Abstract
To date, lacking of a clinically-suitable source of engraftable human stem/progenitor cells with adequate neurogenic potential has been the major setback in developing effective cell-based therapies against a wide range of neurological disorders. Derivation of human embryonic stem cells (hESCs) provides a powerful tool to investigate the molecular controls in human embryonic neurogenesis as well as an unlimited source to generate the diversity of human neuronal cell types in the developing CNS for repair. However, realizing the developmental and therapeutic potential of hESCs has been hindered by conventional multi-lineage differentiation of pluripotent cells, which is uncontrollable, inefficient, highly variable, difficult to reproduce and scale-up. We recently identified retinoic acid (RA) as sufficient to induce the specification of neuroectoderm direct from the pluripotent state of hESCs under defined platform and trigger progression to human neuronal progenitors (hESC-I hNuPs) and neurons (hESC-I hNus) in the developing CNS with high efficiency, which enables hESC neuronal lineage-specific differentiation and opens the door to investigate human embryonic neurogenesis using the hESC model system. In this study, genome-scale profiling of microRNA (miRNA) differential expression patterns in hESC neuronal lineage-specific progression was used to identify molecular signatures of human embryonic neurogenesis. These in vitro neuroectoderm-derived human neuronal cells have acquired a neuron al identity by down-regulating pluripotence-associated miRNAs and inducing the expression of miRNAs linked to regulating human CNS development to high levels in a stage-specific manner, including silencing of the prominent pluripotence-associated hsa-miR-302 family and drastic expression increases of the Hox hsa-miR-10 and let-7 miRNAs. Following transplantation, hESC-I hNuPs engrafted and yielded well-integrated neurons at a high prevalence within neurogenic regions of the brain. In 3D culture, these hESC-I hNuPs proceeded to express subtype neuronal markers, such as dopaminergic and motor neurons, demonstrating their therapeutic potential for CNS repair. Our study provides critical insight into molecular neurogenesis in human embryonic development as well as offers an adequate human neurogenic cell source in high purity and large quantity for scale-up CNS regeneration.
Keywords: microRNA, Genome-scale mapping, Human development, Embryogenesis, Human embryonic stem cell, Pluripotence, Neuronal lineage-specific differentiation, Neurogenesis, Neuroectoderm, Neuronal progenitor, Neuron, Dopaminergic neuron, Motor neuron, miR-302, miR-10, Let-7, CNS disease, CNS repair, Cell therapy
Introduction
Understanding the much more complex human embryonic development has been hindered by the restriction on human embryonic and fetal materials as well as the limited availability of human cell types and tissues for study. In particular, there is a fundamental gap in our knowledge regarding the molecular networks and regulatory pathways underlying the central nervous system (CNS) formation in human embryonic development. Human embryonic stem cells (hESCs) have the unconstrained capacity for long-term stable undifferentiated growth in culture and the intrinsic potential for differentiation into all somatic cell types in the human body [1]. Derivation of hESCs, essentially the in vitro representation of the pluripotent inner cell mass (ICM) or epiblast of the human blastocyst, provides a powerful in vitro model system to investigate the molecular controls in human embryonic neurogenesis as well as an unlimited supply to generate the diversity of human neuronal cell types in the developing CNS for repair [1–3]. However, realizing the developmental and therapeutic potential of hESCs has been hindered by the current state of the art for generating functional cells from pluripotent cells through multi-lineage differentiation, which is uncontrollable, inefficient, instable, highly variable, difficult to reproduce and scale-up [2,3]. Conventional approaches rely on multi-lineage inclination of pluripotent cells through spontaneous germ layer differentiation, which yields mixed populations of cell types that may reside in three embryonic germ layers and often makes desired differentiation not only inefficient, but uncontrollable and unreliable as well [2,3]. Following transplantation, these pluripotent-cell-derived grafts tend to display not only a low efficiency in generating functional neuronal cell types necessary for reconstruction of the damaged CNS structure, but also phenotypic heterogeneity and instability, hence, a high risk of tumorigenicity [2–5]. Without a practical strategy to convert pluripotent cells direct into a specific lineage, previous studies and profiling of pluripotent hESCs and their differentiating multi-lineage aggregates have generated compromised implications to molecular controls in human embryonic development [6,7]. Development of novel strategies for well-controlled efficient differentiation of hESCs into functional lineages is not only crucial for unveiling the molecular and cellular cues that direct human embryogenesis, but also vital to harnessing the power of hESC biology for cell-based therapies.
Although neural lineages appear at a relatively early stage in hESC differentiation, only a small fraction of hESCs (< 5%) undergo spontaneous differentiation into neurons [2]. Under protocols presently employed in the field, these human neural grafts derived from pluripotent cells through multi-lineage differentiation not only yield neurons at a low prevalence following engraftment, but also accompanied by unacceptably high incidents of teratoma and/or neoplasm formation, posing considerable safety concern when administered to humans [2–5]. Similar to primary human neural stem cells (hNSCs) isolated directly from the CNS, those hNSCs derived secondarily from hESCs via multi-lineage differentiation are nestin-positive neuroepithelial-like cells that can spontaneously diffeerentiate into a mixed population of cells containing neurons, astrocytes, and oligodendrocytes in vitro and in vivo [8–10]. Before further differentiation, those hESC-derived hNSCs need to be mechanically isolated or enriched from hESC-differentiating multi-lineage aggregates. Early study of those uncommitted hESC-derived hNSCs showed that the grafted cells not only yielded a small number of dopaminergic (DA) neurons in vivo following transplantation, but could not acquire a DA phenotype in the lesioned brain [11]. Transplanting DA neurons pre-differentiated from those hESC-derived hNSCs ex vivo did not increase the yield of DA neurons in the lesioned brain [12]. Similar to their CNS counterpart, the therapeutic effect of those hESC-derived hNSCs was mediated by neuroprotective or trophic mechanism to rescue dying host neurons, but not related to regeneration from the graft or host remyelination [2,11,12]. A recent report showed that further directed differentiation of those hESC-derived neuroepithelial-like nestin-positive hNSCs into floor-plate precursors of the developing midbrain appeared to increase the efficiency of DA neuron engraftment in Parkinson's disease models [13]. Growing evidences indicate that the poor in vivo performance of those nestin-positive neuroepithelial-like hNSCs derived from hESCs in vitro was due to incomplete neuronal lineage specification [2,3,13]. Therefore, development of well-controlled strategies for efficiently directing hESCs commit into a more specific neuronal lineage in high purity and large quantity is crucial for harnessing the therapeutic potential of pluripotent hESCs for CNS repair.
