Abstract
Breast cancer stem cells (BrCSC) are resistant to common therapeutic modalities including chemotherapy, radiation, and hormonal agents. They are thought to contribute to treatment resistance, relapse, and metastases. This study examines the effect of a monoclonal anti-DR5 antibody (TRA-8) and chemotherapy (adriamycin, taxol) on BrCSC populations from basal-like breast cancer cell lines. Doubly enriched BrCSC (CD44+, CD24−, ALDH+) cells were exposed to TRA-8 and control reagents and examined for cytotoxicity, caspase activation, tumorsphere formation and tumorigenicity. Doubly enriched BrCSC populations expressed cell surface DR5 and were sensitive to TRA-8 mediated cytotoxicity with induction of caspase 8 and 3 activation. TRA-8 at sub-nanomolar concentrations inhibited 2LMP and SUM159 BrCSC tumorsphere formation and was more than 50-fold more inhibitory than TRAIL or anti-DR4 at equimolar concentrations. Chemotherapy treatment of 2LMP and SUM159 cell lines resulted in a relative increase of BrCSC, whereas TRA-8 produced a decrease in the percentage of BrCSC. TRA-8 exposure to 2LMP and SUM159 BrCSC preparations produced significant inhibition of tumorigenicity. DR5 maybe a therapeutic target on the surface of basal-like BrCSC which is amenable to agonistic monoclonal anti-DR5 therapy.
Keywords: Anti-DR5, Tigatuzumab, Basal-like breast cancer, Breast cancer stem cells, Tumor initiating cells, Tumorspheres, Death receptor 5
Introduction
Basal-like breast cancer accounts for about 15% of all breast cancer [1]. It is characterized by a unique mRNA profile with CK5/6 expression, inactivation of BRCA1 and commonly lacks estrogen receptor, progesterone receptor, and HER-2 amplification [1–3]. They are further categorized into basal A and basal B subtypes and appear to commonly have substantial numbers of breast cancer stem cells (BrCSC) or tumor initiating cells [4–6].
The cancer stem cell hypothesis suggests that tumors, similar to normal tissue, are organized in a cellular hierarchy, with cancer stem cells (CSC) at the top, as the only cells with potentially limitless proliferation abilities which are capable of driving tumor growth [7]. The more ‘differentiated’ descendants, which account for the majority or bulk of the tumor population, may also be able to proliferate, but regenerative ability is limited [7]. Cancer stem cells were first described in patients with acute leukemia and subsequently in a variety of solid tumors [8, 9]. In breast cancer, CSC were first reported in 2003 by Muhammad Al-Hajj using CD44+ and CD24− surface expression [10]. Since then BrCSC have been characterized based on other cell surface antigens (EpCAM+, CD133+, CD90+) and by functional activities including enhanced efflux pumping of a Hoechst dye (side population), over-expression of aldehyde dehydrogenase (ALDH, ALDEFLUOR assay), retention of the lipophilic dye PKH26, and tumorsphere-forming ability [11–14]. BrCSC are also called tumor initiating cells that are described as having the ability to self-renew, induce tumors at low cell numbers, have low rates of cell division, exhibit chemotherapy and radiation resistance, and have gene expression profiles which differ from the more differentiated cancer cell counterparts [15]. The concept of solid tumor and particularly BrCSC is controversial with several alternative explanations for stem-like cell behaviors [11, 16].
CSC are generally reported to be resistant to chemotherapy and radiation and BrCSC commonly lack ‘targetable’ receptors like ER or HER2 [17–19]. Thus, there is considerable interest in finding therapeutic agents targeted to BrCSC. The presence of substantial numbers of BrCSC in basal-like breast cancer cell lines [10] provided the opportunity to examine the effects of TRA-8 (anti-DR5) on BrCSC enriched populations in terms of anti-DR5 mediated cytotoxicity, inhibition of tumorsphere formation in vitro, and tumorigenicity in vivo. TRA-8 is an agonistic monoclonal anti-DR5 antibody with cytotoxicity and antitumor activity in a variety of human tumor cell lines and murine tumor xenografts [20–23] including basal-like breast cancer cell lines [24].
Materials and methods
Drugs and antibodies
Adriamycin and taxol were purchased from Sigma Aldrich Chemical Co. (St. Louis, MO) and prepared as 10 mM stock solutions in distilled H2O or DMSO, respectively. Purified TRA-8 (IgG1) mAb was provided by Tong Zhou at the University of Alabama at Birmingham (UAB) as described previously [25]. Isotype-specific IgG1 control antibody was obtained from Southern Biotechnology Associates (Birmingham, AL). Anti-DR4 mAb 2E12 (IgG1, k) was provided by Tong Zhou (UAB). Super Killer TRAIL™ was purchased from Enzo Life Sciences International, Inc. (Plymouth Meeting, PA). Conjugated antibodies APC mouse anti-human CD44, PE-Cy7 rat anti-mouse CD44, and corresponding isotype control antibodies were purchased from BD Pharmingen (San Jose, CA). ALDEFLUOR kit including diethylaminobenzaldehyde (DEAB) negative control was obtained from StemCell Technologies (Durham, NC). Cleaved caspase 8 rabbit mAb and cleaved caspase 3 rabbit mAb were purchased from Cell Signaling (Billerica, MA). Secondary antibodies, Alexa fluor 405 goat anti-rabbit IgG, and Alexa fluor 647 goat anti-mouse IgG1 were purchased from Invitrogen (Carlsbad, CA).
