Abstract
The extracellular matrix (ECM) of the embryonic heart guides assembly and maturation of cardiac cell types and, thus, may serve as a useful template, or blueprint, for fabrication of scaffolds for cardiac tissue engineering. Surprisingly, characterization of the ECM with cardiac development is scattered and fails to comprehensively reflect the spatiotemporal dynamics making it difficult to apply to tissue engineering efforts. The objective of this work was to define a blueprint of the spatiotemporal organization, localization, and relative amount of the four essential ECM proteins, collagen types I and IV (COLI, COLIV), elastin (ELN), and fibronectin (FN) in the left ventricle of the murine heart at embryonic stages E12.5, E14.5, and E16.5 and 2 days postnatal (P2). Second harmonic generation (SHG) imaging identified fibrillar collagens at E14.5, with an increasing density over time. Subsequently, immunohistochemistry (IHC) was used to compare the spatial distribution, organization, and relative amounts of each ECM protein. COLIV was found throughout the developing heart, progressing in amount and organization from E12.5 to P2. The amount of COLI was greatest at E12.5 particularly within the epicardium. For all stages, FN was present in the epicardium, with highest levels at E12.5 and present in the myocardium and the endocardium at relatively constant levels at all time points. ELN remained relatively constant in appearance and amount throughout the developmental stages except for a transient increase at E16.5. Expression of ECM mRNA was determined using quantitative polymerase chain reaction and allowed for comparison of amounts of ECM molecules at each time point. Generally, COLI and COLIII mRNA expression levels were comparatively high, while COLIV, laminin, and FN were expressed at intermediate levels throughout the time period studied. Interestingly, levels of ELN mRNA were relatively low at early time points (E12.5), but increased significantly by P2. Thus, we identified changes in the spatial and temporal localization of the primary ECM of the developing ventricle. This characterization can serve as a blueprint for fabrication techniques, which we illustrate by using multiphoton excitation photochemistry to create a synthetic scaffold based on COLIV organization at P2. Similarly, fabricated scaffolds generated using ECM components, could be utilized for ventricular repair.
Introduction
Cardiomyocytes are the muscular drivers of the beating heart. However, when they are lost as a consequence of disease (e.g., coronary artery disease, arrhythmia), the heart has a limited ability to regenerate these crucial cells. Tissue engineering has been proposed as a means to recover cardiomyocyte mass thereby improving cardiac function. Efforts to date include either driving stem cell differentiation to cardiomyocytes (or their precursors)1–6 or delivering cells to the cardiac microenvironment for therapeutic applications.7–16 Because differentiation and delivery each present unique challenges, they have generally been explored independently. The extracellular matrix (ECM) is a crucial component of the stem cell environment that could effectively address key challenges in both areas.
The ECM provides a scaffold that incorporates cues for cellular proliferation, differentiation, migration, and morphogenesis.13,17–26 Indeed, the composition and arrangement of the ECM change to accommodate varying needs of cells during tissue development and maintenance. Therefore, it is not surprising that efforts to recreate cardiac tissues to repair damage often attempt to mimic the composition and arrangement of the ECM. Recent reports describe delivery of stem cells, or their progeny, to the heart via (1) layered cell sheets (which includes endogenously produced, but unidentified ECM),8,10,27 (2) biologically inert synthetic hydrogels (that typically include synthetic adhesion peptides, but not whole ECM molecules),28,29 (3) decellularized tissues of cardiovascular30,31 or noncardiovascular origin,12 or (4) ECM molecules that spontaneously form 3D structures independent of other ECM proteins (typically type I collagen [COLI] gels).13,16,32 The outcomes of these studies have been modest in terms of long-term survival of engrafted cells and functional heart recovery. Most of these efforts focused on mimicking the adult ECM; however, since the successful recovery of the cardiac tissue is likely to require proliferation and appropriate functional differentiation of cells, the ECM of the adult heart may not provide adequate or appropriate signals for these particular cell behaviors. Hence, a better understanding and mimicry of the heart microenvironment throughout embryonic development may better meet this goal.
Previous studies investigating ECM proteins in the developing heart have contributed to our knowledge of the embryonic cardiac microenvironment. However, they are limited in the number of proteins, regions, and time points assessed and often used methods that precluded a quantitative analysis. These studies identified ECM proteins present in the heart, including those most prevalent during development. The ECM of the developing heart consists of glycosaminoglycans (hyaluronic acid and chondroitin sulfate),33 glycoproteins (fibronectin [FN], laminin [LN], vitronectin, cytotactin, fibulin, fibrillin, and thrombospondin),34–36 COLI, COLIII, COLIV,37,38 and proteoglycans.39 Of these, the most prevalent, functionally relevant ECM proteins in the developing heart include (1) COLI and COLIII, fibrillar proteins that provide structural support,19 (2) COLIV and other basement membrane proteins that serve to align and polarize cell layers,36,40 (3) FN, which attaches to cell surface integrins and other ECM proteins (including COLI, fibrin, heparin, and syndecan) to mediate changes in the structural or mechanical properties of the matrix and phenotype of adhering cells,41,42 and (4) elastin (ELN), which is critical for elasticity, allowing tissue to stretch and recoil, as in the beating heart. A summary of ECM protein expression and distribution in the embryonic mouse and chick hearts was recently reviewed by Jung et al.43 The reviewed studies identify general changes in ECM protein expression and distribution, which likely contribute to important changes in cell behavior, such as proliferation, which is most active between embryonic day (E) 11.5–14,44 and terminal differentiation of cardiac cells, which is most active between E8.5–E13.5.45 Despite the breadth of studies examining the ECM composition during cardiac development, we failed to find any study that directly established a blueprint of essential ECM proteins and quantified changes in relative amount and distribution at different embryonic developmental stages. An experimentally derived and validated ECM blueprint would be a significant tissue engineering milestone as obtaining this information would not only improve our understanding of cardiac developmental biology, but would allow for the potential mimicry of the embryonic microenvironment through tissue scaffolds. These structures have the potential to induce a desirable combination of proliferation and differentiation of engrafted cardiac progenitor cells.
Here we used complementary approaches of SHG imaging, IHC, and real-time quantitative polymerase chain reaction (qPCR) to evaluate the contribution of four essential ECM proteins—COLI, COLIV, FN, and ELN—of the murine heart at E12.5, E14.5, and E16.5, and 2 days postnatal (P). We chose these ECM components not only because of their prevalence in cardiac tissues, but also because they encompass structural, basement membrane, connective and elastic elements, respectively. We assessed only four ECM components and developmental stages using IHC to maintain experimental feasibility and allow for colocalization imaging of each protein in consecutive heart sections. The noninvasive optical capabilities of SHG enabled a high-resolution imaging of the collagen composition in intact tissue sections. The high-throughput capability of qPCR allowed us to include COLIII and LN gene expression as well as P7 hearts in our quantitative analysis. The range of stages was chosen to include the establishment of the functional heart and the period of maximal cell proliferation and differentiation. We focused this study on the expression and localization of the ECM proteins across the left ventricle because this region is most susceptible to damage with disease and, therefore, the highest therapeutic priority for a scaffold template. This study represents an important first step in establishing an ECM blueprint for scaffold fabrication ex vivo that will provide appropriate development-based signals for maximizing regenerative cardiac repair. We demonstrate the feasibility of using such data as a template for fabricating a synthetic scaffold.
Materials and Methods
Expanded details regarding methods of tissue preparation, IHC, microscopy, image analysis, and qPCR can be found in the online supplement; Supplementary Data are available online at www.liebertpub.com/tea.
Fluorescence IHC
Each ECM protein was detected within the paraffin-embedded mouse heart or embryonic sections based on the described methods.16 Following a 30-min incubation at room temperature in a blocking solution, rabbit anti-mouse primary antibodies were incubated at 4°C overnight in a blocking solution containing anti-COLI (1:100, Abcam ab21286), anti-COLIV (1:50, Millipore AB748), anti-ELN (1:200, Millipore AB2039), or anti-FN (1:100, Millipore AB2033).
Microscopy
Second harmonic generation (SHG)
SHG of whole ventricles (ventricle thickness up to ∼1.9 mm thickness; imaging depth up to ∼340 μm) was examined using multiphoton laser scanning microscopy (MPLSM).46–48 Ventricles were imaged using a TE300 inverted microscope (Nikon) equipped with a Plan APO VC 20×(N.A. 0.75; Nikon Instruments) objective lens by using a mode-locked Ti:Sapphire laser (Spectra Physics Mai Tai) using an excitation wavelength of 890 nm with a 445-nm +/−20-nm narrow-band pass emission filter (Thin Film Imaging).
