Abstract
Background
Pluripotent human embryonic stem cells (hESCs) proffer cures for a wide range of neurological disorders by supplying the diversity of human neuronal cell types in the developing CNS for repair. However, realizing the therapeutic potential of hESC derivatives has been hindered by generating neuronal cells from pluripotent cells through uncontrollable and inefficient multi-lineage differentiation. Previously, we used a defined platform to identify retinoic acid as sufficient to induce the specification of neuroectoderm direct from the pluripotent state of hESCs and trigger uniform neuronal lineage-specific progression to human neuronal progenitors (hESC-I hNuPs) and neurons (hESC-I hNus) in the developing CNS with high efficiency.
Methods
Having achieved uniformly conversion of pluripotent hESCs to a neuronal lineage, in this study, the expression and intracellular distribution patterns of a set of chromatin modifiers in hESC-I hNuPs were examined and compared to the two prototypical neuroepithelial-like human neural stem cells (hNSCs) either derived from hESCs or isolated directly from the human fetal neuroectoderm in vivo.
Results
These hESC-I hNuPs expressed high levels of active chromatin modifiers, including acetylated histone H3 and H4, HDAC1, Brg-1, and hSNF2H, retaining an embryonic acetylated globally active chromatin state. Consistent with this observation, several repressive chromatin remodeling factors regulating histone H3K9 methylation, including SIRT1, SUV39H1, and Brm, were inactive in hESC-I hNuPs. These Nurr1-positive hESC-I hNuPs, which did not express the canonical hNSC markers, yielded neurons efficiently and exclusively, as they did not differentiate into glial cells. Following engraftment in the brain, hESC-I hNuPs yielded well-dispersed and well-integrated human neurons at a high prevalence.
Conclusions
These observations suggest that, unlike the prototypical neuroepithelial-like nestin-positive hNSCs, these in vitro neuroectoderm-derived Nurr1-positive hESC-I hNuPs are a more neuronal lineage-specific and plastic human stem cell derivative, providing an engraftable human embryonic neuronal progenitor in high purity and large supply with adequate neurogenic potential for scale-up CNS regeneration as stem cell therapy to be translated to patients in clinical trials.
Keywords: human embryonic stem cells, human, human pluripotent stem cells, human neural stem cells, human neuronal progenitors, human stem cells, neurons, multipotency, pluripotency, epigenome, acetylation, methylation, chromatin, plasticity, differentiation, lineage-specific, derivative, development, cell therapy, CNS repair, regeneration
Introduction
Human pluripotent stem cells (hPSCs), most classically exemplified by human embryonic stem cells (hESCs) derived from the inner cell mass (ICM) or epiblast of the blastocyst [1], have the theoretic ability to differentiate into all somatic cell types, whereas human somatic stem/progenitor/precursor cells (hSSCs) are multipotent, indicating that their potential is restricted to a particular germ layer, organ, or tissue of origin. Although hSSCs have traditionally been isolated directly from such sources of origin in vivo (primary hSSCs), the field is becoming more adept at deriving hSSCs in vitro from pluripotent cells (secondary hSSCs). Heretofore, these two derivation strategies have been regarded as equivalent to generate transplantation materials for therapeutic use despite mounting evidences of their discrepancies. Previous reports indicate that neural progenitors derived from hPSCs give rise to non-teratoma neoplastic cells following transplantation into an adult animal model of Parkinsonism, while neural progenitors derived from the CNS tissue, when transplanted into models of the same disease, do not [2–5]. It has been recognized that hPSCs, when spontaneously differentiating towards a particular lineage, continue to express genes associated with extraneous lineages [5,6]. In view of the growing interest in the use of hPSCs, including artificially-reprogrammed human induced pluripotent cells (hiPS cells) [7–10], teratoma formation and/ or the emergence of inappropriate or undesired cell types remain a constant concern following transplantation [2,3,5]. The growing numbers of identified stem cell populations or derivatives and the escalating concerns for safety and efficacy of these cells towards clinical applications have made it increasingly crucial to assess the likely relative risk-benefit ratio of a given stem cell from a given source for a particular disease. Selection of an optimal source of human stem cells for safe and effective clinical translation requires reliably predictive molecular parameters for comparing not only the plasticity but, more importantly, the specificity of a particular stem cell derivative in its lineage-commitment to the tissue or organ in need of repair prior to transplantation. Similar metrics are required as well for evaluating the protocols for differentiating such human stem cells once chosen towards clinically-relevant lineages for cell-based therapy.
Packaging of the eukaryotic genome into chromatin, a nucleoprotein complex in which the DNA helix is wrapped around an octamer of core histone proteins to fold into nucleosomal DNA structures, confers a higher order of epigenomic structure and control over the unfolding of lineage commitment programs, which goes beyond what might be predicted based solely on profiling a cell’s genomic or proteomic patterns [11]. Discerning the intrinsic plasticity and potential of human stem cell populations might reside in chromatin modifications that shape the respective epigenomes of their derivation routes. Therefore, chromatin states have been used to characterize and compare the intricate plasticity and potential of human stem cell derivatives [11–17]. In general, histone acetylation is associated with a globally active chromatin state, while histone deacetylation and histone H3 K9 methylation are associated with a globally repressed chromatin state [11–17]. However, without a practical strategy to convert pluripotent cells direct into a specific lineage, previous studies and profiling of pluripotent hESCs and their differentiating multi-lineage aggregates have not been able to resolve the epigenomic landscape features of human stem cell populations that might be used to predict their intrinsic plasticity and regenerative potential, hence, safety and efficacy, when deriving the optimal human stem cell preparations for clinical indications.
