Kacem et al., 2001; Milicic et al., 2001; Lee et al., 2002, 2003; Barton et al., 2004; Takeuchi et al., 2006) and this group comprised the Caucasians mostly.
The genotyping of CTLA-4 3′UTR (AT)n repeats revealed a significant association only with SLE patients. The heterozygous genotype of (AT)15/6 showed a significant association with SLE (p=0.01), while RA and OP patients showed a pattern similar to healthy subjects. The allele with (AT)6 repeats was found to be prominent (>85%) in both cases and controls. Three studies were found on CTLA-4 3′UTR (AT)n using SLE samples, of which the Japanese (Ahmed et al., 2001) and Portuguese (Baretto et al., 2004) found an association between the disease and the 106-bp (6 repeats) allele, while another study on Japanese (Matsushita et al., 1999) itself failed to show any significant association. A single study was conducted using RA patients as subjects on CTLA-4 3′UTR (AT)n in a Tunisian population, which even failed to produce any significant association with the disease (Hadj-Kacem et al., 2001). These contradictory results could be explained on the basis of genetic divergence between populations. Several other factors, such as clinical heterogeneity, genotyping and diagnostic errors, differences in statistical strategies, and limitations in the sample size, could also contribute to misleading results. The Kuwaiti population has its roots in the Caucasians, which explains the similarity between their results.
Our study is one of the few to explore the role of CTLA-4 isoforms at the mRNA level. A significant increase in the expression of sCTLA-4 mRNA was observed in SLE patients (p=0.04) and OP (p=0.05) compared to healthy controls. Expression of sCTLA-4 mRNA in SLE and OP were found to be similar probably suggesting the predominance of SLE in OP patients. The majority of OP patients tested in our study were primarily diagnosed with SLE and later with one or more additional autoimmune disorders, such as RA, HT, AA, and diabetes mellitus. It is difficult to classify OP patients to a single disease category since they exhibit a continuous spectrum of clinical features ranging from organ-specific to systemic autoimmune diseases. Additionally, we observed a significant increase in the expression of mCTLA-4 (p=0.01) in OP patients. Of note, higher levels of mCTLA-4 could not restore homeostasis in OP patients. A recent work by Budarf et al. (2011) confirms the existence of multiple risk factors for SLE and supports the notion that some risk factors for SLE are shared with other inflammatory disorders. Hence, it is questionable whether it is adequate to study the CTLA-4 gene alone to depict the autoimmune etiology.
Several studies have previously reported the increased concentration of serum sCTLA-4 in autoimmune patients. Our experimental data further confirm and extend these findings; consistently, 132 out of 134 autoimmune patients had detectable sCTLA-4 in their serum. A significant increase in the serum sCTLA-4 concentration was observed in SLE (p=0.007), RA (p=0.005), and OP (p=0.012) patients compared to healthy controls. The present study is the first to report a statistically significant association between OP patients and serum sCTLA-4. Furthermore, our data confirms the findings of Liu et al. (2003) who showed increased serum levels of sCTLA-4 in SLE patients. Nevertheless, the role played by sCTLA-4 in the pathogenesis of RA is not clearly understood. Since RA is a chronic joint disease characterized by infiltration of synovium by T cells, it is speculated that higher levels of sCTLA may sustain T cell activation leading to autoreactivity. Despite these findings, no correlation was observed between the levels of sCTLA-4 mRNA transcripts and serum proteins in RA patients. The sCTLA-4 mRNA levels (p=0.56) failed to show any significant association, while serum proteins showed a significant difference (p=0.005) between RA patients and healthy controls. This conflicting result could be due to a shorter half-life of mRNA. The precise mechanisms that control the level sCTLA-4 mRNA are not known. The fact that half-lives of mRNA might change over time and the chemical half-life might not reflect its functional form makes it further difficult to reach a decision.
In short, the potential of our study was to explore the role of the CTLA-4 gene at the genetic, mRNA, and protein levels in various autoimmune diseases. Our study indicates that CTLA-4 probably plays a significant role in the pathogenesis of autoimmune diseases, such as SLE, RA, and OP. Increased detection of serum sCTLA-4 in OP and individual autoimmune patients sheds light on the fact that, these autoimmune diseases share a common causative gene or, more probably, polygenes leading to the assumption that a common pathway of etiology leads to the development of autoimmune diseases in genetically susceptible individuals.