To tackle the shortcomings in conventional approaches, previously, we have resolved the elements of a defined culture system necessary and sufficient for sustaining the epiblast pluripotence of hESCs, serving as a platform for de novo derivation of clinically-suitable hESCs and effectively directing such hESCs uniformly towards functional lineages [3,14]. To achieve uniformly conversion of pluripotent hESCs to a specific lineage, we have employed the defined culture system capable of insuring hESC proliferation to screen a variety of small molecules. We recently reported the identification of conditions necessary and sufficient for directing hESCs from the pluripotent stage exclusively towards a lineage-specific fate without an intervening multi-lineage differentiation stage [15–19]. Retinoic acid (RA) was found to induce the specification of neuroectoderm direct from the pluripotent state of hESCs and trigger progression to human neuronal progenitors (hESC-I hNuPs) and neurons (hESC-I hNus) in the developing CNS efficiently by promoting nuclear translocation of the neuronal-specific transcription factor Nurr-1 [3,15,18,19]. Similarly, nicotinamide (NAM) was found to induce the specification of cardiomesoderm direct from the pluripotent state of hESCs and trigger progression to human cardiac precursors and beating cardiomyocytes efficiently by promoting the expression of the earliest cardiac-specific transcription factor Csx/Nkx2.5 [3,16–18]. This technology breakthrough enables lineage-specific differentiation direct from the pluripotent state of hESCs with small molecule induction, which opens the door to investigate human embryonic neurogenesis using an in vitro cellular model system that emulates human embryonic development. Compared to the two prototypical neuroepithelial-like nestin-positive hNSCs either derived from hESCs in vitro or isolated from the CNS in vivo, these in vitro neuroectoderm-derived human neuronal progenitors are a novel more lineage-specific neuronal progenitor, as they yielded neurons exclusively and did not differentiate into other neural cell types such as oligodendrocytes and astrocytes [3,15,19]. The availability of human neuronal progenitors in high purity and large quantity with adequate neurogenic potential will greatly facilitate the development of effective cell-based therapies against a wide range of CNS disorders.
MicroRNAs (miRNAs) are emerging as important regulators of stem cell pluripotence and differentiation [18,20–25]. miRNAs are small, evolutionarily conserved non-coding RNAs that modulate gene expression by inhibiting mRNA translation and promoting mRNA degradation. miRNAs act as the governors of gene expression networks, thereby modify complex cellular phenotypes in development or disorders [26–28]. miRNA expression profiling using microarrays is a powerful high-throughput tool capable of monitoring the regulatory networks of the entire genome and identifying functional elements of developmental processes in high resolution. To identify mechanisms of small molecule induced lineage-specification of pluripotent hESCs, in previous report, we found that NAM induced nuclear translocation of NAD-dependent histone deacetylase SIRT1 and global chromatin silencing in cardiomesoderm specification of hESCs, while RA induced silencing of pluripotence-associated hsa-miR-302 family and drastic up-regulation of neuroectodermal Hox miRNA hsa-miR-10 family to high levels in neuroectoderm specification of hESCs [18]. To uncover molecular signatures in human embryonic neurogenesis, in this study, genome-scale profiling of miRNA differential expression patterns during small-molecule-induced hESC neuronal lineage-specific progression towards neuronal progenitors and neurons was further used to identify stage-specific human embryonic neurogenic miRNAs. A unique set of pluripotence-associated miRNAs was down-regulated, while novel sets of stage-specific neuronal lineage-driving miRNAs were up-regulated during hESC neuronal lineage-specific progression. Following transplantation, these neuroectoderm-derived human neuronal progenitors engrafted and migrated widely, and yielded well-dispersed and well-integrated human neurons at a high prevalence within neurogenic regions of the brain, consistent to our previous observation [19]. In 3D culture, these hESC-derived neuronal cells further proceeded to express subtype neuronal markers associated with ventrally-located neuronal populations, such as dopaminergic and motor neurons, demonstrating their therapeutic potential for CNS repair. Our study provides critical insight into human embryonic neurogenesis as well as offers an adequate human neurogenic cell source in high purity and large quantity for scale-up CNS regeneration.
Materials and Methods
Culture of undifferentiated hESCs
The hESC lines WA01 and WA09 (WiCell Research Institute) and newly-derived biologics-free hESCs (Xcel-hESCs) [17] were used in this study. The defined culture systems consist of DMEM/F-12 or KO-DMEM (knockout-DMEM) (80%), Knockout Serum Replacement (KO) (20%), L-alanyl-L-gln or L-gln (2 mM), MEM nonessential amino acids (MNAA, 1×), β-Mercaptoethanol (β-ME, 100 μM) (all from Invitrogen), human purified laminin (Sigma) or laminin/collagen (growth factor reduced Matrigel, BD Bioscience) as the matrix protein, and bFGF (basic fibroblast growth factor, 20 ng/ml) (PeproTech Inc). The KO can be replaced with defined essential factors containing MEM essential amino acids (MEAA, 1×), human insulin (20 μg/ml) (Sigma), and ascorbic acid (50 μg/ml) (Sigma), in which activin A (50 ng/ml, Sigma), human albumin (10 mg/ml, Sigma), and human transferrin (8 μg/ml, Sigma) were added in order to increase cell survival and maintain normal shape and healthy colonies.
Neuronal lineage-specific differentiation direct from the pluripotent state of hESCs
Undifferentiated hESCs maintained under the defined culture conditions were treated with RA (10 μM) 3 days after seeding for 4–5 days. These neuroectoderm-differentiated hESCs were transferred to a serum-free suspension culture to allow floating neuroblasts (hESC-I hNuPs) to form in the hESC media lacking bFGF for 4–5 days. For further differentiating into a neuronal phenotype, the neuroblasts were then permitted to attach to a tissue culture plate in a defined medium containing DMEM/F-12, N-2 supplement (1%), heparin (8 μg/ml), VEGF (20 ng/ml), NT-3 (10 ng/ml), and BDNF (10 ng/ml). β-III-tubulin- and Map-2-expressing, extensively neurite-bearing cells and pigmented cells, typical of those in the ventral mesencephalon, were observed within 2 weeks of continuous cultivation, increased in numbers with time.
Cellular immunofluorescence
The cells were fixed with 4% paraformaldehyde and blocked in PBS containing 0.2% Triton X-100 and 2% BSA. The cells were incubated with the primary antibody in 0.1% Triton X-100 in PBS at 4°C overnight, and then with secondary antibody (Molecular Probe/Invitrogen) in the same buffer at room temperature for 45 min. After staining with DAPI, cells were visualized under an immunofluorescence and deconvolution microscope, and quantified by the image analysis software (Olympus).