Cells and cell culture
The 2LMP subclone of the human breast cancer cell line MDA-MB-231 was obtained from Dr. Marc Lippman (University of Miami, Coral Gables, FL) and maintained in improved MEM supplemented with 10% FBS (Hyclone, Logan, UT). Basal-like cell lines HCC38, HCC1187, HCC1143, MDA-MB-436, BT-20, and BT-549 were obtained from American Type Culture Collection (Manassas, VA) and cultured according to supplier’s directions with the exception of MDA-MB-436, which was grown in DMEM supplemented with 10 μg/ml insulin, glutathione, and 10% FBS. SUM159 was obtained from Asterand (Detroit, MI) and grown according to supplier’s recommendation. All cell lines were maintained in antibiotic-free medium at 37°C in a 5% CO2 atmosphere and routinely screened for Mycoplasma contamination. Sorted cells and tumorspheres were maintained in MEGM medium (Lonza, Walkersville, MD).
Doubly enriched BrCSC isolation by flow cytometry
Basal-like cell lines were plated in T75 flasks (Costar, Cambridge, MA) in corresponding media and harvested at 75% confluence. Cells were harvested with trypsin and labeled with 1 μl of ALDEFLUOR reagent in 100 μl ALDEFLUOR buffer per 5 × 106 cells and incubated at 37°C for 30 min. Cells were then labeled with APC-CD44 (1:25) and PE-CD24 (1:25) in 200 μl of ALDEFLUOR buffer on ice for 15 min. The ALDEFLUOR positive population was established by using 2 × 106 ALDEFLUOR labeled cells and 5 μl DEAB in 200 μl ALDEFLUOR buffer. The sorting gates were established using negative controls, DEAB and side scatter and forward scatter profiles were used to eliminate cell doublets [10, 11, 16]. Samples were sorted on a Becton–Dickinson-FACSAriaII™ or analyzed on Becton–Dickinson-LSRII™ flow cytometer (Chicago, IL). Data was evaluated using FlowJo software (Tree Star, Inc., Ashland, OR).
DR5 expression and functional caspase activation
2LMP, SUM159, and HCC1143 cell lines were harvested using cell stripper (Mediatech, Manassas, VA) to prevent cleavage of death receptor. Cells were incubated with ALDEFLUOR reagents for 30 min at 37°C. Cells were then labeled on ice with TRA-8 (IgG1) or IgG1 istotype control for 15 min. Cells were then incubated with CD44-PE-Cy7 (1:1,000), CD24-PE (1:100), and secondary antibody (Alexa-647) (1:100) for 15 min on ice. Samples were analyzed by flow cytometry for DR5 expression on the CD44+/CD24−/ALDH+ subpopulation. Analysis of caspase 8 and 3 activation of BrCSC was accomplished by harvesting cells using cell stripper and sorting for the ALDH+ population. Sorted cells were treated for 2 h with TRA-8 or IgG1 control (~1 × 106 cells with 1 μg/ml TRA-8 or IgG1 in MEGM medium + 2% BSA). Cells were fixed with 1% paraformaldehyde for 5 min on ice and labeled with CD44-APC and CD24-PE (1:100) on ice for 15 min. Cells were then permeabilized using 3% BSA, 0.1% saponin in 200 μl PBS on ice for 15 min and labeled with cleaved caspase 3 or 8 (1:500) on ice for 15 min. Cells were incubated with secondary antibody Alexa-405 anti-rabbit (1:100) on ice for 15 min. Samples were kept in 0.1% saponin and analyzed by flow cytometry. Analysis was done on a minimum of three independent experiments.
Cell viability assays using ATPLite
Sorted CD44+/CD24−/ALDH+ cells were plated on ultra-low attachment plates (Costar) at 2,000 cells per 50 μl of MEGM medium. Bulk unseparated cells were collected from total viable gates established by forward and side scatter parameters to control for any variables introduced by sorting the cells. Cells from the bulk unseparated populations were plated in optically clear 96-well black plates (Costar) in corresponding media. Sorted and bulk cells were treated with (0.1, 1, 10, 100, or 1,000 ng/ml) of TRA-8 immediately after plating and incubated for 24 h at 37°C. TRA-8 was diluted in culture medium immediately before use. Cell viability was determined by measurement of cellular ATP levels using the ATPLite luminescence-based assay (Packard Instruments, Meriden, CT) described else-where [26]. The manufacturer’s recommended protocol was followed with the exception that all reaction volumes (culture medium and reagents) were reduced by one-half. All samples were assayed in quadruplicate and IC50 values are reported as the median from a minimum of three independent experiments.
In vitro treatment of tumorspheres
2LMP and SUM159 cell lines were sorted for ALDH+ cells. Approximately ~1 × 106 cells were allowed to form primary spheres at a density of 100,000 cells/ml for 3–4 days in MEGM medium. Tumorspheres were mechanically dissociated and plated in ultra-low attachment 96-well plates (Costar) at 2,000 cells per well. TRA-8 (anti-DR5), 2E12 (anti-DR4), TRAIL, IgG isotype control, adriamycin, or taxol were added to the wells and incubated at 37°C for 48 h in quadruplicate. Tumorspheres were visually counted using a reticle eye piece. Mean tumorsphere inhibition was calculated relative to untreated control spheres. At least three independent experiments were conducted per cell line in quadruplicate.