An overview of our results suggests that, lack of association at the genetic level does not necessarily indicate the noninvolvement of the candidate gene in the pathogenesis of autoimmune disease. Although no significant association was observed at a genetic level, higher levels of mRNA and protein supports the notion that association analysis at the genomic level alone is not sufficiently informative. Therefore, to explore the pathology of a disease, a deep insight into the mRNA and protein level is necessary. A combined genetic and functional approach is mandatory to depict autoimmune etiology.
Acknowledgment
This work was supported by the Kuwait University Research Administration Grant (NM 01/07). I wish to thank Dr. Sukhbir Singh Uppal for his support.
Author Disclosure Statement
No competing financial interests exist.
References
- Aguilar F. Torres B. Sanchez-Roman J, et al. CTLA4 polymorphism in Spanish patients with systemic lupus erythematosus. Hum Immunol. 2003;64:936–940. doi: 10.1016/s0198-8859(03)00171-x. [DOI] [PubMed] [Google Scholar]
- Ahmed S. Ihara K. Kanemitsu S, et al. Association of CTLA-4 but not CD28 gene polymorphisms with systemic lupus Erythematosus in the Japanese population. Rheumatol Oxford. 2001;40:662–667. doi: 10.1093/rheumatology/40.6.662. [DOI] [PubMed] [Google Scholar]
- AlFadhli S. Kharrat N. AlRebai A. Genome scan meta- analysis in systemic lupus erythematosus strong linkage with loci 6p22.3-p21.1 and 2q31.1–34. J Med Genet Genomics. 2010;2:001–009. [Google Scholar]
- Anjos S. Nguyen A. Ounissi-Benkalha H, et al. A common autoimmunity predisposing signal peptide variant of the cytotoxic T-lymphocyte antigen 4. J BiolChem. 2002;277:46478–46486. doi: 10.1074/jbc.M206894200. [DOI] [PubMed] [Google Scholar]
- Ban Y. Davies TF. Greenberg DA, et al. Analysis of the CTLA-4, CD-28, and inducible co-stimulatory (ICOS) gene in thyroid diseases. Genes Immun. 2003;4:586–593. doi: 10.1038/sj.gene.6364018. [DOI] [PubMed] [Google Scholar]
- Baretto M. Santos E. Ferreira R, et al. Evidence for CTLA-4 as a susceptibility gene for systemic lupus erythematosus. Hum Genet. 2004;12:620–626. doi: 10.1038/sj.ejhg.5201214. [DOI] [PubMed] [Google Scholar]
- Barton A. Jury F. Eyre S, et al. Haplotype analysis in simplex families and novel analytic approaches in a case–control cohort reveal no evidence of association of the CTLA-4 gene with rheumatoid arthritis. Arthritis Rheum. 2004;50:748–752. doi: 10.1002/art.20118. [DOI] [PubMed] [Google Scholar]
- Barton A. Myerscough A. John S, et al. A single nucleotide polymorphism in exon 1 of cytotoxic T-lymphocyte-associated-4 (CTLA-4) is not associated with rheumatoid arthritis. Rheumatology. 2000;39:63–66. doi: 10.1093/rheumatology/39.1.63. [DOI] [PubMed] [Google Scholar]
- Bengtsson AA. Rylander L. Hagmar L, et al. Risk factors for developing systemic lupus erythematosus: a case-control study in southern Sweden. Rheumatology. 2002;41:563–571. doi: 10.1093/rheumatology/41.5.563. [DOI] [PubMed] [Google Scholar]
- Budarf ML. Goyette P. Boucher G, et al. A targeted association study in systemic lupus erythematosus identifies multiple susceptibility alleles. Genes Immun. 2011;12:51–58. doi: 10.1038/gene.2010.47. [DOI] [PubMed] [Google Scholar]