Transplantation into mouse brain
Human neuronal progenitors were generated from undifferentiated hESCs maintained under the defined culture by retinoic acid induction. The human neuronal progenitor cells in suspension culture were pulled together and dissociated with a brief treatment (~ 1 min) of Accutase (Invitrogen) followed by gently triturated into single cell suspension, and resuspended in PBS (Mg2+ & Ca2+ free) (Invitrogen) at a density of 50,000/μl. A total of 106 human donor cells (20 μl at 50,000/μl) were injected into the cerebral ventricles of immunocompetent wild type and SOD1 mutant newborn mice (n=54) (The Jackson Laboratory). Controls were similarly transplanted with the same amount of human fibroblast cells (Hs27, ATCC). After at least 3 months post-grafting, the mice were sacrificed and processed for histological and immunocytochemical analysis of the transplanted brain. No graft overgrowth, formation of teratomas or neoplasms, or appearance of non-neuronal cell types was observed in at least 54 transplanted animals and at least 1 year after transplantation (the latest time point examined). Transplanted wild type mice developed hyper-active behavior, such as fast movement and fast spin (see videos at http://www.sdrmi.org). The animal experiments were reviewed and approved by the relevant institutional animal care and use committees.
miRNA microarray analysis
Total RNAs, including miRNAs, were isolated from hESCs and their neuronal derivatives (treated with RA under the defined culture) using the Qiagen miRNeasy kit (Qiagen, www.qiagen.com). The quality of purified total RNAs was verified by electrophoresis analysis. The human miRNA expression profiling was done and the microarray data analysis from at least two biological replicate sets (e.g., WA01 and WA09) was provided by LC Sciences (Huston, TX, www.lcsciences.com) as part of miRNA microarray service. Multiple redundant regions and control probes were included in the content of each chip. Each region further comprises a miRNA probe region, which detects miRNA transcripts listed in Sanger miRBase Release 17.0 (http://www.sanger.ac.uk/Software/Rfam/mirna/). Statistic tests and clustering analysis were provided by LC Sciences as part of miRNA microarray service. The signal values were derived by background subtraction and normalization. Detectable transcripts were subjected to data processing statistics and signal intensities were listed in average values of repeating spots.
Results and Discussion
Neuroectoderm specification of hESCs induces uniform neuronal lineage-specific progression
In previous report, we observed that RA was rendered sufficient to induce the pluripotent hESCs maintained in the defined culture system to transition from a pluripotent state exclusively and uniformly towards a neuroectoderm phenotype by promoting nuclear translocation of the neuronal specific transcription factor Nurr-1, a member of the orphan nuclear hormone receptor super-family implicated in ventral neuronal development, particularly ventral mesencephalic development and activation of the tyrosine hydroxylase (TH) gene, the rate-limiting step in DA neuronal differentiation [3,15,18,19]. These neuroectoderm-specified hESCs further progressed to human neuronal progenitors (hESC-I hNuPs) and neurons (hESC-I hNus) in the developing CNS with high efficiency [3,15,18,19] (as illustrated in Figure 1A). Compared to the two prototypical neuroepithelial-like nestin-positive hNSCs either derived from hESCs or CNS [2,8–10,29], hESC-I hNuPs, which have acquired a neuroectodermal identity through RA induction of pluripotent hESCs in vitro [3,15,18,19], did not express nestin, but assumed uniformly strong expression and nuclear localization of Nurr-1 (Figures 1B and 1C) [19]. Although CNS-derived hNSCs, which have acquired their neurectodermal identity through in vivo developmental processes [2,29], show moderate expression and nuclear localization of Nurr-1, in hESC-derived hNSCs [8–10], Nurr-1 localizes to the cell-surface and cytoplasm, suggesting its being inactive (Figure 1B) [19]. To further differentiate into neurons, hESC-I hNuPs were permitted to attach in a defined media containing neurotrophic factors without bFGF [15]. Under these neuronal differentiation conditions, hESC-I hNuPs yielded exclusively neurons that expressed neuronal marker β-III-tubulin and co-expressed Map-2 (Figure 1D), with a drastic increase in efficiency (~ 94%) when compared to the yields of β-III-tubulin-positive neurons differentiated under similar conditions from hESC-derived hNSCs (~ 6%) or CNS-derived hNSCs (~ 13%) as the control (Figure 1E). These observations suggest that these neuroectoderm-derived Nurr1-positive hESC-I hNuPs are a novel more lineage-specific neuronal progenitor than the prototypical neuroepithelial-like nestin-positive hNSCs either isolated from the CNS tissue in vivo or derived from hESC in vitro, consistent with our previous observations [19]. This lineage-specific differentiation approach by small molecule induction may dramatically increase the clinical efficacy of graft-dependent neuron replacement and safety of hESC-derived cellular products for CNS repair.
Figure 1. Neuroectoderm specification of hESCs induces uniform neuronal lineage-specific progression.
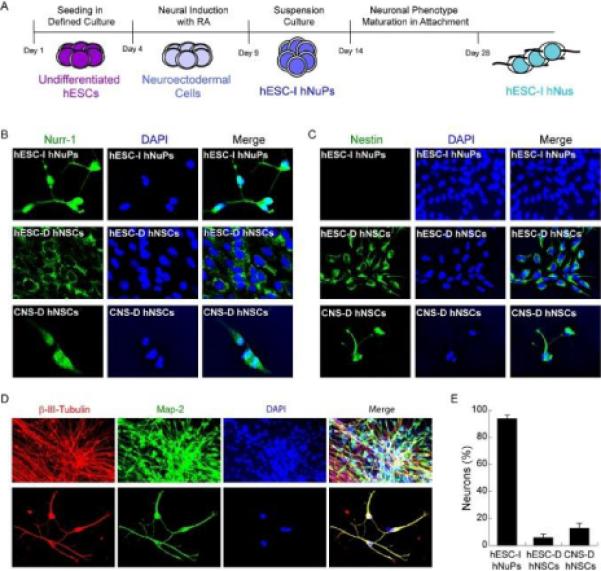
(A) Schematic depicting of the protocol time line of directed neuronal differentiation of hESCs by small molecule induction. (B) Neuroectoderm-derived hESC-I hNuPs display strong expression and nuclear localization of Nurr-1 (green), compared to fetal CNS-derived hNSCs (CNS-D hNSCs) that show moderate expression and nuclear localization of Nurr-1 and hESC-derived hNSCs (hESC-D hNSCs) that show cell-surface and cytoplasm localization of Nurr-1. (C) Neuroectoderm-derived hESC-I hNuPs did not express nestin (green), compared to the two neuroepithelial-like nestin-positive hNSCs either derived from hESCs or CNS. (D) After permitting to attach, neuroectoderm-derived hESC-I hNuPs yielded exclusively neurons that expressed neuronal marker β-III-tubulin (red) and co-expressed Map-2 (green). Bottom panel better visualizes individual hESC-derived neuronal cells (hESC-I hNus). (E) Neuroectoderm-derived hESC-I hNuPs differentiated towards a neuronal lineage with a drastic increase in efficiency (~ 94%) when compared to the yields of neurons differentiated under similar conditions from hESC-D hNSCs (~ 6%) or CNS-D hNSCs (~ 13%) as the control, as assessed by the percentages of cells that expressed β-III-tubulin. Quantitative data are mean values from at least 2 different cell lines and at least three separate experiments were conducted with each cell line.