Effect of drug treatment of breast cancer cells on BrCSC population
2LMP and SUM159 breast cancer cells were plated in 6-well well culture plates at 80,000 cells/well (Costar 3516). Cells were treated with adriamycin (200 nM) or taxol (200 nM) for 48 h, or TRA-8 (10 ng/ml) for 24 h. Cells in suspension after treatment along with attached cells were harvested and incubated with Aldefluor reagent for 30 min at 37°C following manufacturer’s protocol. Cells were analyzed using a LSRII™ flow cytometer (Becton–Dickinson). Cells were gated based on forward and side scatter properties for single viable cells. The signal from autofluorescence of drug treatment was accounted for in the final analysis of BrCSC ALDH marker expression.
Ex vivo treatment of BrCSC and tumor implantation
CD44+/CD24−/ALDH+ 2LMP and SUM159 cells (1 × 106) were sorted and allowed to recover for 13 h in MEGM medium in ultra-low attachment plates at 37°C. Cells (2 × 104) were separated into treatment groups and drug or antibody was added (IgG, 20 nM), 2E12 (20 nM), TRA-8 (20 nM), and adriamycin (500 nM). Cells were treated for 3 h at 37°C and then aliquoted in 200 μl (1:1 Matrigel) and injected into the mammary fat pad of 4 week old NOD/SCID mice (Harlan, Prattville, AL). Tumor size was determined by the product of two largest diameters. Two independent animal experiments were conducted for the 2LMP and SUM159 cell lines.
Statistical analysis
The IC50 is the drug concentration producing the median effect of 50% cell killing which was estimated based on the Hill Equation with nonlinear regression model for each assay [27]. Due to small number of replicate experiments, a nonparametric statistical method with Kruskal–Wallis test was used for the comparison between two groups, e.g., IC50, percentage of tumorsphere number and ATP level [28]. The Generalized Linear Model (GLM) with PROC MIXED was used to compare the tumor size over time among experimental groups. Main effect and interaction between treatment groups and measurement time point were fitted in the model with appropriate variance and covariance structure selected. The statistical analysis was carried out with Statistical Analysis Software (SAS) version 9.2.
Results
Anti-DR5 (TRA-8) induced cytotoxicity to BrCSC enriched cells
Eight basal-like cell lines underwent dual BrCSC enrichment (CD44+/CD24−/ALDH+) and these BrCSC enriched populations were compared with their unseparated parental cells in regard to sensitivity to anti-DR5 (TRA-8) mediated cytotoxicity (Table 1). As reported previously [24], the basal-like cell lines were quite sensitive (IC50 < 100 ng/ml) to TRA-8 mediated cytotoxicity except for HCC1143 which was moderately resistant (IC50 of 101–1,000 ng/ml). All the cell line BrCSC enriched cell preparations were very sensitive to anti-DR5 mediated cytotoxicity including cell line HCC1143. In 6/8 instances, the BrCSC enriched cells were significantly more sensitive than their parental cells.
Table 1.
Sensitivity of sorted BrCSC and unseparated parental cells to TRA-8 mediated cytotoxicity
| Phenotype | Cell line | Sorted CD44+CD24− ALDH+ IC50 TRA-8 (ng/ml) |
Unseparated parental IC50 TRA-8 (ng/ml) |
P value sorted vs. unsorted |
|---|---|---|---|---|
| Basal B | HCC38 | 0.10a | 0.74 | 0.127 |
| 2LMP | 0.65 | 1.06 | 0.008 | |
| SUM159 | 0.88 | 5.54 | 0.049 | |
| MDA-MB-436 | 0.62 | 0.31 | 0.248 | |
| BT-549 | 0.63 | 5.55 | 0.02 | |
| Basal A | HCC1187 | 0.85 | 24.72 | 0.049 |
| BT-20 | 7.24 | 16.49 | 0.275 | |
| HCC1143 | 77.12 | 628.43 | 0.049 |
All samples were assayed in quadruplicate and are reported as the median from a minimum of three independent experiments
DR5 expression of BrCSC enriched cells
The dual separated (CD44+/CD24−/ALDH+) and unseparated cell preparations from 2LMP, SUM159, and HCC1143 underwent flow cytometry for analysis of cell membrane expression of DR5. As shown in Fig. 1, cells from all cell lines were strongly positive and the unseparated (blue lines) and dual enriched (red lines) had comparable expression of DR5.
Fig. 1.