- Chan IH. Tang NL. Leung TF, et al. Association of plasma soluble CTLA-4 with lung function and gene polymorphism in Chinese asthmatic children. Int Arch Allergy Immunol. 2010;152:113–121. doi: 10.1159/000265532. [DOI] [PubMed] [Google Scholar]
- Chang WW. Zhang L. Yao YS, et al. Association between CTLA-4 exon-1+49A/G polymorphism and systemic lupus erythematosus: an updated analysis. Mol Biol Rep. 2012;39:9159–9165. doi: 10.1007/s11033-012-1788-4. [DOI] [PubMed] [Google Scholar]
- Chua KH. Puah SM. Chew CH, et al. Study of the CTLA-4 gene polymorphisms in systemic lupus erythematosus (SLE) samples from Malaysia. Ann Hum Biol. 2010;37:274–280. doi: 10.3109/03014460903325185. [DOI] [PubMed] [Google Scholar]
- Conne B. Stutz A. Vassali J. The 3′ untranslated region of messenger RNA: a molecular ‘hotspot’ for pathology? Nat Med. 2000;6:637–641. doi: 10.1038/76211. [DOI] [PubMed] [Google Scholar]
- D'Alfonso S. Rampi M. Bocchio D, et al. Systemic lupus erythematosus candidate genes in the Italian population: evidence for a significant association with interleukin-10. Arthritis Rheum. 2000;43:120–128. doi: 10.1002/1529-0131(200001)43:1<120::AID-ANR15>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Daroszewski J. Pawlak E. Karabon L, et al. Soluble CTLA-4 receptor an immunological marker of Graves' disease and severity of ophthalmopathy is associated with CTLA-4 Jo31 and CT60 gene polymorphisms. Eur J Endocrinol. 2009;161:5787–5793. doi: 10.1530/EJE-09-0600. [DOI] [PubMed] [Google Scholar]
- Donner H. Braun J. Seidl C. Codon 17 polymorphism of the cytotoxic T lymphocyte antigen 4 gene in Hashimoto's thyroiditis and Addison's disease. J Clin Endocrinol Metab. 1997a;82:4130–4132. doi: 10.1210/jcem.82.12.4406. [DOI] [PubMed] [Google Scholar]
- Donner H. Rau H. Walfish PG. CTLA4 alanine-17 confers genetic susceptibility to Graves' disease and to type 1 diabetes mellitus. J Clin Endocrinol Metab. 1997b;182:143–146. doi: 10.1210/jcem.82.1.3699. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Escribano MF. Rodriguez R. Valenzuela A, et al. CTLA4 polymorphisms in Spanish patients with rheumatoid arthritis. Tissue Antigens. 1999;53:296–300. doi: 10.1034/j.1399-0039.1999.530311.x. [DOI] [PubMed] [Google Scholar]
- Hadj-Kacem H. Kaddour N. Adyel FZ, et al. HLA-DQB1 CAR1/CAR2, TNFa IR2/IR4 and CTLA-4 polymorphisms in Tunisian patients with rheumatoid arthritis and Sjogren's syndrome. Rheumatology. 2001;40:1370–1374. doi: 10.1093/rheumatology/40.12.1370. [DOI] [PubMed] [Google Scholar]
- Harley JB. Moser KL. Gaffney PM, et al. The genetics of human systemic lupus erythematosus. Curr Opin Immunol. 1998;10:690–696. doi: 10.1016/s0952-7915(98)80090-3. [DOI] [PubMed] [Google Scholar]
- Heward J. Gordon C. Allahabadia A, et al. The A–G polymorphism in exon 1 of the CTLA-4 gene is not associated with systemic lupus erythematosus. Ann Rheum Dis. 1999;58:193–195. doi: 10.1136/ard.58.3.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson LL. Rocca K. Song YW, et al. CTLA-4 gene polymorphisms in systemic lupus erythematosus: a highly significant association with a determinant in the promoter region. Hum Genet. 2002;111:452–455. doi: 10.1007/s00439-002-0807-2. [DOI] [PubMed] [Google Scholar]