Silencing of prominent pluripotence-associated miRNAs in hESC neuronal lineage specific progression
To uncover molecular signatures of human embryonic neurogenesis, we performed miRNA microarray analysis (LC Sciences, www.lcsciences.com) to profile the differential expression patterns of miRNAs in the process of hESC neuronal lineage specific progression to neuronal progenitors (hESC-I hNuPs) and neurons (hESC-I hNus) (Figures 2A and 3). Identified signature miRNAs were further grouped according to their differential expression patterns in progressive developmental stages of pluripotent hESCs, hESC-I hNuPs, and hESC-I hNus (Figures 2B and 4–6) (Table 1). We found that a group of human pluripotence-associated miRNAs was significantly down-regulated in hESC neuronal lineage-specific progression (Figures 2B and 4) (Table 1). In particular, the expression of the prominent cluster of human pluripotence-associated miRNA hsa-miR-302 family, including hsa-miR-302b, 302a, 302a*, 302d, 302c, was silenced (~ 100-500-fold of down-regulation) upon hESC neuronal lineage specification (Figures 2, 3A, 3B, 4A and 4E) (Table 1). The human pluripotence-associated miRNA group 1, consisting of the cluster of hsa-miR-302 family, had a profile of the highest expression in pluripotent hESCs and drastic expression down-regulation of ~ 100-500-fold in both hESC-I hNuPs and hESC-I hNus (Figures 2B, 4A and 4E) (Table 1).
Figure 2. Genome-scale profiling of miRNA differential expression in hESCs neuronal lineage-specific progression.
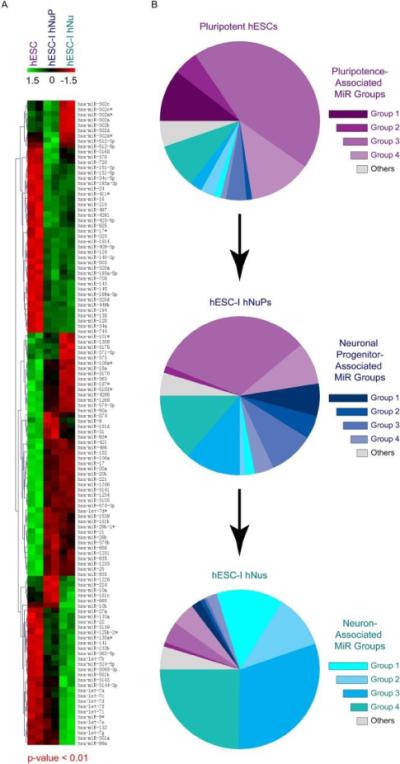
(A) Hierarchal clustering of differentially expressed miRNAs in pluripotent hESCs, hESC-I hNuPs, and hESC-I hNus. Statistically significant p values and clustering analysis were provided by LC Sciences as part of miRNA microarray service. Microarray data from two biological replicate sets were shown. (B) Pie charts showing decreased contribution of a set of pluripotence-associated miRNAs (purple) and increased contribution of distinct sets of neuronal progenitor-associated miRNAs (blue) and neuron-associated miRNAs (cyan) to the entire miRNA populations during hESC neuronal lineage-specific progression. Note that the expression of pluripotence-associated hsa-miR-302 clusters (dark purple) was silenced and the expression of Hox miRNA hsa-miR-10 cluster (dark blue) was induced to high levels in these in vitro neuroectoderm-derived hESC-I hNuPs. See human pluripotence-associated miRNA groups in figure 4, human neuronal progenitor-associated miRNA groups in figure 5, and human neuronal-associated miRNA groups in figure 6. Aslo see Table 1. The numbers for the color bar are Z values, showing the extents of down-regulation (green, −1.5) or up-regulation (red, 1.5) of the expression.
Figure 3. Comparing miRNA differential expression patterns in pluripotent hESCs, hESC-I hNuPs, and hESC-I hNus.
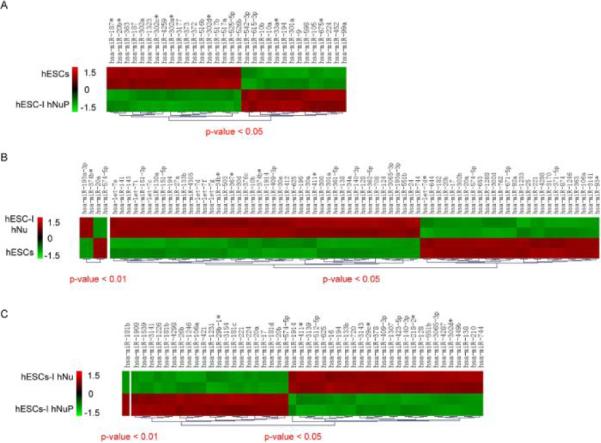
(A) Hierarchal clustering of differentially expressed miRNAs in pluripotent hESCs and hESC-I hNuPs. (B) Hierarchal clustering of differentially expressed miRNAs in pluripotent hESCs and hESC-I hNus. (C) Hierarchal clustering of differentially expressed miRNAs in hESC-I hNuPs and hESC-I hNus. Statistically significant p values and clustering analysis were provided by LC Sciences as part of miRNA microarray service. Microarray data from two biological replicate sets were shown. The numbers for the color bar are Z values, showing the extents of down-regulation (green, −1.5) or up-regulation (red, 1.5) of the expression.
Figure 4. Down-regulation of a set of human pluripotence-associated miRNAs in hESC neuronal lineage specific progression.

(A) The expression of the human pluripotence-associated miRNA group 1, consisting of the prominent cluster of pluripotence-associated miRNAs hsa-miR-302, was silenced upon hESC neuronal lineage specification. (B) The expression of the human pluripotence-associated miRNA group 2 was significantly down-regulated in both hESC-I hNuPs and hESC-I hNus. (C) The expression of the human pluripotence-associated miRNA group 3 was significantly down-regulated in hESC-I hNus. (D) The expression of the human pluripotence-associated miRNA group 4 was gradually down-regulated upon hESC neuronal lineage-specific differentiation, albeit to less extents. (E) Log2 ratios of down-regulation of the human pluripotence-associated miRNA group 1–3. The levels of miRNA expression down-regulation in (A)–(D): * 5-fold ---10-fold, ** 10-fold ---100-fold, and *** 100-fold --- 500-fold. All microarray data are mean values from at least two biological replicate sets, standard derivations were not shown.
Figure 6. A unique set of human embryonic neurogenic miRNAs contributes to the neuronal identity of the developing CNS.
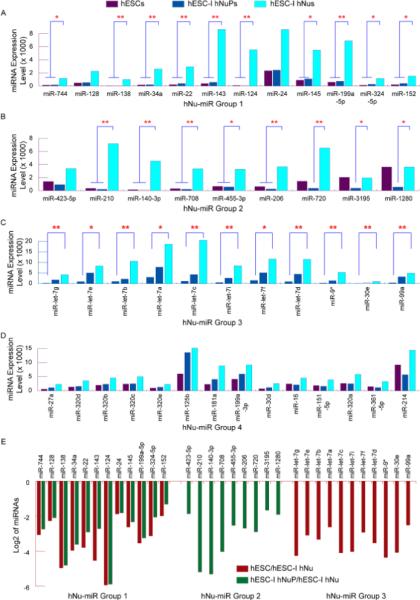
(A) The expression of the human neuron-associated miRNA group 1 was expressed at low levels in pluripotent hESCs, slightly up-regulated in hESC-I hNuPs, and displayed drastic increases of expression to high levels in further differentiated hESC-I hNus. (B) The expression of the human neuron-associated miRNA group 2 was down-regulated to low levels of expression in hESC-I hNuPs, and then significantly up-regulated to high levels of expression in further differentiated hESC-I hNus. (C) The expression of the human neuron-associated miRNA group 3, including the let-7 miRNA family, was expressed at low levels in pluripotent hESCs, and significantly up-regulated in the progression of hESC neuronal lineage specific differentiation to high levels of expression in hESC-I hNus. (D) The expression of the human neuron-associated miRNA group 4 was up-regulated in hESC-I hNus, albeit to less extents. (E) Log2 ratios of up-regulation of human neuron-associated miRNA group 1–3. The levels of miRNA expression up-regulation in (A)–(D): * 5-fold ---10-fold, ** 10-fold ---100-fold, and *** 100-fold --- 500-fold. All microarray data are mean values from at least two biological replicate sets, standard derivations were not shown.