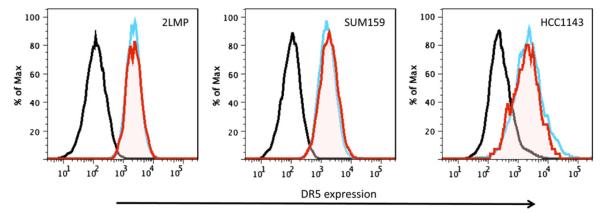
Flow cytometry analysis of DR5 membrane expression on the CD44+/CD24−/ALDH+ subpopulation of basal-like breast cancer cells. 2LMP, SUM159, and HCC1143 cells were labeled with ALDEFLUOR (FITC), CD44 (PE, Cy7), CD24 (PE), and anti-DR5 (TRA-8 Alexa-647) then analyzed by flow cytometry. DR5 membrane expression on total unsorted bulk population (blue line) or CD44+/CD24−/ALDH+ (red line) subpopulation compared with isotype control (black line). 2LMP, SUM159, and HCC1143 CD44+/CD24−/ALDH+ subpopulations had similar DR5 expression compared with the total unsorted cell population
Apoptosis of anti-DR5 treated BrCSC enriched cell populations
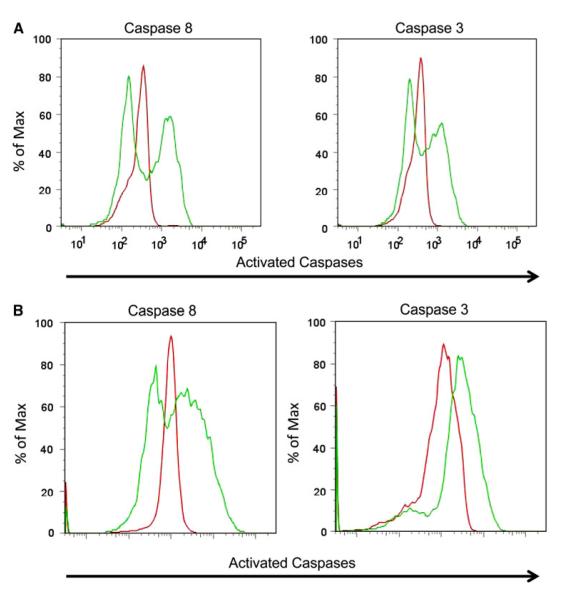
To determine that anti-DR5 can mediate apoptosis of the BrCSC enriched cells, the 2LMP and SUM159 ALDH+ populations were incubated with anti-DR5 or control IgG for 2 h, and the CD44+ CD24− cell population were tested for cellular expression of activated caspase 8 and activated caspase 3. Figure 2 illustrates that a substantial portion of the 2LMP (Fig. 2a) and SUM159 (Fig. 2b) BrCSC underwent caspase 8 and 3 activation (green lines) as compared with BrCSC exposed to control IgG (red line). This delineates that anti-DR5 triggers caspase activation and apoptosis of BrCSC doubly enriched cells over even short durations of 2–3 h.
Fig. 2.
Flow cytometry analysis of caspase activation in CD44+/CD24−/ALDH+ subpopulation of 2LMP and SUM159 cells after treatment with TRA-8. 2LMP (a) and SUM159 (b) cells were sorted for the ALDH+ subpopulation and then treated with TRA-8 or control IgG for 2 h. Cells were then fixed and stained for CD44 (APC), CD24 (PE), and activated caspases 3 or 8 (secondary Alexa 405). 2LMP and SUM159 BrCSC enriched cells had caspase 3 and caspase 8 activation (green line) after incubation with TRA-8 compared with IgG control (red line)
Analysis of anti-DR5 effect on BrCSC tumorsphere formation
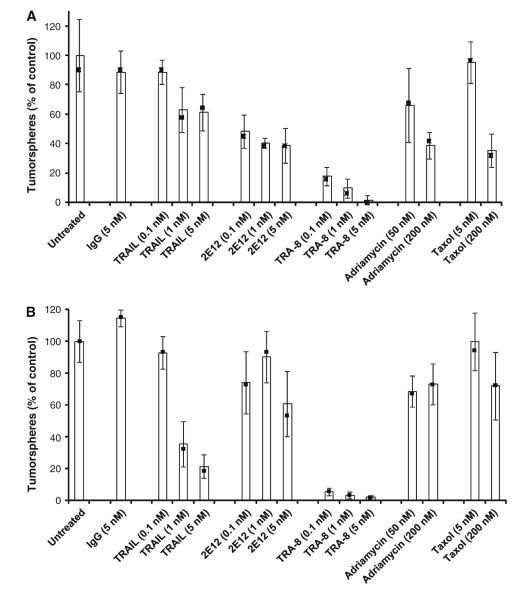
Tumorsphere formation has been reported as a measure of BrCSC presence in enriched cell populations [29]. Figure 3 provides the effects of anti-DR5 (TRA-8), anti-DR4 (2E12), TRAIL, adriamycin, taxol, control IgG, and control media on secondary tumorsphere formation of 2LMP (Fig. 3a), and SUM159 (Fig. 3b) cell lines. Impressive inhibition of secondary tumorsphere formation was caused by anti-DR5 at doses as low as 0.1 nM with 80% inhibition of 2LMP cells (P = 0.019) and 95% inhibition of SUM159 cells (P = 0.02). Anti-DR5 produced significantly more inhibition of tumorsphere formation at 0.1 nM than 5.0 nM TRAIL (P = 0.019) or 5.0 nM anti-DR4 (P = 0.028) in 2LMP cells. The SUM159 cells had comparable observations with 0.1 nM anti-DR5 producing more inhibition than 5.0 nM TRAIL (P = 0.019) or 5.0 nM anti-DR4 (P = 0.02). Adriamycin and taxol had modest or no inhibitory effects. Thus, anti-DR5 appears to be superior to other DR-mediated agents at tumorsphere formation inhibition.
Fig. 3.