- Kouki T. Sawai Y. Gardine CA. CTLA-4 gene polymorphism at position 49 in exon 1 reduces the inhibitory function of CTLA-4 and contributes to the pathogenesis of Graves' disease. J Immunol. 2000;165:6606–6611. doi: 10.4049/jimmunol.165.11.6606. [DOI] [PubMed] [Google Scholar]
- Kuersten S. Goodwin E. The power of the 3 UTR: translational control and development. Nat Rev Genet. 2003;4:626–637. doi: 10.1038/nrg1125. [DOI] [PubMed] [Google Scholar]
- Lee CS. Lee YJ. Liu HF. Association of CTLA4 gene A-G polymorphism with rheumatoid arthritis in Chinese. Clin Rheumatol. 2003;22:221–224. doi: 10.1007/s10067-003-0720-7. [DOI] [PubMed] [Google Scholar]
- Lee YH. Choi SJ. Ji JD, et al. No association of polymorphisms of the CTLA-4 exon 1 (+49) and promoter (–318) genes with rheumatoid arthritis in the Korean population. Scand J Rheumatol. 2002;31:266–270. doi: 10.1080/030097402760375142. [DOI] [PubMed] [Google Scholar]
- Lesage S. Goodnow CC. Organ-specific autoimmune disease: a deficiency of tolerogenic stimulation. J Exp Med. 2001;194:31–36. doi: 10.1084/jem.194.5.f31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MF. Wang CR. Chen PC, et al. Increased expression of soluble Cytotoxic T- Lymphocyte associated antigen-4 molecule in patients with systemic lupus erythematosus. Scand J Immunol. 2003;57:568–572. doi: 10.1046/j.1365-3083.2003.01232.x. [DOI] [PubMed] [Google Scholar]
- Liu MF. Wang CR. Lin LC, et al. CTLA-4 gene polymorphism in promoter and exon-1 regions in Chinese patients with systemic lupus erythematosus. Lupus. 2001;10:647–649. doi: 10.1191/096120301682430249. [DOI] [PubMed] [Google Scholar]
- Lowe RM. Graham J. Sund G. The length of the CTLA-4 microsatellite (AT)n-repeat affects the risk for type 1 diabetes Diabetes Incidence in Sweden Study Group. Autoimmunity. 2000;32:173–180. doi: 10.3109/08916930008994090. [DOI] [PubMed] [Google Scholar]
- Magistrelli G. Jeannin P. Herbault N, et al. A soluble form of CTLA-4 generated by alternative splicing is expressed by nonstimulated human T cells. Eur J Immunol. 1999;29:3596–3062. doi: 10.1002/(SICI)1521-4141(199911)29:11<3596::AID-IMMU3596>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Matsushita M. Tsuchiya N. Shiota M, et al. Lack of a strong association of CTLA-4 exon 1 polymorphism with the susceptibility to rheumatoid arthritis and systemic lupus erythematosus in Japanese: an association study using a novel variation screening method. Tissue Antigens. 1999;54:578–584. doi: 10.1034/j.1399-0039.1999.540607.x. [DOI] [PubMed] [Google Scholar]
- Mehrian R. Quismorio FP., Jr. Strassmann G, et al. Synergistic effect between IL-10 and bcl-2 genotypes in determining susceptibility to systemic lupus erythematosus. Arthritis Rheum. 1998;41:596–602. doi: 10.1002/1529-0131(199804)41:4<596::AID-ART6>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Milicic A. Brown MA. Wordsworth BP. Polymorphism in codon 17 of the CTLA-4 gene (49 A/G) is not associated with susceptibility to rheumatoid arthritis in British. Caucasians Tissue Antigens. 2001;58:50–54. doi: 10.1034/j.1399-0039.2001.580110.x. [DOI] [PubMed] [Google Scholar]
- Oaks MK. Hallett KM. Cutting edge: a soluble form of CTLA-4 in patients with autoimmune thyroid disease. J Immunol. 2000;164:5015–5018. doi: 10.4049/jimmunol.164.10.5015. [DOI] [PubMed] [Google Scholar]