Table 1.
Stage-Specific Human Embryonic microRNAs in hESC Neuronal Lineage-Specific Progression.
| miRNA Group | Highest Expression in Cells | Expression Levels in hESCs | Expression Levels in hNuPs | Expression Levels in hNus | miRNAs | |
|---|---|---|---|---|---|---|
|
| ||||||
| Human Pluripotence-Associated miRNAs | Group 1 | hESC | +++ | − | − | hsa-miR-302b, 302a, 302a*, 302d, 302c |
|
| ||||||
| Group 2 | hESC | +++ | + | + | hsa-miR-373, 3178, 363, 3170 | |
|
| ||||||
| Group 3 | hESC | +++ | +++/++ | + | hsa-miR-574-5p, 20a, 20b, 17, 4298, 106a, 3141, 1246, 1268, 221, 92a, 574-3p, 182, 466, 638, 3185, 762, 4281 | |
|
| ||||||
| Group 4 | hESC | +++ | ++ | + | hsa-miR-3196, 1915, 222, 107, 663, 92b, 15b | |
|
| ||||||
| Human Neuronal Progenitor-Associated miRNAs | Group 1 | hNuP | − | +++ | ++/+ | hsa-miR-10b, 10a, 224 |
|
| ||||||
| Group 2 | hNuP | + | +++ | + | hsa-miR-9, 125a-5p, 181d, 4324 | |
|
| ||||||
| Group 3 | hNuP | ++ | +++ | + | hsa-miR-26b, 181b, 25, 31, 21, 335, 1275 | |
|
| ||||||
| Group 4 | hNuP | ++ | +++ | ++ | hsa-miR-27b, 93, 99b, 100 | |
|
| ||||||
| Human Neuron-Associated miRNAs | Group 1 | hNu | +/− | + | +++ | hsa-miR-744, 128, 138, 34a, 22, 143, 124, 24, 145, 199a-5p, 324-5p, 152 |
|
| ||||||
| Group 2 | hNu | + | +/− | +++ | hsa-miR-423-5p, 210, 140-3p, 708, 455-3p, 206, 720, 3195, 1280 | |
|
| ||||||
| Group 3 | hNu | + | ++ | +++ | hsa-miR-let-7g, let-7e, let-7b, let-7a, let-7c, let-7i, let-7f, let-7d, 9*, 30e, 99a | |
|
| ||||||
| Group 4 | hNu | ++ | +++/++ | +++ | hsa-miR-27a, 320d, 320b, 320c, 320e, 125b, 181a, 199a-3p, 30d, 16, 151-5p, 320a, 361-5p, 214 | |
The human pluripotence-associated miRNA group 2, including hsa-miR-373, 3178, 363, 3170, was found to be significantly down-regulated in both hESC-I hNuPs and hESC-I hNus (Figures 2, 3A, 3B, 4B and 4E) (Table 1). The human pluripotence-associated miRNA group 3, including hsa-miR-574-5p, 20a, 20b, 17, 4298, 106a, 3141, 1246, 1268, 221, 92a, 574-3p, 182, 466, 638, 3185, 762, 4281, was found to be significantly down-regulated in hESC-I hNus (Figures 2, 3A, 3B, 4C and 4E) (Table 1). The clusters of hsa-miR-17 and hsa-miR-20, which were strongly expressed in pluripotent hESCs and which have near-identical seed sequences with hsa-miR-302 family, have been implicated in cell proliferation previously [21,22]. The human pluripotence-associated miRNA group 4, including hsamiR-3196, 1915, 222, 107, 663, 92b, 15b, was found to be gradually down-regulated upon hESC neuronal lineage-specific progression, albeit to less extents (Figures 2, 3A, 3B and 4D) (Table 1). Our miRNA profiling data suggested that these in vitro neuroectoderm-derived Nurr1-positve human neuronal progenitors and neurons have acquired a neuronal lineage-specific identity by down-regulating pluripotence-associated miRNAs, including silencing of the prominent pluripotence-associated hsa-miR-302 family (Figures 2, 3A, 3B and 4) (Table 1).
The hESC-derived Nurr1-positive human neuronal progenitors continue to express high levels of neuroectodermal miRNAs
A group of human neuronal progenitor-associated miRNAs, which had a profile of the highest expression in hESC-I hNuPs, displayed an expression pattern of significant up-regulation in hESC-I hNuPs and down-regulation upon further neuronal differentiation (Figures 2B and 5) (Table 1). The expression of the human neuronal progenitor-associated miRNA group 1, including hsa-miR-10b, 10a, 224, was silenced in pluripotent hESCs and displayed drastic increases to high levels in neuroectoderm-derived hESC-I hNuPs (Figures 2, 3A, 3C, 5A and 5E) (Table 1). Notably, the expression of hsa-miR-10b was completely silenced in pluripotent hESCs and had an average increase of 223-fold in neuroectoderm-derived hESC-I hNuPs (Figures 2B, 5A and 5E). The miR-10 genes locate within the Hox clusters of developmental regulators [30]. In several species, miR-10 is coexpressed with a set of Hox genes and has been found to repress the translation of Hox transcripts [30,31]. The enhancer of the mouse Hoxb-1 gene, which controls the RA response and regulates expression predominantly in neuroectoderm, contains a retinoic acid response element (RARE) that is not only involved in the ectopic response to RA, but is also essential for establishing the early Hoxb-1 expression pattern [32]. Our miRNA profiling data suggested that Hox miRNA hsa-miR-10 that regulates gene expression predominantly in neuroectoderm continued to express at a high level in these in vitro neuroectoderm-derived hESC-I hNuPs (Figures 2B, 5A and 5E), consistent with our previous observation in hESC-derived neuroectodermal cells induced by RA [18]. The drastic expression increase of hsa-miR-10 upon exposure of hESCs to RA suggested that RA might induce the expression of Hox genes and co-expression of Hox miRNA hsa-miR-10 to silence pluripotence-associated genes and miRNA hsa-miR-302 to drive pluripotent hESCs towards a neuronal fate [3,15,18,19].
Figure 5. Up-regulation of a novel set of human neuronal progenitor-associated miRNAs in neuroectoderm-derived hESC-I hNuPs.