Secondary tumorsphere formation inhibition by TRA-8. 2LMP cells (a) and SUM159 (b) cells were sorted using flow cytometry for ALDH+ cells and allowed to form primary tumorspheres for 3 days. After tumorspheres were mechanically dissociated, single cells (2,000 cells/well) were plated in low attachment plates and treated with IgG, TRAIL, 2E12 (anti-DR4), TRA-8, adriamycin or taxol. After 48 h, tumorspheres ranging from 40 to 120 μm in size were visually counted using a reticle eye piece. Mean tumorsphere inhibition was calculated relative to untreated controls (blue bars) (filled square) represent median values. Error bars represent SD of the samples run in quadruplicate
Effect of drug exposure of breast cancer cell lines on BrCSC population
Drug or TRA-8 treatment of breast cancer cell lines may change the percentage of ALDH+ cells (BrCSC) in the total cell population. Flow cytometry analysis of 2LMP basal-like cells after adriamycin or taxol treatment showed a 4.6-fold or 2.2-fold increase of ALDH+ cells, respectively (Fig. 4). SUM159 cells treated with adriamycin or taxol had a 3.3-fold and 1.9-fold increase in the percentage of ALDH+ cells, respectively. By contrast, 2LMP and SUM159 cells treated with TRA-8 resulted in a 1.8-fold and 1.7-fold decrease in ALDH+ tumor cells compared with untreated cells, respectively. Thus, BrCSC appear to be resistant to chemotherapy but sensitive to anti-DR5.
Fig. 4.
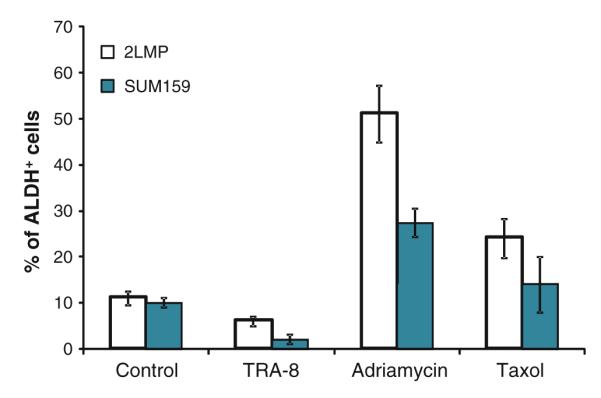
In vitro treatment with TRA-8 decreased the percentage of ALDH+ cells in 2LMP and SUM159 breast cancer cell lines. 2LMP and SUM159 cells (80,000 cells/well) were treated with TRA-8 (10 ng/ml) for 24 h, and adriamycin (200 nM) or taxol (200 nM) for 48 h in 6-well cell culture plates. Mean ALDH+ cells after treatment from a minimum of three independent experiments are shown. Error bars denote SE
Effect of anti-DR5 exposure on BrCSC enriched cell population tumorigenicity
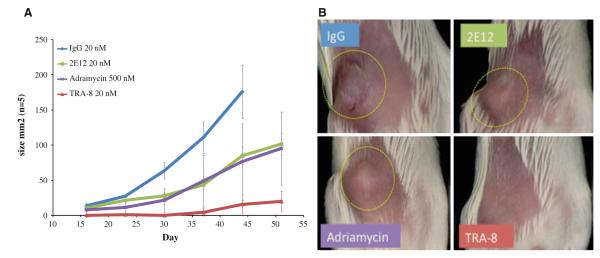
The major requirement of BrCSC is the ability to generate fully constituted human breast cancer in immuno-compromised mice. BrCSC enriched cell populations were exposed to anti-DR5 (TRA-8), anti-DR4 (2E12), control IgG, or adriamycin for 3 h before injection of treated cells into the mammary fat pads of NOD/SCID mice (n = 5 for each group). Figure 5a illustrates that the control IgG, anti-DR4, and adriamycin treated cells generated 5/5 tumors. The IgG control mice were sacrificed on day 44 to comply with IACUC guidelines with tumor sizes exceeding 175 mm2. The anti-DR4 and adriamycin treated tumors were somewhat slower growing but not statistically different than the control. In contrast, the anti-DR5 treated cells developed measurable tumors in only 2/5 animals by day 50 and even these tumors were small and had severely retarded growth compared with the IgG control treated tumors (P = 0.0001). Figure 5b illustrates examples of the 2LMP tumors in the control and treated NOD/SCID mice. The tumors that grew in the four treated groups of mice had similar histology with poorly differentiated cells, high mitotic rates, and focal areas of necrosis. Similar studies with SUM159 cell line showed that the IgG treated BrCSC enriched cells generated 5/5 tumors compared with 0/5 mice observed in the TRA-8 treated group at day 105. Thus, it appears that TRA-8 can seriously impair the tumorigenicity of BrCSC enriched cell populations.
Fig. 5.
Effect of ex vivo treatment of BrCSC enriched cells on tumorgenicity in NOD/SCID mice. 2LMP cells were sorted using flow cytometry for CD44+/CD24−/ALDH+ BrCSC markers and the cells were allowed to recover for 13 h. Cells were treated with TRA-8, 2E12, adriamycin or IgG control for 3 h and implanted into the MFP of groups of five NOD/SCID mice. a Graph represents the average tumor size and number of tumors formed. Only 2/5 small, slow growing tumors were observed to develop within 50 days with TRA-8 treated cells while 5/5 tumors developed in the IgG, 2E12, and adriamycin treatment groups (P value <0.0001). b These are images taken of one representative mouse in each group at day 30 after implantation (dotted circle shows the tumor)
Discussion
There is considerable interest in finding therapeutic agents that could be targeted to CSC to enhance the efficacy of treatment regimens and potentially reduce tumor resistance and relapse. We have previously shown the anti-tumor activity of an agonistic monoclonal anti-DR5 antibody (TRA-8) to 2LMP and other basal-like cell lines in vitro and in vivo [24]. Others have shown DR-mediated cytotoxicity to basal B but not basal A breast cancer cell lines [30]. Given that basal-like cell lines are enriched in CSC [5, 18], it represented an opportunity to examine the effect of TRA-8 on BrCSC populations.