Fluorescence
Sections were imaged on an IX71 inverted deconvolution fluorescence microscope (Olympus) using a 20×UPlanFluor objective (NA=0.5) and Slidebook software (Intelligent Imaging Innovations).
Image analysis
SHG image analysis
SHG image stacks (100 images) from the apical region of two or three ventricles from each developmental stage were compiled as sum z-projections using ImageJ (Fiji distribution; open source software, http://pacific.mpi-cbg.de/wiki/index.php/Fiji). A 200-μm line was drawn in each of the four quadrants (horizontal in upper left and lower right, vertical in upper right and lower left) of each image. The number of fibers crossing each of these lines was counted manually to assess the fiber density.
Fluorescence image analysis
Image gating and quantitation of the mean intensity per pixel was performed using ImageJ.49 This gating was performed to quantitatively compare the mean intensity per pixel among time points for each separate ECM protein in a consistent manner. Following gating, a custom-developed macro (“Threshold and Calculate Average Intensity with Brush Tools Macro for IHC,” http://loci.wisc.edu/software/home) was used to calculate the mean intensities per pixel per region of the ventricle (e.g., epicardium, myocardium, and endocardium) for each fluorescent image.
Quantitative real-time PCR
Total RNA was isolated using the RNeasy Mini Kit (Qiagen, 74104) following the animal tissue protocol. Complementary DNA (cDNA) was synthesized following the instructions from the ThermoScript RT-PCR System (Invitrogen, 11146–016). Primer efficiencies were extracted from StepOne Software v2.2.2 and verified with melting curves. The standard curve method was employed to determine copy numbers of each ECM and GAPDH genes. The ECM gene expression level was determined as the copy number of the ECM gene of interest normalized to that of GAPDH at each time point.
Statistical analyses
Mean fluorescence intensities of the detected ECM proteins were compared with the difference in the least squares means (LSM) test. Each section yielded three mean intensity values from the three locations, and thus, those three values were dependent on the section. For COLI, the residuals of the mean intensities were not normally distributed, so the natural logs of the mean intensities were used in the difference in the LSM test. Distribution of the variance of expression of ECM mRNA and SHG between samples was assessed using the Levene's test. For data sets lacking equal variance, one-way ANOVA with the Tamhane's post hoc test (α=0.01) was performed. For data sets with equal variance, one-way ANOVA with the Tukey's post hoc test (α=0.01) was performed. All tests were performed using Statistical Analysis System (SAS Institute Inc.).
Fabrication based on ECM blueprint
Multiphoton excited photochemistry was used to fabricate a 3D model of the fibrillar morphology in the blueprint, where the overall method has been described previously.50 The same region was imaged in 20 serial sections from a postnatal day 2 ventricle labeled with the anti-COLIV antibody. Antibody labeling and subsequent imaging were performed as described above. To improve the reconstruction, the brightness and contrast of each image was adjusted to enhance the labeled structures and extraneous-labeled debris was removed. The resulting stack of images was reconstructed into a 3D model (Materialize Mimics Software; Materialize). A threshold was applied to discriminate the background and make the collagen fibers clearly visible for reconstruction. A sterolithographic (STL) file was generated from the 3D model, and then sliced using Materialize Magics Software (Materialize) into z sections for nanofabrication. AutoCAD software (Autodesk) was then used to hatch each slice as coordinates for the scanning unit. The fabrication solution used for the structure was the synthetic compound trimethylolpropanetriacrylate (TMPTA, neat) (Sigma-Aldrich) in combination with the coinitiator triethanolamine (0.1M) (Sigma-Aldrich) and Rose Bengal (1 mM) (Sigma-Aldrich) as the photoactivator, where the photochemistry has been previously described.51 The fabrication instrument consisted of an Axioskop 2 microscope (Zeiss), a homebuilt laser scanning system,52 a 10×objective (NA of 0.5), and 780 nm laser wavelength. LabVIEW software (National Instruments) was used to control the galvo mirrors in the laser scanning system to fabricate the hatched files generated for the 3D structure. The 3D structure was rinsed with ethanol to remove the solution around the structure. Multiphoton imaging was performed (Fluoview300 laser scanning microscope, Olympus) with a 20×objective (0.75 NA) using 890-nm excitation and 500–650-nm emission filters to capture the fluorescence arising from Rose Bengal trapped within the structure.
Results
Maturation of fibrous structure of the ventricle with development
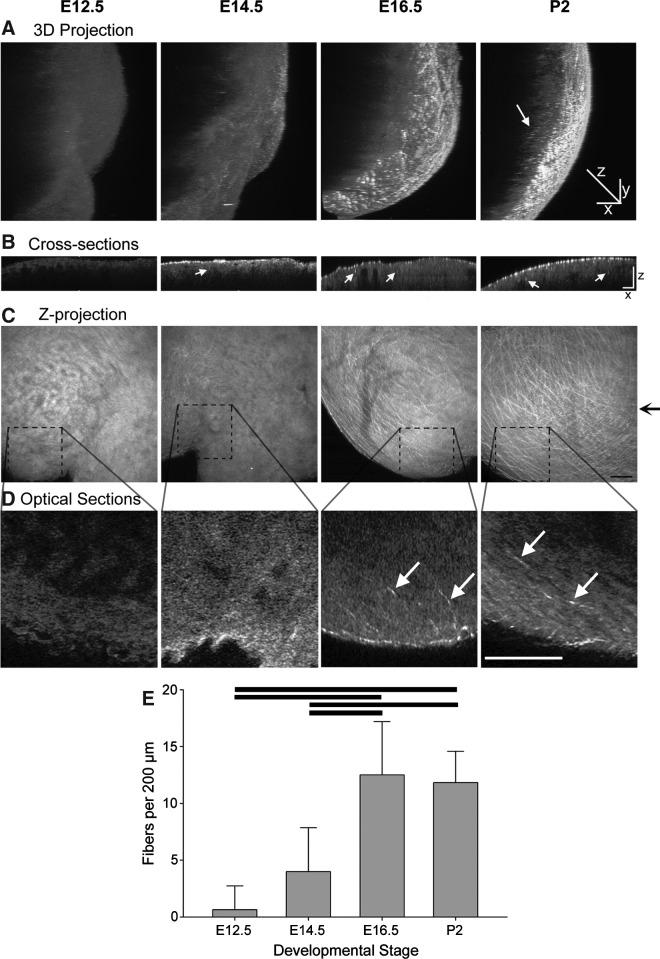
To assess changes in the fiber density with development, SHG from whole ventricles was examined. The SHG signal is generated when two photons of incident light interact with the noncentrosymmetric structure of collagen fibers, so that the resulting photons are half the wavelength of the incident photons.46 This signal is generated in the absence of exogenous label and, because MPLSM allows for deep sectioning of 3D dimensional tissues,46 we used SHG collected in the backward direction53,54 to assess the 3D organization of collagen fibers in whole mouse ventricles from developmental stages E12.5, E14.5, E16.5, and P2 (Fig. 1A, Supplementary Videos S1–S4). We found that E12.5 ventricles were essentially devoid of distinct detectable fiber structures, while a few very fine fibers could be identified in some regions of the E14.5 ventricles. By the later stages, the fibers are clearly evident and occupy the entire field of view with a meshwork pattern (Fig. 1C). This network was most apparent in the epicardium, but fibers extending into the myocardium were also observed (Fig. 1B, D). The density of the fibers was significantly greater at the later stages (E16.5 and P2) than in the earlier stages (Fig. 1E, Supplementary Table S2). These data indicate an increase in the density and organizational complexity of collagen fibers with developmental maturation of the ventricles.
FIG. 1.
Changes in density of collagen fibers of the ventricle during cardiac development. (A) Three-dimensional reconstruction of z-stacks of SHG images, collected as optical sections spaced 2 μm apart, of whole mouse ventricles from the apical region at different developmental stages. These reconstructions are rotated 110° from the plane of imaging. Epicardium is toward the right. The z-axis scale is doubled from the x-axis and y-axis to more clearly show any infiltrating fibers (white arrow). (B) Cross-sectional slices of the ventricles shown in (A), reconstructed from z-stacks and taken at the level indicated by black arrow to right of (C). The z-axis scale is doubled compared to the x-axis to show detail. Examples of fiber cross sections in the myocardial region are indicated by white arrows. (C) Z-projections of 100 SHG images, spaced 2 μm apart, of the ventricles in (A). (D) Although many of the collagen fibers detected with SHG are located in the epicardium, single optical sections, enlarged from regions depicted by dotted lines in (C), illustrate fibers extending away from the epicardium into the myocardium (white arrows) in the later stage ventricles. (E) Quantitation of the fiber density present at each developmental stage as detected by SHG and counted in 100 section z-projections. Bars show statistical differences (p<0.01, mean±SD). Scale bars in C=50 μm; all other scale bars=100 μm. SHG, second harmonic generation; E, embryonic day; P, postnatal day.