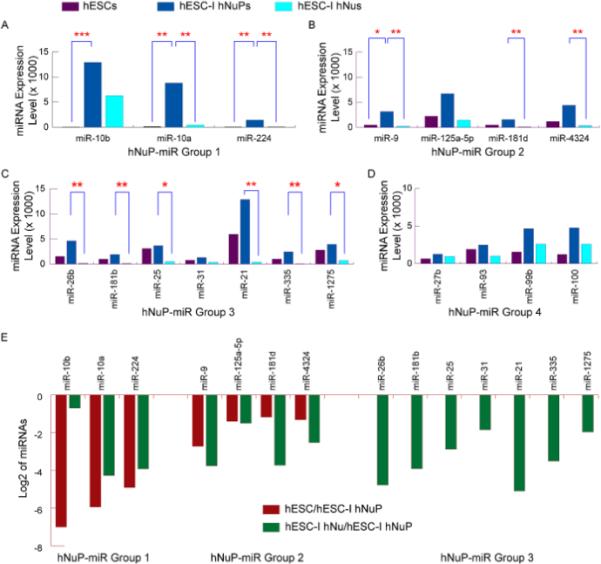
(A) The expression of the human neuronal progenitor-associated miRNA group 1 was silenced in pluripotent hESCs and displayed drastic increases in neuroectoderm-derived hESC-I hNuPs. Notably, the expression of hsa-miR-10b was completely silenced in pluripotent hESCs and had an average increase of 223-fold in hESC-I hNuPs. (B) The expression of the human neuronal progenitor-associated miRNA group 2 was significantly up-regulated in hESC-I hNuPs, and then significantly down-regulated in further differentiated hESC-I hNus. (C) The expression of the human neuronal progenitor-associated miRNA group 3 was up-regulated in hESC-I hNuPs, and then significantly down-regulated in further differentiated hESC-I hNus. (D) The expression of the human neuronal progenitor-associated miRNA group 4 was up-regulated in hESC-I hNuPs, and then down-regulated in further differentiated hESC-I hNus, albeit to less extents. (E) Log2 ratios of up-regulation of human neuronal progenitor-associated miRNA group 1–3. The levels of miRNA expression up-regulation in (A)–(D): * 5-fold ---10-fold, ** 10-fold ---100-fold, and *** 100-fold --- 500-fold. All microarray data are mean values from at least two biological replicate sets, standard derivations were not shown.
The human neuronal progenitor-associated miRNA group 2, including hsa-miR-9, 125a-5p, 181d, 4324, was found to be significantly up-regulated in hESC-I hNuPs, and then significantly down-regulated in further differentiated hESC-I hNus (Figures 2, 3A, 3C, 5B and 5E) (Table 1). The miR-9 is implicated in regulation of spinal motor neuron function previously [26]. Knockdown of miR-9 inhibits hESC-derived hNSC proliferation, delays maturation, and promotes migration in vitro and in vivo [33]. The human neuronal progenitor-associated miRNA group 3, including hsa-miR-26b, 181b, 25, 31, 21, 335, 1275, was found to be up-regulated in hESC-I hNuPs, and then significantly down-regulated in further differentiated hESC-I hNus (Figures 2, 3A, 3C, 5C and 5E) (Table 1). The human neuronal progenitor-associated miRNA group 4, including hsa-miR-27b, 93, 99b, 100, was found to be up-regulated in hESC-I hNuPs, and then down-regulated in further differentiated hESC-I hNus, albeit to less extents (Figures 2, 3A, 3C and 5D) (Table 1). Our miRNA profiling data suggested that a distinct set of human embryonic miRNAs, many of which were not previously linked to neuronal development, contributes to the identity of in vitro neuroectoderm-derived hESC-I hNuPs (Figures 2, 3A, 3C and 5) (Table 1). Therefore, these in vitro neuroectoderm-derived Nurr1-positve hESC-I hNuPs have acquired a neuronal identity by silencing pluripotence-associated miRNAs and inducing the expression of human neuroectodermal miRNAs to high levels (Figures 2–5) (Table 1).
A unique set of human embryonic neurogenic miRNAs contributes to the neuronal identity of the developing CNS
A group of human neuron-associated miRNAs, which had a profile of the highest expression in hESC-I hNus, displayed an expression pattern of up-regulation during the progression of neuronal lineage-specific differentiation of hESCs induced by RA (Figures 2B and 6) (Table 1). The expression of the human neuron-associated miRNA group 1, including hsa-miR-744, 128, 138, 34a, 22, 143, 124, 24, 145, 199a-5p, 324-5p, 152, was expressed at low levels in pluripotent hESCs, slightly up-regulated in hESC-I hNuPs, and displayed drastic increases of expression to high levels in further differentiated hESC-I hNus (Figures 2, 3B, 3C, 6A and 6E) (Table 1). The human neuron-associated miRNA group 2, including hsa-miR-423-5p, 210, 140-3p, 708, 455-3p, 206, 720, 3195, 1280, was found to be down-regulated to low levels of expression in hESC-I hNuPs, and then significantly up-regulated to high levels of expression in further differentiated hESC-I hNus (Figures 2, 3B, 3C, 6B and 6E) (Table 1). The human neuron-associated miRNA group 3, including the let-7 miRNA family (hsa-miR-let-7g, let-7e, let-7b, let-7a, let-7c, let-7i, let-7f, and let-7d), 9*, 30e, 99a, was expressed at low levels in pluripotent hESCs, and significantly up-regulated in hESC neuronal lineage specific progression to high levels of expression in hESC-I hNus (Figures 2, 3B, 3C, 6C and 6E) (Table 1). The let-7 miRNAs silence the ESC self-renewal program in vivo and in culture, down-regulating pluripotence-associated genes such as myc and lin28, an RNA binding protein that inhibits let-7 processing [24]. Previous report suggested that Let-7b was expressed in mammalian brains and exhibited increased expression during neural differentiation [34]. The human neuron-associated miRNA group 4, including hsa-miR-27a, 320d, 320b, 320c, 320e, 125b, 181a, 199a-3p, 30d, 16, 151-5p, 320a, 361-5p, 214, was found to be up-regulated in hESC-I hNus, albeit to less extents (Figures 2, 3B, 3C and 6D) (Table 1). Our miRNA profiling study suggested that distinct sets of stage-specific human embryonic neurogenic miRNAs, many of which were not previously linked to neuronal development and function, contribute to the development of neuronal identity in human CNS formation (Figures 2–6) (Table 1).