Doubly enriched BrCSC subpopulations (CD44+/CD24−/ALDH+) of both basal-like A and B type were sensitive to TRA-8 mediated cytotoxicity. Further, DR5 expression on BrCSC subpopulations were identical to their unseparated parental population and brief interaction with TRA-8 triggered caspase 8 and 3 activation. Thus, it appears that basal-like BrCSC subpopulations share sensitivity to anti-DR5 mediated cytotoxicity similar to their parental cells and in some instances even have increased sensitivity. In vitro treatment of parental breast cancer cell lines with adriamycin or taxol increased the percentage of ALDH+ cells, while TRA-8 produced a decrease in the percentage of ALDH+ cells. These results indicate that the bulk cells were more sensitive to chemotherapy treatment than the BrCSC, whereas the BrCSC were more sensitive to TRA-8 treatment than the bulk cells. Similarly, anti-DR5 treatment decreased the percentage of CSC in pancreatic cancer [31].
A prior study had reported that TRAIL was able to mediate cytotoxicity to colon cancer CSC (dye efflux side population) and that this population was enriched for expression of DR4 [32]. We thus contrasted the effects of TRAIL, anti-DR4, and anti-DR5 on BrCSC tumorsphere formation. These studies demonstrated the superiority of TRA-8 over TRAIL and anti-DR4 in terms of inhibition of BrCSC tumorsphere formation. Similarly, TRA-8 was superior to anti-DR4 in inhibition of BrCSC tumorigenicity. This may reflect differences among CSC of different tumor types. Indeed, CSC from glioblastoma cell lines have been reported to be resistant to TRAIL mediated cytotoxicity [33].
These observations suggest that DR5 maybe a target on the surface of basal-like breast cancer cell lines and BrCSC by which an agonistic monoclonal anti-DR5 antibody could mediate anti-tumor activity/efficacy. Tigatuzumab is the CDR grafted, humanized version of TRA-8 which has entered clinical trials [34]. Because of these studies and others, the Translational Breast Cancer Research Consortium has recently opened a randomized phase II trial of abraxane ± tigatuzumab for metastatic triple negative breast cancer (ClinicalTrials.gov NCT01307891).
Acknowledgments
Supported in part by NIH SPORE in Breast Cancer 5P50 CA089019-08, Komen for the Cure Promise Grant KG090969, Breast Cancer Research Foundation of Alabama, and DOD Training grant W81XWH-11-1-0151. The authors thank Dr. William Grizzle for histologic analysis of tumor grafts. Technical support was provided by Andres Aristizabal and Enid Keyser. D. J. Buchsbaum and Albert F. LoBuglio: intellectual property interest in TRA-8, Daiichi Sankyo.
Footnotes
Portions of this manuscript were presented as a poster (Late-Breaking Abstract, Tumor Biology 2 # LB-260) at the 101st AACR Annual Meeting, Washington DC, 17–21 April 2010.
Contributor Information
Angelina I. Londoño-Joshi, Department of Molecular and Cellular Pathology, University of Alabama at Birmingham, 1825 University Blvd. SHEL 771, Birmingham, AL 35294-2182, USA
Patsy G. Oliver, Department of Radiation Oncology, University of Alabama at Birmingham, 1825 University Blvd; SHEL 701, Birmingham, AL 35294-2182, USA
Yufeng Li, Department of Medicine, University of Alabama at Birmingham Cancer Center, Birmingham, AL, USA.
Choo Hyung Lee, Department of Medicine, University of Alabama at Birmingham Cancer Center, Birmingham, AL, USA.
Andres Forero-Torres, Department of Medicine, University of Alabama at Birmingham Cancer Center, Birmingham, AL, USA.
Albert F. LoBuglio, Department of Medicine, University of Alabama at Birmingham Cancer Center, Birmingham, AL, USA
Donald J. Buchsbaum, Department of Radiation Oncology, University of Alabama at Birmingham, 1825 University Blvd; SHEL 701, Birmingham, AL 35294-2182, USA
References
- 1.Liedtke C, Gonzalez-Angulo A-M, Pusztai L. Definition of triple-negative breast cancer and relationship to basal-like molecular subtype. PPO Updat Prin Prac Oncol. 2010;24:1–6. [Google Scholar]
- 2.Nielsen TO, Hsu FD, Jensen K, Cheang M, Karaca G, Hu Z, Hernandez-Boussard T, Livasy C, Cowan D, Dressler L, Akslen LA, Ragaz J, Gown AM, Gilks CB, van de Rijn M, Perou CM. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10(16):5367–5374. doi: 10.1158/1078-0432.CCR-04-0220. doi:10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- 3.Lakhani SR, Reis-Filho JS, Fulford L, Penault-Llorca F, van der Vijver M, Parry S, Bishop T, Benitez J, Rivas C, Bignon YJ, Chang-Claude J, Hamann U, Cornelisse CJ, Devilee P, Beckmann MW, Nestle-Kramling C, Daly PA, Haites N, Varley J, Lalloo F, Evans G, Maugard C, Meijers-Heijboer H, Klijn JG, Olah E, Gusterson BA, Pilotti S, Radice P, Scherneck S, Sobol H, Jacquemier J, Wagner T, Peto J, Stratton MR, McGuffog L, Easton DF. Prediction of BRCA1 status in patients with breast cancer using estrogen receptor and basal phenotype. Clin Cancer Res. 2005;11(14):5175–5180. doi: 10.1158/1078-0432.CCR-04-2424. doi:10.1158/1078-0432.CCR-04-2424. [DOI] [PubMed] [Google Scholar]