Spatiotemporal organization of multiple ECM proteins in the left ventricle of the developing heart
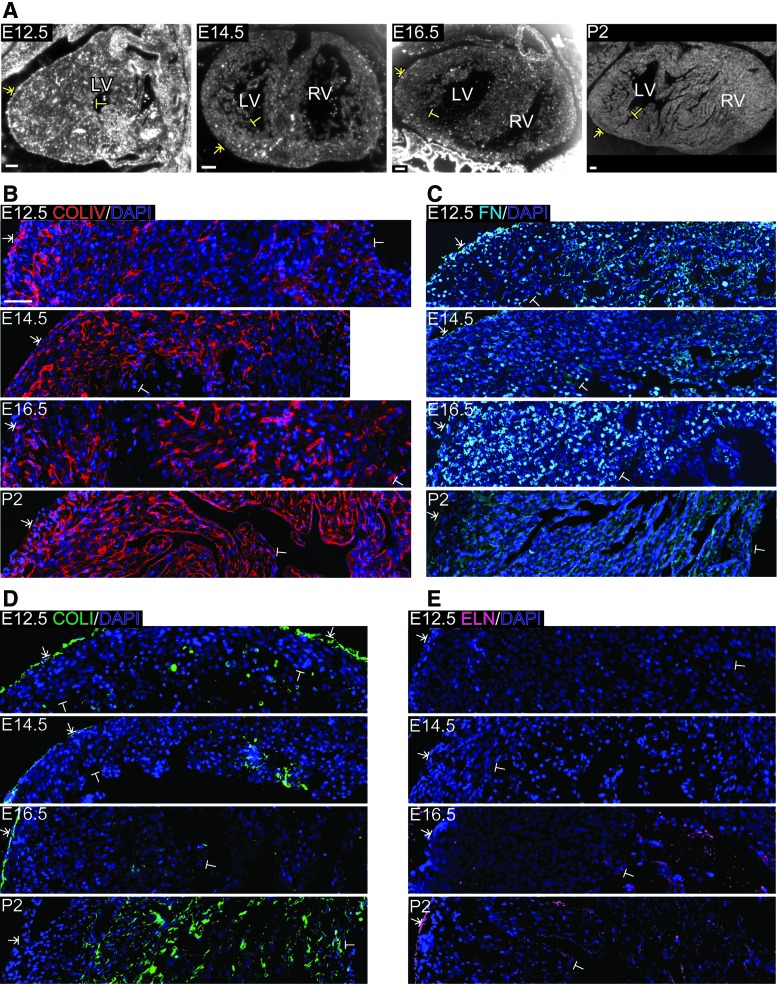
Because the fibrous scaffold of the ventricle changes with cardiac development, we examined the distribution of specific ECM proteins over time. To visualize the organization of several ECM proteins at each developmental stage, we utilized fluorescence IHC. Images were collected across the left ventricle and stitched together to form a composite mosaic. The location of the mosaic relative to the histologic section of the entire heart is shown in Figure 2A. COLIV, a basement membrane component, was mainly present in the epicardium and myocardium at E12.5, with a sporadic appearance in the endocardium lining the trabeculae (Fig. 2B). COLIV staining was punctate in appearance except that the myocardial tissue in close proximity to the epicardium contained more fibril-like staining. By E14.5, COLIV remained localized to the epicardium and myocardium, with more fibrils and a disconnected circular organization often surrounding myocytes. At E16.5, COLIV was not detected in the epicardium as in the other stages, but had a similar fibril-like appearance and circular organization in the myocardium, as observed at E14.5. The most dramatic change in the spatial organization occurred between E16.5 and P2, with COLIV becoming distinctly present in the endocardium lining the trabeculae. At this postnatal stage, COLIV was more fibrillar in appearance and formed a more interconnected network throughout the myocardium than seen during the embryonic stages.
FIG. 2.
Spatiotemporal organization of ECM in the left ventricle of E12.5, E14.5, E16.5, and P2 murine hearts. (A) Brightfield images of sagittal (E12.5) and coronal (E14.5, E16.5, and P2) sections indicate example regions of the left ventricle imaged, beginning in the epicardium (T-arrow) and extending through the myocardium to the endocardium (T-bar). (B–D) Fluorescence micrographs showing COLIV (red, B), FN (cyan, C), COLI (green, D), and ELN (magenta, E) labeled via IHC in separate sections at different stages in development. For these images, FIJI software was used to adjust the intensity of all fluorescent pixels for each individual protein and developmental stage to maximize visibility of protein organization. Scale bars (A)=100 μm (B)=50 μm. ECM, extracellular matrix; IHC, immunohistochemistry; COLIV, collagen IV; COLI, collagen I; FN, fibronectin; ELN, elastin; T-arrow, epicardium; T-bar, endocardium; LV, left ventricle; RV, right ventricle. Color images available online at www.liebertpub.com/tea
Glycoproteins of the myocardium associated with mechanical strength (COLI), tissue elasticity (ELN), and protein crosslinking (FN) were also assessed. For all stages, FN was present in the epicardium and endocardium (Fig. 2C). By E14.5, FN began to form thin, isolated fibrils within the myocardium, which increased in density until P2, when the fibrils formed a more interconnected network, somewhat similar to the progression and spatial distribution of COLIV. Interestingly, FN staining also colocalized with some nuclei. Nuclear staining was clearly present at the fetal stages and was particularly prominent at E16.5. Similar to what was observed using SHG, from E12.5 through E16.5, COLI was localized to the epicardium lining the left ventricular wall (Fig. 2D). At P2, COLI was sporadically seen as thin fibrils lining the trabeculae, but most often formed in separate structures associated with cells in the myocardium. ELN was not detected until E16.5, at which time it formed thin fibrils in the endocardium without lining the trabeculae (Fig. 2E). By P2, ELN was organized in fibrils along the epicardium, myocardium, and endocardium lining the outer ventricular wall and the trabeculae, respectively, and also lined the blood vessel walls (images containing vessels not shown). To provide a positive control and benchmark comparison for each ECM protein, we labeled and imaged the adult murine myocardium (Supplementary Fig. S1).
Quantitative comparison of ECM proteins in the left ventricle of the developing heart
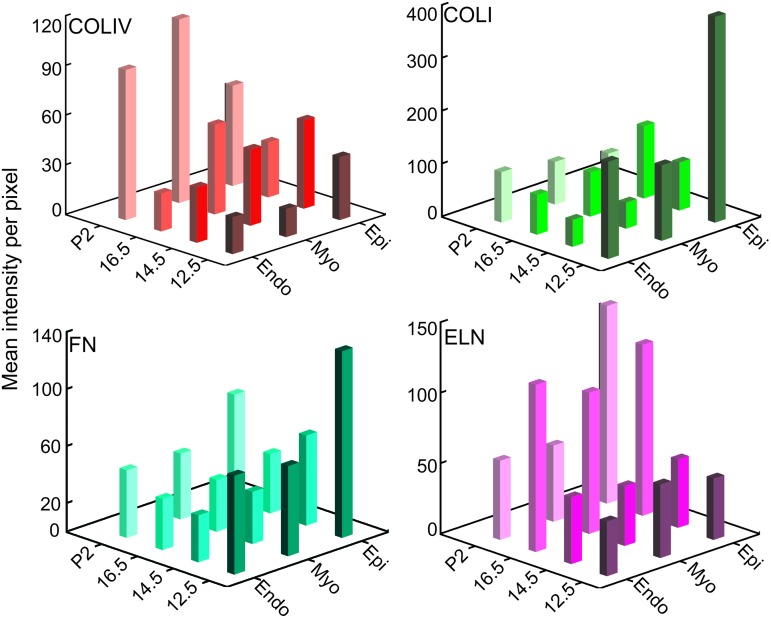
To compare the relative amount of each ECM protein across the left ventricle at each developmental stage, we quantified the mean intensity per pixel in the epicardium, myocardium, and endocardium of equivalently exposed and gated images for each individual protein. Comparisons were conducted between mean intensity values for a single ECM protein at each location and developmental stage. Statistical comparisons identified the significant changes in ECM expression between time points and between regions at particular developmental stages for each ECM protein (Supplementary Figure S2, all p-values can be found in Supplementary Table 3). In addition, 3D bar graphs of these data were created to show spatial and temporal changes at a glance (Fig. 3). The mean intensity of COLIV was relatively constant in the epicardium, increased from E12.5 to P2 in the myocardium, and increased from E16.5 to P2 in the endocardium (significant differences p<0.01: P2 epi vs. P2 myo; E12.5 myo vs. P2 myo; E16.5 endo vs. P2 endo). In contrast, the COLI mean intensity was highest at E12.5 in all locations, with the E12.5 epicardium intensity distinguishably greater than the others (significant differences p<0.01: E12.5 epi vs. E12.5 myo; E12.5 epi vs. E12.5 endo; E12.5 epi vs. P2 epi; E14.5 epi vs. E14.5 endo; E16.5 epi vs. E16.5 endo). For FN, the mean intensity was generally stable in the myocardium and endocardium over time. The most notable change was seen in the epicardium over time, decreasing significantly during fetal development and increasing slightly in the postnatal period (significant differences p<0.01: E12.5 epi vs.E14.5epi; E12.5 epi vs. E16.5 epi). Additionally, at E12.5, the levels of FN in the epicardium were higher than in the myocardium or endocardium (significant differences p<0.01: E12.5 epi vs.E12.5 myo; E12.5 epi vs. E12.5 endo). Interestingly, ELN mean intensity was greatest at E16.5 across all locations, with the exception of the P2 epicardium (significant differences p<0.01: P2 epi vs. P2 myo; P2 epi vs. P2 endo).