The neuroectoderm-derived human neuronal progenitors specified from hESCs are safely engraftable and highly neurogenic following transplantation and in 3D culture
To address whether the neuroectoderm-derived Nurr1-positive hESC-I hNuPs could be safely engrafted in the brain and could migrate and retain their neurogenic ability in vivo, hESC-I hNuPs were transplanted into the cerebral ventricles of newborn mice. This route allows excellent access to the subventricular zone (SVZ), a secondary germinal zone from which cells widely migrate and respond to appropriate regional developmental cues [29]. After at least 3 months post-grafting, the mice were sacrificed and processed for histological analysis. To evaluate their therapeutic potential for CNS repair, both wild type and SOD1 mutant newborn mice were used for transplantation. The animal experimental protocols were reviewed and approved by the relevant institutional animal care and use committees. The transgenic SOD1 mouse model expresses mutated forms of the enzyme copper-zinc superoxide dismutase 1 (SOD1) that replicate with fidelity the onset and progression of the disease of Amyotrophic Lateral Sclerosis (ALS) and have been largely used to test therapies to be translated to patients in clinical trials [35–37]. Transplanted hESC-I hNuPs survived, engrafted, and migrated widely and yielded well-dispersed and well-integrated human neurons at a high prevalence, including Nurr1-positve and tyrosine hydroxylase (TH)-positive DA neurons, within neurogenic regions of the brain (Figure 7A), consistent with our previous observations [19]. Transplanted wild type mice developed hyper-active behavior, such as fast movement and fast spin (see videos at http://www.sdrmi.org), further suggesting that the transplanted human neuronal cells had survived, migrated, and integrated into the mouse brain to function and dominate the mouse behavior. Therefore, this neuronal lineage-specific differentiation approach by small molecule induction may dramatically increase the clinical efficacy of graft-dependent neuron replacement and safety of hESC-derived cellular products for scale-up CNS regeneration.
Figure 7. The neuroectoderm-derived human neuronal progenitors specified from hESCs are highly neurogenic following transplantation and in 3D culture.

(A) hESC-I hNuPs were injected into the cerebral ventricles of newborn mice. Histological analysis of transplanted mice showed well-dispersed and well-integrated human neurons at a high prevalence, indicated by anti-human mitochondrial antibody (hMit) (red) and their immunoreactivity to Map-2 (green), including Nurr1-positve (green) and TH-positive (green) DA neurons, within neurogenic regions of the brain. DAPI nuclear marker (blue) stains all cells in the field. (B) Under neuronal subtype specification conditions in 3D culture, these hESC-derived neuronal cells (expressing Map-2 and co-expressing βIII-tubulin) further proceeded to express subtype neuronal markers associated with ventrally-located neuronal populations, such as TH (DA neurons) and Hb9/Lim3/Isl1 (motor neurons) (shown in 3D matrix). All cells are indicated by DAPI staining of their nuclei (blue).
Thus far, testing potential therapeutic strategies have largely relied on animal models for behavior, safety, and efficacy evaluation of therapeutic candidates and human cell therapy products. However, because of interspecies differences, conventional studies using animal models are often poor predictors of human efficacy and safety. Animal models are xeno-hosts for transplantation of human cells, not ideal for testing the behavior, safety, and efficacy of therapeutic outcomes of human stem cells. Large primate models are very costly and often taken years to obtain results. In addition, the results of animal studies can be highly variable and difficult to reproduce, making them unreliable as benchmarks for decisions on humans in clinical trials. Development and utilization of multi-cellular 3-dimentional (3D) human embryonic models using hESCs will provide an authentic and reliable in vitro tool targeted for rapid and high fidelity safety and efficacy evaluation of human therapeutic candidates and products, and thus reduce the reliance on animal models to test potential therapeutic strategies [3]. Therefore, as an authentic and reliable alternative to animal models, we sought to develop multi-cellular 3D models of human nervous system using hESC-derived human embryonic neuronal cells for rapid and high fidelity evaluation of their therapeutic potential against CNS disorders. We found that, under neuronal subtype specification conditions in 3D culture, these hESC-derived neuronal cells (expressing Map-2 and co-expressing β-III-tubulin) further proceeded to express subtype neuronal markers associated with ventrally-located neuronal populations, such as TH (DA neurons) and Hb9/Lim3/Isl1 (motor neurons) (Figure 7B), demonstrating their therapeutic potential for regeneration of CNS neuronal cell types and subtypes in vivo as stem cell therapy to be translated to patients in clinical trials.
Conclusions
Previously, we found that pluripotent hESCs maintained under defined conditions can be uniformly converted into a neuronal or cardiac lineage by simple provision of small molecules [3,15–19]. RA induces the specification of neuroectoderm direct from the pluripotent state of hESCs and triggers progression to neuronal progenitors and neurons efficiently by promoting nuclear translocation of the neuronal-specific transcription factor Nurr-1 (Figures 1A and 1B) [3,15,18,19]. Compared to the prototypical neuroepithelial-like nestin-positive hNSCs either derived from hESCs in culture or isolated from the human fetal CNS tissue, these in vitro neuroectoderm-derived Nurr1-positive human neuronal cells have acquired drastically increased neurogenic potential in vitro and in vivo (Figures 1 and 7), providing a novel neuronal lineage-specific engraftable human embryonic progenitor in high purity and large quantity as a potentially adequate source for scale-up CNS regeneration. Genome-scale profiling of miRNA differential expression patterns identified novel sets of stage-specific human embryonic neurogenic miRNAs during hESC neuronal lineage-specific progression (Figures 2–6). A unique set of pluripotence-associated miRNAs was down-regulated, while novel sets of neuronal lineage-driving miRNAs were up-regulated during hESC neuronal lineage-specific progression, including silencing of the prominent pluripotence-associated hsa-miR-302 family and drastic expression increases of hsa-miR-10 and the let-7 miRNAs (Figures 2–6). Our findings suggest that these in vitro neuroectoderm-derived Nurr1-positive human neuronal cells have acquired a neuronal lineage-specific identity by silencing pluripotence-associated miRNAs and inducing the expression of miRNAs linked to regulating human CNS development to high levels, therefore, highly neurogenic in vitro and in vivo. Our study provides critical insight into molecular neurogenesis in human embryonic development as well as offers an adequate human neurogenic cell source in high purity and large quantity for developing effective stem cell therapy to be translated to patients in clinical trials to restore the lost nerve tissue and function for a wide range of devastating and untreatable neurological disorders.
Acknowledgements
XHP has been supported by National Institute of Health (NIH) grants from National Institute on Aging (NIHK01AG024496) and The Eunice Kennedy Shriver National Institute of Child Health and Human Development (NIHR21HD056530).
Footnotes
Disclosures
The authors declare competing interests. XHP is the founder of Xcelthera and has intellectual properties related to hESCs.