- 4.Honeth G, Bendahl PO, Ringner M, Saal LH, Gruvberger-Saal SK, Lovgren K, Grabau D, Ferno M, Borg A, Hegardt C. The CD44+/CD24− phenotype is enriched in basal-like breast tumors. Breast Cancer Res. 2008;10(3):R53. doi: 10.1186/bcr2108. doi:10.1186/bcr2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Charafe-Jauffret E, Ginestier C, Iovino F, Wicinski J, Cervera N, Finetti P, Hur MH, Diebel ME, Monville F, Dutcher J, Brown M, Viens P, Xerri L, Bertucci F, Stassi G, Dontu G, Birnbaum D, Wicha MS. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 2009;69(4):1302–1313. doi: 10.1158/0008-5472.CAN-08-2741. doi:10.1158/0008-5472.CAN-08-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giatromanolaki A, Sivridis E, Fiska A, Koukourakis MI. The CD44+/CD24− phenotype relates to ‘triple-negative’ state and unfavorable prognosis in breast cancer patients. Med Oncol. 2010 doi: 10.1007/s12032-010-9530-3. doi:10.1007/s12032-010-9530-3. [DOI] [PubMed] [Google Scholar]
- 7.Dontu G. Breast cancer stem cell markers—the rocky road to clinical applications. Breast Cancer Res. 2008;10(5):110. doi: 10.1186/bcr2130. doi:10.1186/bcr2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3(7):730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 9.Ailles LE, Weissman IL. Cancer stem cells in solid tumors. Curr Opin Biotechnol. 2007;18(5):460–466. doi: 10.1016/j.copbio.2007.10.007. doi:10.1016/j.copbio.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 10.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100(7):3983–3988. doi: 10.1073/pnas.0530291100. doi:10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG, Liu S, Schott A, Hayes D, Birnbaum D, Wicha MS, Dontu G. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1(5):555–567. doi: 10.1016/j.stem.2007.08.014. doi:10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donnenberg VS, Donnenberg AD, Zimmerlin L, Landreneau RJ, Bhargava R, Wetzel RA, Basse P, Brufsky AM. Localization of CD44 and CD90 positive cells to the invasive front of breast tumors. Cytom B Clin Cytom. 2010;78(5):287–301. doi: 10.1002/cyto.b.20530. doi:10.1002/cyto.b.20530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meyer MJ, Fleming JM, Lin AF, Hussnain SA, Ginsburg E, Vonderhaar BK. CD44posCD49fhiCD133/2hi defines xenograft-initiating cells in estrogen receptor-negative breast cancer. Cancer Res. 2010;70(11):4624–4633. doi: 10.1158/0008-5472.CAN-09-3619. doi:10.1158/0008-5472. CAN-09-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pece S, Tosoni D, Confalonieri S, Mazzarol G, Vecchi M, Ronzoni S, Bernard L, Viale G, Pelicci PG, Di Fiore PP. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell. 2010;140(1):62–73. doi: 10.1016/j.cell.2009.12.007. doi:10.1016/j.cell.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 15.Fillmore CM, Kuperwasser C. Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res. 2008;10(2):R25. doi: 10.1186/bcr1982. doi:10.1186/bcr1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Croker AK, Goodale D, Chu J, Postenka C, Hedley BD, Hess DA, Allan AL. High aldehyde dehydrogenase and expression of cancer stem cell markers selects for breast cancer cells with enhanced malignant and metastatic ability. J Cell Mol Med. 2009;13(8B):2236–2252. doi: 10.1111/j.1582-4934.2008.00455.x. doi:10.1111/j.1582-4934.2008.00455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Debeb BG, Xu W, Woodward WA. Radiation resistance of breast cancer stem cells: Understanding the clinical framework. J Mammary Gland Biol Neoplasia. 2009;14(1):11–17. doi: 10.1007/s10911-009-9114-z. doi:10.1007/s10911-009-9114-z. [DOI] [PubMed] [Google Scholar]
- 18.Ricardo S, Vieira AF, Gerhard R, Leitao D, Pinto R, Cameselle-Teijeiro JF, Milanezi F, Schmitt F, Paredes J. Breast cancer stem cell markers CD44, CD24 and ALDH1: expression distribution within intrinsic molecular subtype. J Clin Pathol. 2011 doi: 10.1136/jcp.2011.090456. doi:10.1136/jcp.2011.090456. [DOI] [PubMed] [Google Scholar]