FIG. 3.
Quantitative comparison of relative ECM protein in the developing ventricle. Fluorescent images of E12.5, E14.5, E16.5, and P2 mouse left ventricles labeled with anti-COLIV, anti-COLI, anti-FN, or anti-ELN. Mean intensities per pixel of equivalently gated, background subtracted, fluorescent antibody-labeled sections across developmental time points. This orthogonal view provides spatiotemporal trend visualization. Epi, epicardium; Myo, myocardium; Endo, endocardium. Color images available online at www.liebertpub.com/tea
Location of ECM proteins relative to other ECM proteins
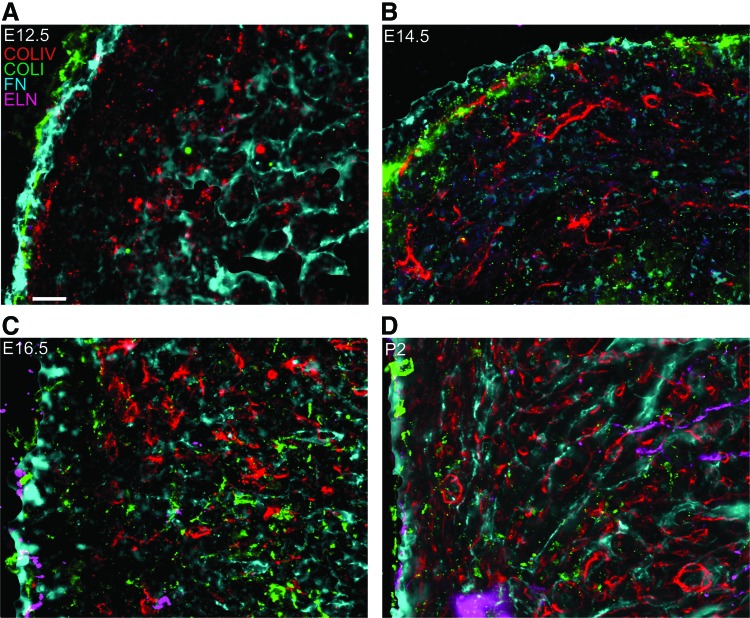
To visualize the organization and localization of each ECM protein relative to each other, we labeled four consecutive sections (at least 10 μm apart) with each of the four antibodies, and then imaged the same location within the myocardium of the left ventricle for all of the labels. FIJI software49 was used to adjust the intensity of all fluorescent pixels for each individual protein and developmental stage to maximize visibility of protein organization. Images for each protein in these consecutive sections were overlaid, generating two-dimensional z-projection images as well as isometric 3D videos depicting each individual section (Fig. 4; separate channels in Supplementary Fig. S3). At E12.5, COLI, COLIV, and FN colocalize in the epicardium lining the ventricular wall. FN and COLIV are both present in the endocardium, with FN distinctly lining the inner ventricular wall and COLIV localizing in endocardial tissue in close proximity to the trabeculae carneae lining (Fig. 4A, Supplementary Video S5). At E14.5, COLI, COLIV, and FN are still colocalized in the outer ventricular wall. COLIV and FN are present sporadically in the myocardium, with no apparent alignment relative to each other (Fig. 4B, Supplementary Video S6). At E16.5, COLI, ELN, and FN are colocalized in the epicardium. COLIV, COLI, and FN are present throughout the myocardium without a clear organization relative to each other (Fig. 4C, Supplementary Video S7). At P2, all four proteins are localized in the outer ventricular wall, while the myocardium contains COLIV and FN with a similar fibril-like appearance and organization (Fig. 4D, Supplementary Video S8).
FIG. 4.
Relative locations of ECM proteins in the developing murine ventricle. (A–D) Overlays of consecutive sections labeled via IHC for COLIV (red), COLI (green), FN (cyan), or ELN (magenta) in the murine ventricle. Each developmental time point is represented, including E12.5 (A), E14.5 (B), E16.5 (C), and P2 (D). For these images, FIJI software was used to adjust the intensity of all fluorescent pixels for each individual protein and developmental stage to maximize visibility of protein organization. The same locations, consisting of the epicardium and myocardium, from each antibody label were overlaid. Scale bar=20 μm. Color images available online at www.liebertpub.com/tea
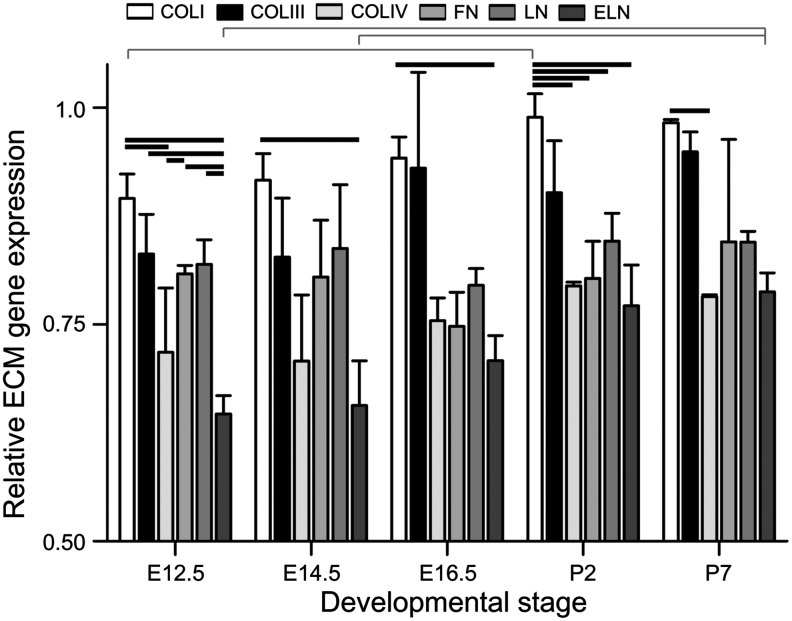
Changes in mRNA expression of ECM components
To further quantitatively assess changes in the ECM composition during cardiac development, the mRNA expression of ECM genes from ventricles was compared using qPCR (Fig. 5). Different ECM gene expression levels were compared within each developmental stage and levels of the same ECM gene expression were compared across developmental stages. In addition to the ECM proteins and stages assessed via IHC, the high-throughput nature of qPCR allowed us to add COLIII and LN gene expression, as well as the P7 developmental stage to our analysis. The P7 stage was added in an effort to identify postnatal changes in ECM gene expression. We chose P7 since previous reports suggest the regenerative potential of neonatal cardiomyocytes is lost by this stage, which could coincide with altered ECM gene expression.55 The expression levels of COLI, COLIII, COLIV, and ELN mRNA generally increased from E12.5 to P2, although this trend did not yield statistically significant differences except in the relative amounts of COLI expression between E12.5 and P2 and of ELN expression between E12.5/E14.5 and P7. In addition, there were a number of significant differences detected among the mRNA expression levels of the ECM genes within each time point, with COLI gene expression being the highest at every developmental stage. ([p-values listed in Supplementary Tables 4 and 5] significant differences p<0.01:E12.5 COLI vs. E12.5 COLIV; E12.5 COLI vs. E12.5 ELN; E12.5 COLI vs. P2 COLI; E12.5 COLIII vs. E12.5 ELN; E12.5 COLIV vs. E12.5 FN; E12.5 FN vs. E12.5 ELN; E12.5 LN vs. E12.5 ELN; E12.5 ELN vs. P7 ELN; E14.5 COLI vs. E14.5 ELN; E14.5 ELN vs. P7 ELN; E16.5 COLI vs. E16.5 ELN; P2 COLI vs. P2 COLIV; P2 COLI vs. P2 FN; P2 COLI vs. P2 LN; P2 COLI vs. P2 ELN; P7 COLI vs. P7 COLIV).