- Parks CG. Hudson LL. Cooper GS, et al. CTLA-4 gene polymorphisms and systemic lupus erythematosus in a population-based study of whites and African-Americans in the southeastern United States. Lupus. 2004;13:784–791. doi: 10.1191/0961203304lu1085oa. [DOI] [PubMed] [Google Scholar]
- Sato S. Fujimoto M. Hasegawa M, et al. Serum soluble CTLA-4 levels are increased in diffuse cutaneous systemic sclerosis. Rheumatology. 2004;43:1261–1266. doi: 10.1093/rheumatology/keh303. [DOI] [PubMed] [Google Scholar]
- Saverino D. Brizzolara Simone R, et al. Soluble CTLA-4 in autoimmune thyroid disease; Relationship with clinical status and possible role in the immune regulation. Clin Immunol. 2007;123:190–198. doi: 10.1016/j.clim.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Seidl C. Donner H. Fischer B, et al. CTLA4 codon 17 dimorphism in patients with rheumatoid arthritis. Tissue Antigen. 1998;51:62–65. doi: 10.1111/j.1399-0039.1998.tb02947.x. [DOI] [PubMed] [Google Scholar]
- Simone R. Brizzolara R. Chiappori A, et al. A functional soluble form of CTLA-4 is present in serum of celiac patients and correlates with mucosal injury and iTG antibody production. Intl Immunol. 2009;21:1037–1042. doi: 10.1093/intimm/dxp069. [DOI] [PubMed] [Google Scholar]
- Takeuchi F. Kawasugi K. Mori M, et al. The genetic contribution of CTLA-4 dimorphisms in promoter and exon 1 regions in Japanese patients with rheumatoid arthritis. Scand J Rheumatol. 2006;35:154–155. doi: 10.1080/03009740500407651. [DOI] [PubMed] [Google Scholar]
- Takeuchi F. Kawasugi K. Nabeta H, et al. CTLA-4 dimorphisms in Japanese patients with systemic lupus erythematosus. Clin Exp Rheumatol. 2003;21:527–528. [PubMed] [Google Scholar]
- Umemura T. Ota M. Hamano H. Association of autoimmune Pancreatis with cytotoxic T-lymphocyte Antigen 4. Gastroenterology. 2008;103:588–594. doi: 10.1111/j.1572-0241.2007.01750.x. [DOI] [PubMed] [Google Scholar]
- Vyse TJ. Kotzin BL. Genetic basis of systemic lupus erythematosus. Curr Opin Immunol. 1996;8:843. doi: 10.1016/s0952-7915(96)80014-8. [DOI] [PubMed] [Google Scholar]
- Wan B. Nie H. Liu A, et al. Aberrant regulation of synovial T cell activation by soluble costimulatory molecules in rheumatoid arthritis. J Immunol. 2006;177:8844–8850. doi: 10.4049/jimmunol.177.12.8844. [DOI] [PubMed] [Google Scholar]
- Wang XB. Kakoulidou M. Giscombe R. Abnormal expression of CTLA-4 T cells from patients with Myasthenia gravis: effect of an AT rich gene sequence. J Neuroimmunol. 2002;130:224–228. doi: 10.1016/s0165-5728(02)00228-x. [DOI] [PubMed] [Google Scholar]
- Wong CK. Lit LC. Tam LS, et al. Aberrant production of soluble co-stimulatory molecules CTLA-4,CD-28, CD-80 and CD-86 in patients with Systemic Lupus Erythematosus. Rheumatology. 2005;44:989–994. doi: 10.1093/rheumatology/keh663. [DOI] [PubMed] [Google Scholar]
- Xu Y. Graves PN. Tomer Y, et al. CTLA-4 and autoimmune thyroid disease: lack of influence of the A49G signal peptide polymorphism on functional recombinant human CTLA-4. Cell Immunol. 2002;215:133–140. doi: 10.1016/s0008-8749(02)00018-7. [DOI] [PubMed] [Google Scholar]
- Yanagawa T. Gomi K. Nakao E, et al. CTLA4 gene polymorphism in Japanese patients with rheumatoid arthritis. J Rheumatol. 2000;27:2740–2742. [PubMed] [Google Scholar]