References
- 1.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Parsons XH, Teng YD, Snyder EY. Important precautions when deriving patient-specific neural elements from pluripotent cells. Cytotherapy. 2009;11:815–824. doi: 10.3109/14653240903180092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parsons XH, Teng YD, Moore DA, Snyder EY. Patents on technologies of human tissue and organ regeneration from pluripotent human embryonic stem cells. Rec Pat Regen Med. 2011;1:142–163. doi: 10.2174/2210297311101020142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aubry L, Bugi A, Lefort N, Rousseau F, Peschanski M, et al. Striatal progenitors derived from human ES cells mature into DARPP32 neurons in vitro and in quinolinic acid-lesioned rats. Proc Natl Acad Sci USA. 2008;105:16707–16712. doi: 10.1073/pnas.0808488105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roy NS, Cleren C, Singh SK, Yang L, Beal MF, et al. Functional engraftment of human ES cell-derived dopaminergic neurons enriched by coculture with telomerase-immortalized midbrain astrocytes. Nat Med. 2006;12:1259–1268. doi: 10.1038/nm1495. [DOI] [PubMed] [Google Scholar]
- 6.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 7.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang S, Wernig M, Duncan ID, Brustle O, Thomson JA. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat Biotechnol. 2001;19:1129–1133. doi: 10.1038/nbt1201-1129. [DOI] [PubMed] [Google Scholar]
- 9.Elkabetz Y, Panagiotakos G, Al Shamy G, Socci ND, Tabar V, et al. Human embryonic stem cell-derived neural rosettes reveal a functionally distinct early neural stem cell stage. Genes Dev. 2008;22:152–165. doi: 10.1101/gad.1616208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koch P, Opitz T, Steinbeck JA, Ladewig J, Brüstle O. A rosette-type, self-renewing human embryonic stem cell-derived neural stem cell with potential for in vitro instruction and synaptic integration. Proc Natl Acad Sci USA. 2009;106:3225–3230. doi: 10.1073/pnas.0808387106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ben-Hur T, Idelson M, Khaner H, Pera M, Reinhartz E, et al. Transplantation of human embryonic stem cell-derived neural progenitors improves behavioral deficit in Parkinsonian rats. Stem Cells. 2004;22:1246–1255. doi: 10.1634/stemcells.2004-0094. [DOI] [PubMed] [Google Scholar]
- 12.Perrier AL, Tabar V, Barberi T, Rubio ME, Bruses J, et al. Derivation of midbrain dopamine neurons from human embryonic stem cells. Proc Natl Acad Sci USA. 2004;101:12543–12548. doi: 10.1073/pnas.0404700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kriks S, Shim JW, Piao J, Ganat YM, Wakeman DR, et al. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson's disease. Nature. 2011;480:547–551. doi: 10.1038/nature10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parsons XH. The dynamics of global chromatin remodeling are pivotal for tracking the normal pluripotency of human embryonic stem cells. Anatom Physiol. 2012;S3:002. [PMC free article] [PubMed] [Google Scholar]
- 15.Parsons XH, Teng YD, Parsons JF, Snyder EY, Smotrich DB, et al. Efficient derivation of human neuronal progenitors and neurons from pluripotent human embryonic stem cells with small molecule induction. J Vis Exp. 2011;56:e3273. doi: 10.3791/3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parsons XH, Teng YD, Parsons JF, Snyder EY, Smotrich DB, et al. Efficient derivation of human cardiac precursors and cardiomyocytes from pluripotent human embryonic stem cells with small molecule induction. J Vis Exp. 2011;57:e3274. doi: 10.3791/3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parsons JF, Smotrich DB, Gonzalez R, Snyder EY, Moore DA, et al. Defining conditions for sustaining epiblast pluripotence enables direct induction of clinically-suitable human myocardial grafts from biologics-free human embryonic stem cells. J Clin Exp Cardiolog. 2012;S9:001. doi: 10.4172/2155-9880.s9-001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parsons XH. MicroRNA profiling reveals distinct mechanisms governing cardiac and neural lineage-specification of pluripotent human embryonic stem cells. J Stem Cell Res Ther. 2012;2:124. doi: 10.4172/2157-7633.1000124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parsons XH. An engraftable human embryonic stem cell neuronal lineage-specific derivative retains embryonic chromatin plasticity for scale-up CNS regeneration. J Reg Med & Tissue Eng. 2012;1:1–12. doi: 10.7243/2050-1218-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ivey KN, Muth A, Arnold J, King FW, Yeh RF, et al. MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell Stem Cell. 2008;2:219–229. doi: 10.1016/j.stem.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laurent LC, Chen J, Ulitsky I, Mueller FJ, Lu C, et al. Comprehensive microRNA profiling reveals a unique human embryonic stem cell signature dominated by a single seed sequence. Stem Cells. 2008;26:1506–1516. doi: 10.1634/stemcells.2007-1081. [DOI] [PubMed] [Google Scholar]
- 22.Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martinez NJ, Gregory RI. MicroRNA gene regulatory pathways in the establishment and maintenance of ESC identity. Cell Stem Cell. 2010;7:31–35. doi: 10.1016/j.stem.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463:621–626. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137:647–658. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 26.Haramati S, Chapnik E, Sztainberg Y, Eilam R, Zwang R, et al. MicroRNA malfunction causes spinal motor neuron disease. Proc Natl Acad Sci USA. 2010;107:13111–13116. doi: 10.1073/pnas.1006151107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu N, Olson EN. MicroRNA regulatory networks in cardiovascular development. Dev Cell. 2010;18:510–525. doi: 10.1016/j.devcel.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams AH, Liu N, van Rooij E, Olson EN. MicroRNA control of muscle development and disease. Curr Opin Cell Biol. 2009;21:461–469. doi: 10.1016/j.ceb.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Redmond DE, Jr, Bjugstad KB, Teng YD, Ourednik V, Ourednik J, et al. Behavioral improvement in a primate Parkinson's model is associated with multiple homeostatic effects of human neural stem cells. Proc Natl Acad Sci USA. 2007;104:12175–12180. doi: 10.1073/pnas.0704091104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yekta S, Tabin CJ, Bartel DP. MicroRNAs in the Hox network: an apparent link to posterior prevalence. Nat Rev Genet. 2008;9:789–796. doi: 10.1038/nrg2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lund AH. MiR-10 in development and cancer. Cell Death Differ. 2010;17:209–214. doi: 10.1038/cdd.2009.58. [DOI] [PubMed] [Google Scholar]
- 32.Marshall H, Studer M, Pöpperl H, Aparicio S, Kuroiwa A, et al. A conserved retinoic acid response element required for early expression of the homeobox gene Hoxb-1. Nature. 1994;370:567–571. doi: 10.1038/370567a0. [DOI] [PubMed] [Google Scholar]
- 33.Delaloy C, Liu L, Lee J, Su H, Shen F, et al. MicroRNA-9 coordinates proliferation and migration of human embryonic stem cell-derived neural progenitors. Cell Stem Cell. 2010;6:323–335. doi: 10.1016/j.stem.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao C, Sun G, Li S, Lang MF, Yang S, et al. MicroRNA let-7b regulates neural stem cell proliferation and differentiation by targeting nuclear receptor TLX signaling. Proc Natl Acad Sci USA. 2010;107:1876–1881. doi: 10.1073/pnas.0908750107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Contestabile A. Amyotrophic lateral sclerosis: from research to therapeutic attempts and therapeutic perspectives. Curr Med Chem. 2011;18:5655–5665. doi: 10.2174/092986711798347289. [DOI] [PubMed] [Google Scholar]
- 36.Papadeas ST, Kraig SE, O'Banion C, Lepore AC, Maragakis NJ. Astrocytes carrying the superoxide dismutase 1 (SOD1G93A) mutation induce wild-type motor neuron degeneration in vivo. Proc Natl Acad Sci USA. 2011;108:17803–17808. doi: 10.1073/pnas.1103141108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haidet-Phillips AM, Hester ME, Miranda CJ, Meyer K, Braun L, et al. Astrocytes from familial and sporadic ALS patients are toxic to motor neurons. Nat Biotechnol. 2011;29:824–828. doi: 10.1038/nbt.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]