FIG. 5.
Quantitative comparison of relative ECM gene expression in the developing heart. Messenger RNA for gene expression quantitation was obtained from whole ventricles and assessed via qPCR. Red brackets indicate statistically significant differences between developmental stages for the same ECM gene. Black bars indicate statistically significant differences at the same developmental stage between two ECM genes (p<0.01, mean±SD). qPCR, quantitative polymerase chain reaction; LN, laminin.
Fabrication of synthetic scaffold based on ECM blueprint
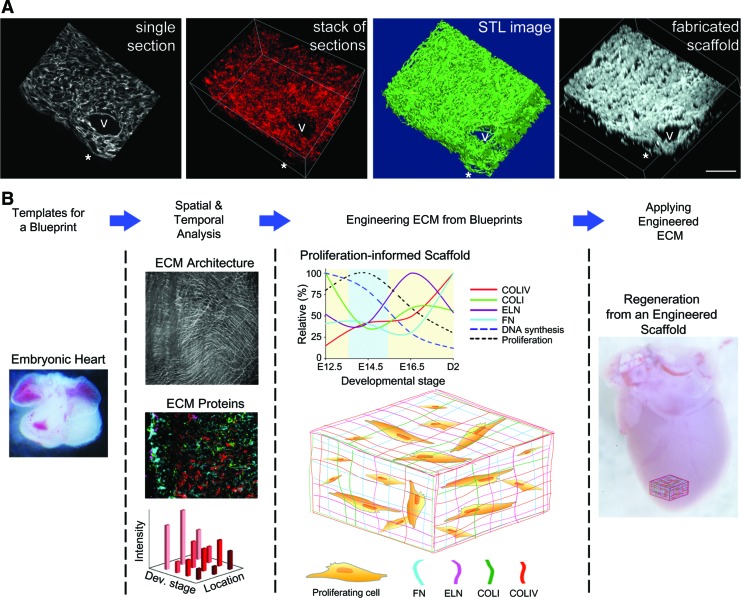
To illustrate the applicability of the antibody data to serve as a blueprint for engineered matrices, we created a scaffold from a synthetic polymer using COLIV immunolabeled serial sections as the template. This scaffold was generated via Computer Aided Design (CAD) control coupled to multiphoton excitation (MPE) fabrication50 of TMPTA. To accomplish this, the same region was imaged in 20 serial sections (∼100 μm total thickness) from a day 2 postnatal ventricle labeled with the COLIV antibody. The resulting image stack was used to create a 3D rendering that shows the 3D organization of these structures (Fig. 6A). This image stack was also used to create a STL file (Fig. 6A) that could then be used by AutoCad software to generate the coordinates necessary to drive the scanning unit for the MPE fabrication. The MPE fabrication process, guided by the COLIV template, was used to create a scaffold from the synthetic polymer TMPTA (Fig. 6A). The synthetic scaffold mimics the original 3D reconstructed template, exhibiting similar large structural components, such as a blood vessel, as well as finer structural elements, such as fiber undulations on the epicardial surface. The STL file was intentionally designed to generate a space-filling reconstruction that exaggerated the thickness of the fibers as compared to the original data set. This was necessary to maintain the structural stability of the fabricated scaffold. In the heart, and with more elaborate fabrication schemes, the empty space would contain additional ECM components, thus adding to the structural stability of the construct and thereby permitting more native fiber thickness.
FIG. 6.
Fabrication of a scaffold based on 3D reconstruction of COLIV images from a postnatal day 2 ventricle and illustration of the theoretical application of the ECM developmental blueprint to the fabrication of ECM-based biomaterials for cardiac regeneration. (A) A single section, from a data set of 20 serial sections, showing COLIV immunolabel in a postnatal day 2 ventricle. This is the fourth section in the series. The stack of images was used to create a 3D virtual reconstruction of the 20 anti-COLIV-labeled serial sections and a solid sterolithographic rendering was generated from the stack of serial sections. Multiphoton excitation-based fabrication was then used to create a 3D physical reconstruction using trimethylolpropanetriacrylate. Rose Bengal, used as the photoactivator, provides the fluorescence for the image shown. Construct was imaged as a z-stack using multiphoton microscopy and reconstructed using Imaris. Scale bar=50 μm. V indicates blood vessel; *Identifies representative structural mimics. (B) To understand the environment required to support the proliferation and differentiation of cardiomyocytes, we examined the distribution and relative amounts of four ECM components in the ventricles of developing mouse hearts using complementary methods of analysis, including SHG imaging of intact ventricles, antibody labeling of left ventricles, and qPCR of both ventricles. These data can be quantified to generate profiles of changes in the ECM composition during heart development. This ECM developmental profile can guide to select a desired cell behavior such as rapid cellular proliferation, encouraged differentiation or limit migration. The appropriate ECM blueprint could serve as a template to build a matrix supportive of the desired cell behaviors. The stage-optimized matrix could be probed to understand cell–matrix interactions better or directly delivered to the site of injury for therapeutic intervention. Color images available online at www.liebertpub.com/tea
Discussion
New fabrication technologies for generating and modifying ECM-based biomaterials motivated this study to reassess the changing composition of the native heart ECM with development. The primary goal of this effort was to more clearly define a natural blueprint that might be used to create replacement tissues. With this motivation, we analyzed the compositional and spatial arrangement of four ECM proteins simultaneously over developmental stages most relevant for cardiomyocyte maturation. The literature provides a general sense for the changes of ECM components over time (as noted in the discussion below), but the continuity and quantitative nature of our study allows a more accurate representation of these developmental changes, which is critical for mimicking tissue ex vivo. Furthermore, we utilize one of these new fabrication technologies, MPE photochemistry, to demonstrate that such compositional data can indeed serve as a blueprint for creating scaffolds. In analogy with multiphoton excited fluorescence microscopy, this method has intrinsic 3D capabilities with a submicron resolution or minimum feature sizes. While the example here used a synthetic polymer, we have previously used this method to crosslink many ECM proteins, with excellent biocompatibility.56–58 The images and quantitative analyses generated within this work will better inform efforts in cardiac tissue engineering and regenerative medicine.
COLIV was prevalent throughout the developing embryo, rather than localized to the basement membrane or blood vessels, and progressed in expression, amount, and organization from E12.5 to P2. While previous studies have not investigated the presence or absence of COLIV specifically between E12.5 and P2, COLIV was identified as one of the main cardiac ECM components before E1159 and in fine fibrils in the cardiac jelly between the endothelium and myocardium, and the basement membrane at E11.36 In the adult heart, COLIV is mainly found in a reticulated organization in blood vessels and the basement membrane.60 Considering the role of COLIV in cell adhesion, migration, and differentiation and that cardiomyocytes (in addition to smooth muscle cells61 and endothelial cells) secrete COLIV,62 the progression and localization differences in COLIV throughout the developing heart and the adult heart may correspond to progenitor cells infiltrating into the myocardium and differentiating into cardiomyocytes as the heart develops.45 It should be noted that mRNA expression did not necessarily coincide with protein expression profiles for COLIV (as well as COLI and ELN). This result is not entirely surprising as mRNA expression does not directly dictate protein synthesis, especially for ECM proteins, wherein an extracellular assembly might be required for antibody recognition.