- 19.Kakarala M, Wicha MS. Implications of the cancer stem-cell hypothesis for breast cancer prevention and therapy. J Clin Oncol. 2008;26(17):2813–2820. doi: 10.1200/JCO.2008.16.3931. doi:10.1200/JCO.2008.16.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeRosier LC, Vickers SM, Zinn KR, Huang Z, Wang W, Grizzle WE, Sellers JC, Stockard CR, Jr, Zhou T, Oliver PG, Arnoletti JP, LoBuglio AF, Buchsbaum DJ. TRA-8 anti-DR5 monoclonal antibody and gemcitabine induce apoptosis and inhibit radiologically validated orthotopic pancreatic tumor growth. Mol Cancer Ther. 2007;6:3198–3207. doi: 10.1158/1535-7163.MCT-07-0299. doi:10.1158/1535-7163.MCT-07-0299. [DOI] [PubMed] [Google Scholar]
- 21.Oliver PG, LoBuglio AF, Zinn KR, Kim H, Nan L, Zhou T, Wang W, Buchsbaum DJ. Treatment of human colon cancer xenografts with TRA-8 anti-death receptor 5 antibody alone or in combination with CPT-11. Clin Cancer Res. 2008;14:2180–2189. doi: 10.1158/1078-0432.CCR-07-1392. doi:10.1158/1078-0432.CCR-07-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amm HM, Zhou T, Steg AD, Kuo H, Li Y, Buchsbaum DJ. Mechanisms of drug sensitization to TRA-8, an agonistic death receptor 5 antibody, involve modulation of the intrinsic apoptotic pathway in human breast cancer cells. Mol Cancer Res. 2011;9:403–417. doi: 10.1158/1541-7786.MCR-10-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bevis KS, McNally LR, Sellers JC, Della Manna D, Londoño-Joshi A, Amm H, Straughn JJM, Buchsbaum DJ. Anti-tumor activity of an anti-DR5 monoclonal antibody, TRA-8, in combination with taxane/platinum-based chemotherapy in an ovarian cancer model. Gynecol Oncol. 2011;121(1):193–199. doi: 10.1016/j.ygyno.2010.11.046. [DOI] [PubMed] [Google Scholar]
- 24.Forero-Torres A, Oliver PG, Joshi AIL, Zhou T, LoBuglio AF, Buchsbaum DJ. Death receptor 5, a therapeutic target for triple negative breast cancer. The Breast Cancer Symposium 2010; San Antonio, Texas. 8–12 December 2010.2010. [Google Scholar]
- 25.Ichikawa K, Liu W, Zhao L, Wang Z, Liu D, Ohtsuka T, Zhang H, Mountz JD, Koopman WJ, Kimberly RP, Zhou T. Tumoricidal activity of a novel anti-human DR5 monoclonal antibody without hepatocyte cytotoxicity. Nat Med. 2001;7:954–960. doi: 10.1038/91000. doi:10.1038/91000. [DOI] [PubMed] [Google Scholar]
- 26.Buchsbaum DJ, Zhou T, Grizzle WE, Oliver PG, Hammond CJ, Zhang S, Carpenter M, LoBuglio AF. Antitumor efficacy of TRA-8 anti-DR5 monoclonal antibody alone or in combination with chemotherapy and/or radiation therapy in a human breast cancer model. Clin Cancer Res. 2003;9:3731–3741. [PubMed] [Google Scholar]
- 27.Hill AV. The possible effects of the aggregation of the molecules of haemoglobin on its dissociation curves. J Physiol. 1910;40:4–7. [Google Scholar]
- 28.Gibbons JD. Nonparametric statistical inference. McGraw-Hill; New York: 1971. [Google Scholar]
- 29.Grimshaw MJ, Cooper L, Papazisis K, Coleman JA, Bohnenkamp HR, Chiapero-Stanke L, Taylor-Papadimitriou J, Burchell JM. Mammosphere culture of metastatic breast cancer cells enriches for tumorigenic breast cancer cells. Breast Cancer Res. 2008;10(3):R52. doi: 10.1186/bcr2106. doi:10.1186/bcr2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rahman M, Davis SR, Pumphrey JG, Bao J, Nau MM, Meltzer PS, Lipkowitz S. TRAIL induces apoptosis in triple-negative breast cancer cells with a mesenchymal phenotype. Breast Cancer Res Treat. 2009;113:217–230. doi: 10.1007/s10549-008-9924-5. doi:10.1007/s10549-008-9924-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rajeshkumar NV, Rasheed ZA, Garcia-Garcia E, Lopez-Rios F, Fujiwara K, Matsui WH, Hidalgo M. A combination of DR5 agonistic monoclonal antibody with gemcitabine targets pancreatic cancer stem cells and results in long-term disease control in human pancreatic cancer model. Mol Cancer Ther. 2010;9(9):2582–2592. doi: 10.1158/1535-7163.MCT-10-0370. doi:10.1158/1535-7163.MCT-10-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sussman RT, Ricci MS, Hart LS, Sun SY, El-Deiry WS. Chemotherapy-resistant side-population of colon cancer cells has a higher sensitivity to TRAIL than the non-SP, a higher expression of c-Myc and TRAIL-receptor DR4. Cancer Biol Ther. 2007;6(9):1490–1495. doi: 10.4161/cbt.6.9.4905. [DOI] [PubMed] [Google Scholar]
- 33.Capper D, Gaiser T, Hartmann C, Habel A, Mueller W, Herold-Mende C, von Deimling A, Siegelin MD. Stem-cell-like glioma cells are resistant to TRAIL/Apo2L and exhibit down-regulation of caspase-8 by promoter methylation. Acta Neuropathol. 2009;117(4):445–456. doi: 10.1007/s00401-009-0494-3. doi:10.1007/s00401-009-0494-3. [DOI] [PubMed] [Google Scholar]
- 34.Forero-Torres A, Shah J, Wood T, Posey J, Carlisle R, Copigneaux C, Luo FR, Wojtowicz-Praga S, Percent I, Saleh M. Phase I trial of weekly tigatuzumab, an agonistic humanized monoclonal antibody targeting death receptor 5 (DR5) Cancer Biother Radiopharm. 2010;25:13–19. doi: 10.1089/cbr.2009.0673. doi:10.1089/cbr.2009.0673. [DOI] [PMC free article] [PubMed] [Google Scholar]