COLI was present mainly in the epicardium at all stages except for P2 when it was present sporadically in the myocardium and endocardium. E12.5 hearts exhibited the highest levels of COLI-associated fluorescence in all locations compared to the other stages, while COLI gene expression steadily increased from E12.5 to P2. Considering the fact that COLI is one of the most prevalent proteins in the heart, the results are initially surprising. However, our observations were consistent with previous studies that identified COLI in the pericardium and endocardial cushions at E12.5,63 subepicardium at E13,64 pericardial membranes at E14.5,63 connective tissue compartments within the ventricle and epicardium at E15,65 and in trace amounts in the ventricle at P2.66 Perhaps, the presence of COLI in the epicardium during the developmental stages is crucial for providing the necessary structural stability of the heart wall as well as cues to encourage attachment and infiltration of cells to the epicardium. The lack of COLI in the myocardium and endocardium in the developmental periods may allow for the necessary tissue elasticity as the heart expands and begins to develop the functional capacity to sustain the blood circulation of the developing embryo. This is also consistent with the SHG analysis, which revealed collagens at image depths corresponding primarily to the epicardium.
For all stages, FN was most prevalent in the epicardium. By E14.5, FN was present in thin, isolated fibrils within the myocardium, which increased in density until at least P2, where the fibrils formed a more interconnected network. In addition, FN staining colocalized with nuclei at fetal time points, most distinctly at E16.5. Although the developmental relevance of this observation is unclear, FN has been detected in the nuclear matrix,67 including nuclei of cardiac cells.68 FN gene expression was fairly consistent with protein expression, decreasing at E16.5 and increasing by P2. This is generally similar to previous studies that detected FN gene expression in the endocardium and myocardium at E14,69,70 FN protein in the endocardium at E12,71 the epicardium at E16,71 in fibrils in the subepicardium at E18,64 and in small amounts around myocytes and the endocardium at P2.66 The presence of FN throughout development may be indicative of its roles in adhesion, migration, and differentiation as a primary cell communicator during stages in which cardiac precursors, fibroblasts, and differentiated myocytes are infiltrating the myocardium from the epicardium and endocardium.41
ELN remained relatively constant in appearance and fluorescent intensity throughout the developmental stages except for E16.5, when its fluorescence intensity increased in all assessed locations. Notably, ELN gene expression also increased at E16.5 and again at P2. In the postnatal ventricle, ELN appeared to be relegated to the lining of vessels of the arterial circulation. ELN was previously identified in the E12 myocardium and E14 epicardium, subendocardium, and blood vessels of the embryonic chick heart, while we did not detect ELN during these stages.72 In assessing our results, perhaps, the increase in ELN at E16.5 is necessary for providing the elasticity for heart recoil as the heart is required to exert more ejection force with an increasing size of the heart and fetal circulation. Soon after birth, the relative decrease in ELN may be the result of, or the stimulus for, complete maturation of cardiomyocytes and corresponding maturation of functional sarcomeres.73
We sought to establish a representative blueprint of the spatiotemporal organization, colocalization, and relative amount of four ECM proteins, COLIV, COLI, ELN, and FN, in the developing mouse heart. While experimental feasibility limited our IHC experiments to four ECM proteins, we chose some of the most prevalent proteins in the heart that encompass the key structural and functional characteristics of the myocardium. This work provides an important foundation for generating a developmentally informed blueprint of a stem cell supportive scaffold for cardiac repair. Future studies could include an examination of various mechanical properties of the developing heart that would provide analytical guideposts for testing scaffolds designed based on these blueprints. Furthermore, investigations into other regions of the heart, such as the aorta and right ventricle, could provide a blueprint for scaffold generation geared toward cell therapy in those regions. Additionally, high-throughput protein quantitation technologies, such as protein arrays or mass spectrometry,74 could be used to determine changes in protein amounts among the ECM components during development. These techniques could identify differences in the regulation of ECM components not expressed at the mRNA level (i.e., glycosylated structures, crosslinks, or bound soluble factors). Although such quantitative protein information might supplement the qPCR data presented here, the IHC data provides additional critical spatial details not obtainable with high-throughput methodologies. We demonstrated that this spatial information can be used in conjunction with a controlled fabrication technique to create a synthetic scaffold that imitates the organization of the COLIV matrix in the postnatal day 2 mouse ventricle. Because this fabrication technology can be utilized with ECM proteins,50 this methodology could create ECM protein scaffolds mimicking the in vivo microenvironment of the heart, or other region of interest, during a developmental stage at which the microenvironment elicits cues to its occupying cells to perform a desired function. For example, research groups interested in maintaining, propagating, and encouraging differentiation of cardiac progenitors might consider utilizing the ECM blueprint defined at E16.5. At this time point, the ECM includes structural, elastic, and connective properties, which likely encourage a moderate level of proliferation of cardiomyocytes or their precursors and may serve to align cardiomyocytes to promote mature coupling (Fig. 6B). With the proper technology and chemistry, such a construct might serve to understand cardiac regeneration ex vivo or as a delivery vehicle for cellular transplantation to recover cardiac damage.
Supplementary Material
Acknowledgments
The authors thank Jimmy Fong for image analysis assistance and Peter Crump for statistical assistance. This work was supported by American Heart Association, Innovative Research Grant, 11IRG5570039, and National Institutes of Health, Challenge Grant, 1RC1HL100014.
Disclosure Statement
No competing financial interests exist.
References
- 1.Kehat I. Kenyagin-Karsenti D. Snir M. Segev H. Amit M. Gepstein A., et al. Human embryonic stem cells can differentiate into myocytes with structural and functional properties of cardiomyocytes. J Clin Invest. 2001;108:407. doi: 10.1172/JCI12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang J. Wilson G.F. Soerens A.G. Koonce C.H. Yu J. Palecek S.P., et al. Functional cardiomyocytes derived from human induced pluripotent stem cells. Circ Res. 2009;104:e30. doi: 10.1161/CIRCRESAHA.108.192237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takahashi K. Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 4.Yang L. Soonpaa M.H. Adler E.D. Roepke T.K. Kattman S.J. Kennedy M., et al. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453:524. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- 5.Freund C. Davis R.P. Gkatzis K. Ward-van Oostwaard D. Mummery C.L. The first reported generation of human induced pluripotent stem cells (iPS cells) and iPS cell-derived cardiomyocytes in the Netherlands. Neth Heart J. 2010;18:51. [PMC free article] [PubMed] [Google Scholar]
- 6.Graichen R. Xu X. Braam S.R. Balakrishnan T. Norfiza S. Sieh S., et al. Enhanced cardiomyogenesis of human embryonic stem cells by a small molecular inhibitor of p38 MAPK. Differentiation. 2008;76:357. doi: 10.1111/j.1432-0436.2007.00236.x. [DOI] [PubMed] [Google Scholar]
- 7.Radisic M. Yang L. Boublik J. Cohen R.J. Langer R. Freed L.E., et al. Medium perfusion enables engineering of compact and contractile cardiac tissue. Am J Physiol Heart Circ Physiol. 2004;286:H507. doi: 10.1152/ajpheart.00171.2003. [DOI] [PubMed] [Google Scholar]
- 8.Shimizu T. Yamato M. Isoi Y. Akutsu T. Setomaru T. Abe K., et al. Fabrication of pulsatile cardiac tissue grafts using a novel 3-dimensional cell sheet manipulation technique and temperature-responsive cell culture surfaces. Circ Res. 2002;90:e40. doi: 10.1161/hh0302.105722. [DOI] [PubMed] [Google Scholar]
- 9.Furuta A. Miyoshi S. Itabashi Y. Shimizu T. Kira S. Hayakawa K., et al. Pulsatile cardiac tissue grafts using a novel 3-dimensional cell sheet manipulation technique functionally integrates with the host heart, in vivo. Circ Res. 2006;98:705. doi: 10.1161/01.RES.0000209515.59115.70. [DOI] [PubMed] [Google Scholar]
- 10.Bel A. Planat-Bernard V. Saito A. Bonnevie L. Bellamy V. Sabbah L., et al. Composite cell sheets: a further step toward safe and effective myocardial regeneration by cardiac progenitors derived from embryonic stem cells. Circulation. 2010;122:S118. doi: 10.1161/CIRCULATIONAHA.109.927293. [DOI] [PubMed] [Google Scholar]
- 11.Stevens K.R. Pabon L. Muskheli V. Murry C.E. Scaffold-free human cardiac tissue patch created from embryonic stem cells. Tissue Eng Part A. 2009;15:1211. doi: 10.1089/ten.tea.2008.0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robinson K.A. Li J. Mathison M. Redkar A. Cui J. Chronos N.A., et al. Extracellular matrix scaffold for cardiac repair. Circulation. 2005;112:I135. doi: 10.1161/CIRCULATIONAHA.104.525436. [DOI] [PubMed] [Google Scholar]
- 13.Zimmermann W.H. Schneiderbanger K. Schubert P. Didie M. Munzel F. Heubach J.F., et al. Tissue engineering of a differentiated cardiac muscle construct. Circ Res. 2002;90:223. doi: 10.1161/hh0202.103644. [DOI] [PubMed] [Google Scholar]
- 14.Radisic M. Park H. Gerecht S. Cannizzaro C. Langer R. Vunjak-Novakovic G. Biomimetic approach to cardiac tissue engineering. Philos Trans R Soc Lond B Biol Sci. 2007;362:1357. doi: 10.1098/rstb.2007.2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singelyn J.M. DeQuach J.A. Seif-Naraghi S.B. Littlefield R.B. Schup-Magoffin P.J. Christman K.L. Naturally derived myocardial matrix as an injectable scaffold for cardiac tissue engineering. Biomaterials. 2009;30:5409. doi: 10.1016/j.biomaterials.2009.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kouris N.A. Squirrell J.M. Jung J.P. Pehlke C.A. Hacker T. Eliceiri K.W., et al. A nondenatured, noncrosslinked collagen matrix to deliver stem cells to the heart. Regen Med. 2011;6:569. doi: 10.2217/rme.11.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baharvand H. Azarnia M. Parivar K. Ashtiani S.K. The effect of extracellular matrix on embryonic stem cell-derived cardiomyocytes. J Mol Cell Cardiol. 2005;38:495. doi: 10.1016/j.yjmcc.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 18.Battista S. Guarnieri D. Borselli C. Zeppetelli S. Borzacchiello A. Mayol L., et al. The effect of matrix composition of 3D constructs on embryonic stem cell differentiation. Biomaterials. 2005;26:6194. doi: 10.1016/j.biomaterials.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 19.Weber K.T. Cardiac interstitium in health and disease: the fibrillar collagen network. J Am Coll Cardiol. 1989;13:1637. doi: 10.1016/0735-1097(89)90360-4. [DOI] [PubMed] [Google Scholar]
- 20.Baudino T.A. McFadden A. Fix C. Hastings J. Price R. Borg T.K. Cell patterning: interaction of cardiac myocytes and fibroblasts in three-dimensional culture. Microsc Microanal. 2008;14:117. doi: 10.1017/S1431927608080021. [DOI] [PubMed] [Google Scholar]
- 21.Bugaisky L.B. Zak R. Differentiation of adult rat cardiac myocytes in cell culture. Circ Res. 1989;64:493. doi: 10.1161/01.res.64.3.493. [DOI] [PubMed] [Google Scholar]
- 22.Chen S.S. Fitzgerald W. Zimmerberg J. Kleinman H.K. Margolis L. Cell-cell and cell-extracellular matrix interactions regulate embryonic stem cell differentiation. Stem Cells. 2007;25:553. doi: 10.1634/stemcells.2006-0419. [DOI] [PubMed] [Google Scholar]
- 23.Czyz J. Wobus A. Embryonic stem cell differentiation: the role of extracellular factors. Differentiation. 2001;68:167. doi: 10.1046/j.1432-0436.2001.680404.x. [DOI] [PubMed] [Google Scholar]
- 24.Eschenhagen T. Fink C. Remmers U. Scholz H. Wattchow J. Weil J., et al. Three-dimensional reconstitution of embryonic cardiomyocytes in a collagen matrix: a new heart muscle model system. FASEB J. 1997;11:683. doi: 10.1096/fasebj.11.8.9240969. [DOI] [PubMed] [Google Scholar]
- 25.Macfelda K. Kapeller B. Wilbacher I. Losert U.M. Behavior of cardiomyocytes and skeletal muscle cells on different extracellular matrix components—relevance for cardiac tissue engineering. Artif Organs. 2007;31:4. doi: 10.1111/j.1525-1594.2007.00334.x. [DOI] [PubMed] [Google Scholar]
- 26.Santiago J.A. Pogemiller R. Ogle B.M. Heterogeneous differentiation of human mesenchymal stem cells in response to extended culture in extracellular matrices. Tissue Eng Part A. 2009;15:3911. doi: 10.1089/ten.tea.2008.0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sekine H. Shimizu T. Dobashi I. Matsuura K. Hagiwara N. Takahashi M., et al. Cardiac cell sheet transplantation improves damaged heart function via superior cell survival in comparison with dissociated cell injection. Tissue Eng Part A. 2011;17:2973. doi: 10.1089/ten.tea.2010.0659. [DOI] [PubMed] [Google Scholar]
- 28.Kenar H. Kose G.T. Hasirci V. Design of a 3D aligned myocardial tissue construct from biodegradable polyesters. J Mater Sci Mater Med. 2010;21:989. doi: 10.1007/s10856-009-3917-8. [DOI] [PubMed] [Google Scholar]
- 29.Martens T.P. Godier A.F. Parks J.J. Wan L.Q. Koeckert M.S. Eng G.M., et al. Percutaneous cell delivery into the heart using hydrogels polymerizing in situ. Cell Transplant. 2009;18:297. doi: 10.3727/096368909788534915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ott H.C. Matthiesen T.S. Goh S.K. Black L.D. Kren S.M. Netoff T.I., et al. Perfusion-decellularized matrix: using nature's platform to engineer a bioartificial heart. Nat Med. 2008;14:213. doi: 10.1038/nm1684. [DOI] [PubMed] [Google Scholar]
- 31.Duan Y. Liu Z. O'Neill J. Wan L.Q. Freytes D.O. Vunjak-Novakovic G. Hybrid gel composed of native heart matrix and collagen induces cardiac differentiation of human embryonic stem cells without supplemental growth factors. J Cardiovasc Transl Res. 2011;4:605. doi: 10.1007/s12265-011-9304-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simpson D. Liu H. Fan T.H. Nerem R. Dudley S.C., Jr A tissue engineering approach to progenitor cell delivery results in significant cell engraftment and improved myocardial remodeling. Stem Cells. 2007;25:2350. doi: 10.1634/stemcells.2007-0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manasek F.J. Reid M. Vinson W. Seyer J. Johnson R. Glycosaminoglycan synthesis by the early embryonic chick heart. Dev Biol. 1973;35:332. doi: 10.1016/0012-1606(73)90028-6. [DOI] [PubMed] [Google Scholar]
- 34.Armstrong P.B. Armstrong M.T. A role for fibronectin in cell sorting. J Cell Sci. 1984;69:179. doi: 10.1242/jcs.69.1.179. [DOI] [PubMed] [Google Scholar]
- 35.Kitten G.T. Markwald R.R. Bolender D.L. Distribution of basement membrane antigens in cryopreserved early embryonic hearts. Anat Rec. 1987;217:379. doi: 10.1002/ar.1092170409. [DOI] [PubMed] [Google Scholar]
- 36.Little C.D. Piquet D.M. Davis L.A. Walters L. Drake C.J. Distribution of laminin, collagen type IV, collagen type I, and fibronectin in chicken cardiac jelly/basement membrane. Anat Rec. 1989;224:417. doi: 10.1002/ar.1092240310. [DOI] [PubMed] [Google Scholar]
- 37.Johnson R.C. Manasek F.J. Vinson W.C. Seyer J.M. The biochemical and ultrastructural demonstration of collagen during early heart development. Dev Biol. 1974;36:252. doi: 10.1016/0012-1606(74)90049-9. [DOI] [PubMed] [Google Scholar]
- 38.Thompson R.P. Fitzharris T.P. Denslow S. LeRoy E.C. Collagen synthesis in the developing chick heart. Tex Rep Biol Med. 1979;39:305. [PubMed] [Google Scholar]
- 39.Gallagher J.T. Lyon M. Steward W.P. Structure and function of heparan sulphate proteoglycans. Biochem J. 1986;236:313. doi: 10.1042/bj2360313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tryggvason K. The laminin family. Curr Opin Cell Biol. 1993;5:877. doi: 10.1016/0955-0674(93)90038-r. [DOI] [PubMed] [Google Scholar]
- 41.Linask K.K. Lash J.W. A role for fibronectin in the migration of avian precardiac cells. II. Rotation of the heart-forming region during different stages and its effects. Dev Biol. 1988;129:324. doi: 10.1016/0012-1606(88)90379-x. [DOI] [PubMed] [Google Scholar]
- 42.Schwarzbauer J.E. Fibronectin: from gene to protein. Curr Opin Cell Biol. 1991;3:786. doi: 10.1016/0955-0674(91)90051-y. [DOI] [PubMed] [Google Scholar]
- 43.Jung J.P. Squirrell J.M. Lyons G.E. Eliceiri K.W. Ogle B.M. Imaging cardiac extracellular matrices: a blueprint for regeneration. Trends Biotechnol. 2012;30:233. doi: 10.1016/j.tibtech.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kirby M.L. Cardiac Development. Oxford, NY: Oxford University Press; 2007. [Google Scholar]
- 45.Sedmera D. Reckova M. DeAlmeida A. Coppen S.R. Kubalak S.W. Gourdie R.G., et al. Spatiotemporal pattern of commitment to slowed proliferation in the embryonic mouse heart indicates progressive differentiation of the cardiac conduction system. Anat Rec A Discov Mol Cell Evol Biol. 2003;274:773. doi: 10.1002/ar.a.10085. [DOI] [PubMed] [Google Scholar]
- 46.Mohler W. Millard A.C. Campagnola P.J. Second harmonic generation imaging of endogenous structural proteins. Methods. 2003;29:97. doi: 10.1016/s1046-2023(02)00292-x. [DOI] [PubMed] [Google Scholar]
- 47.Chen X.Y. Nadiarynkh O. Plotnikov S. Campagnola P.J. Second harmonic generation microscopy for quantitative analysis of collagen fibrillar structure. Nat Protoc. 2012;7:654. doi: 10.1038/nprot.2012.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Provenzano P.P. Inman D.R. Eliceiri K.W. Knittel J.G. Yan L. Rueden C.T., et al. Collagen density promotes mammary tumor initiation and progression. BMC Med. 2008;6:11. doi: 10.1186/1741-7015-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schneider C.A. Rasband W.S. Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cunningham L.P. Veilleux M.P. Campagnola P.J. Freeform multiphoton excited microfabrication for biological applications using a rapid prototyping CAD-based approach. Opt Express. 2006;14:8613. doi: 10.1364/oe.14.008613. [DOI] [PubMed] [Google Scholar]
- 51.Campagnola P.J. Delguidice D.M. Epling G.A. Hoffacker K.D. Howell A.R. Pitts J.D., et al. 3-dimensional submicron polymerization of acrylamide by multiphoton excitation of xanthene dyes. Macromolecules. 2000;33:1511. [Google Scholar]
- 52.Sridhar M. Basu S. Scranton V.L. Campagnola P.J. Construction of a laser scanning microscope for multiphoton excited optical fabrication. Rev Sci Instrum. 2003;74:3474. [Google Scholar]
- 53.Lacomb R. Nadiarnykh O. Campagnola P.J. Quantitative second harmonic generation imaging of the diseased state osteogenesis imperfecta: experiment and simulation. Biophys J. 2008;94:4504. doi: 10.1529/biophysj.107.114405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lacomb R. Nadiarnykh O. Townsend S.S. Campagnola P.J. Phase matching considerations in second harmonic generation from tissues: effects on emission directionality, conversion efficiency and observed morphology. Opt Commun. 2008;281:1823. doi: 10.1016/j.optcom.2007.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Porrello E.R. Mahmoud A.I. Simpson E. Hill J.A. Richardson J.A. Olson E.N., et al. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Basu S. Cunningham L.P. Pins G.D. Bush K.A. Taboada R. Howell A.R., et al. Multiphoton excited fabrication of collagen matrixes cross-linked by a modified benzophenone dimer: bioactivity and enzymatic degradation. Biomacromolecules. 2005;6:1465. doi: 10.1021/bm049258y. [DOI] [PubMed] [Google Scholar]
- 57.Chen X.Y. Brewer M.A. Zou C.P. Campagnola P.J. Adhesion and migration of ovarian cancer cells on crosslinked laminin fibers nanofabricated by multiphoton excited photochemistry. Integr Biol. 2009;1:469. doi: 10.1039/b906310b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Basu S. Campagnola P.J. Properties of crosslinked protein matrices for tissue engineering applications synthesized by multiphoton excitation. J Biomed Mater Res Part A. 2004;71A:359. doi: 10.1002/jbm.a.30175. [DOI] [PubMed] [Google Scholar]
- 59.Drake C.J. Davis L.A. Walters L. Little C.D. Avian vasculogenesis and the distribution of collagens I, IV, laminin, and fibronectin in the heart primordia. J Exp Zool. 1990;255:309. doi: 10.1002/jez.1402550308. [DOI] [PubMed] [Google Scholar]
- 60.Speiser B. Riess C.F. Schaper J. The extracellular matrix in human myocardium: Part I: Collagens I, III, IV, and VI. Cardioscience. 1991;2:225. [PubMed] [Google Scholar]
- 61.Corda S. Samuel J.L. Rappaport L. Extracellular matrix and growth factors during heart growth. Heart Fail Rev. 2000;5:119. doi: 10.1023/A:1009806403194. [DOI] [PubMed] [Google Scholar]
- 62.Lundgren E. Gullberg D. Rubin K. Borg T.K. Terracio M.J. Terracio L. In vitro studies on adult cardiac myocytes: attachment and biosynthesis of collagen type IV and laminin. J Cell Physiol. 1988;136:43. doi: 10.1002/jcp.1041360106. [DOI] [PubMed] [Google Scholar]
- 63.Niederreither K. D'Souza R. Metsaranta M. Eberspaecher H. Toman P.D. Vuorio E., et al. Coordinate patterns of expression of type I and III collagens during mouse development. Matrix Biol. 1995;14:705. doi: 10.1016/s0945-053x(05)80013-7. [DOI] [PubMed] [Google Scholar]
- 64.Tidball J.G. Distribution of collagens and fibronectin in the subepicardium during avian cardiac development. Anat Embryol (Berl) 1992;185:155. doi: 10.1007/BF00185916. [DOI] [PubMed] [Google Scholar]
- 65.Hendrix M.J.C. Localization of collagen types in the embryonic heart and aorta using immunohistochemistry. Perspect Cardiovasc Res. 1981;5:213. [Google Scholar]
- 66.Borg T.K. Gay R.E. Johnson L.D. Changes in the distribution of fibronectin and collagen during development of the neonatal rat heart. Coll Relat Res. 1982;2:211. doi: 10.1016/s0174-173x(82)80015-0. [DOI] [PubMed] [Google Scholar]
- 67.Zerlauth G. Wesierska-Gadek J. Sauermann G. Fibronectin observed in the nuclear matrix of HeLa tumour cells. J Cell Sci. 1988;89:415. doi: 10.1242/jcs.89.3.415. [DOI] [PubMed] [Google Scholar]
- 68.van Dijk A. Niessen H.W. Ursem W. Twisk J.W. Visser F.C. van Milligen F.J. Accumulation of fibronectin in the heart after myocardial infarction: a putative stimulator of adhesion and proliferation of adipose-derived stem cells. Cell Tissue Res. 2008;332:289. doi: 10.1007/s00441-008-0573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Samuel J.L. Farhadian F. Sabri A. Marotte F. Robert V. Rappaport L. Expression of fibronectin during rat fetal and postnatal development: an in situ hybridisation and immunohistochemical study. Cardiovasc Res. 1994;28:1653. doi: 10.1093/cvr/28.11.1653. [DOI] [PubMed] [Google Scholar]
- 70.Kim H. Yoon C.S. Rah B. Expression of extracellular matrix components fibronectin and laminin in the human fetal heart. Cell Struct Funct. 1999;24:19. doi: 10.1247/csf.24.19. [DOI] [PubMed] [Google Scholar]
- 71.Bouchey D. Drake C.J. Wunsch A.M. Little C.D. Distribution of connective tissue proteins during development and neovascularization of the epicardium. Cardiovasc Res. 1996;31:E104. [PubMed] [Google Scholar]
- 72.Hurle J.M. Kitten G.T. Sakai L.Y. Volpin D. Solursh M. Elastic extracellular matrix of the embryonic chick heart: an immunohistological study using laser confocal microscopy. Dev Dyn. 1994;200:321. doi: 10.1002/aja.1002000407. [DOI] [PubMed] [Google Scholar]
- 73.Jacot J.G. McCulloch A.D. Omens J.H. Substrate stiffness affects the functional maturation of neonatal rat ventricular myocytes. Biophys J. 2008;95:3479. doi: 10.1529/biophysj.107.124545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Barallobre-Barreiro J. Didangelos A. Schoendube F.A. Drozdov I. Yin X. Fernandez-Caggiano M., et al. Proteomics analysis of cardiac extracellular matrix remodeling in a porcine model of ischemia/reperfusion injury. Circulation. 2012;125:789. doi: 10.1161/CIRCULATIONAHA.111.056952. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.