Human stem cell transplantation represents a promising therapeutic approach closest to provide a cure to restore the lost nerve tissue and function for a wide range of devastating and untreatable neurological disorders. However, to date, lack of a clinically-suitable source of engraftable human stem/ progenitor cells with adequate neurogenic potential has been the major setback in developing effective cell-based therapy as a treatment option for restoring the damaged or lost central nervous system (CNS) structure and circuitry. The traditional sources of engraftable human stem cells with neural potential for transplantation therapies have been multipotent human neural stem cells (hNSCs) isolated directly from the human fetal CNS [4,5,18–21]. However, cell therapy based on CNS tissue-derived hNSCs has encountered supply restriction and difficulty to use in the clinical setting due to their limited expansion ability and declining plasticity with aging, potentially restricting the tissue-derived hNSC as an adequate source for graft material [4,5,18–21]. Alternatively, the pluripotent hESCs proffer cures for a wide range of neurological disorders by supplying the diversity of human neuronal cell types in the developing CNS for regeneration and repair [5,22–26]. However, realizing the therapeutic potential of hESC derivatives has been hindered by the current state of the art for generating functional cells through multi-lineage differentiation of pluripotent cells, which is uncontrollable, inefficient, instable, highly variable, difficult to reproduce and scale-up, and often causes phenotypic heterogeneity and instability, hence, a high risk of tumorigenicity following transplantation [2,3,5,22–26]. Under protocols presently employed in the field, the prototypical neuroepithelial-like nestin-positive hNSCs, either isolated from CNS in vivo or derived from pluripotent cells in vitro via conventional multi-lineage differentiation, appear to exert their therapeutic effects primarily by their non-neuronal progenies through producing trophic and/or neuroprotective molecules to rescue endogenous dying host neurons, but not related to regeneration from the graft or host remyelination [5,26]. In previous reports, we found that pluripotent hESCs maintained under the defined culture conditions can be uniformly converted into a specific neural or cardiac lineage by small molecule induction [26–30]. Retinoic acid (RA) was found to induce the specification of neuroectoderm direct from the pluripotent state of hESCs and trigger uniform progression to human neuronal progenitors (hESC-I hNuPs) and neurons (hESC-I hNus) in the developing CNS efficiently by promoting nuclear translocation of the neuronal-specific transcription factor Nurr-1 [26,27,30]. Similarly, nicotinamide (NAM) was found to induce the specification of cardiomesoderm direct from the pluripotent state of hESCs and trigger uniform progression to human cardiac precursors and beating cardiomyocytes efficiently by promoting the expression of the earliest cardiac-specific transcription factor Csx/Nkx2.5 [26,28–30]. This technology breakthrough enables well-controlled generation of a large supply of neuronal lineage-specific derivatives across the spectrum of developmental stages direct from the pluripotent state of hESCs with small molecule induction.
Having achieved uniformly conversion of pluripotent hESCs to a neuronal lineage, in this study, the expression and intracellular distribution patterns of a set of chromatin modifiers in these in vitro neuroectoderm-derived Nurr1-positive hESC-I hNuPs were examined and compared to the prototypical neuroepithelial-like nestin-positive hNSCs either isolated directly from the fetal CNS, which acquire their neurectodermal identity in vivo through primary in vivo developmental processes, or derived secondarily from pluripotent hESCs in culture. These in vitro neuroectoderm-derived hESC-I hNuPs expressed high levels of active chromatin modifiers that have been associated with the chromatin states of embryonic stem cells and their derivatives [11–17], including acetylated histones H3 and H4 (acH3 and acH4), histone deacetylase 1 (HDAC1), ATP-dependent active chromatin-remodeling factors (Brg-1 and hSNF2H), suggesting that hESC-I hNuPs retain an embryonic acetylated globally active chromatin state. Consistent with this observation, several repressive chromatin remodeling factors regulating histone H3K9 methylation and mediating chromatin-silencing [31–33], including the NAD+-dependent histone deacetylase SIRT1, the histone methyltransferase (HMT) SUV39H1, and the chromatin remodeling factors Brm, were localized to the cytoplasm, suggesting that they were inactive in hESC-I hNuPs. These Nurr1-positive hESC-I hNuPs, which did not express the canonical hNSC markers, yielded neurons efficiently (> 90%) and exclusively, as they did not differentiate into glial cells, such as astrocytes, and oligodendrocytes. Following engraftment in the brain, hESC-I hNuPs yielded well-dispersed and well-integrated human neurons at a high prevalence. By contrast, the prototypical neuroepithelial-like nestin-positive hNSCs derived either from hESCs or CNS can spontaneously differentiate into a mixed population of cells containing undifferentiated hNSCs, neurons (10–30%), astrocytes, and oligodendrocytes in vitro and in vivo [4,5,18–26]. These observations suggest that, unlike the prototypical neuroepithelial-like nestin-positive hNSCs, these in vitro neuroectoderm-derived Nurr1-positive hESC-I hNuPs are a more neuronal lineage-specific and plastic human stem cell derivative in the developing CNS, providing an engraftable human embryonic neuronal progenitor in high purity and large supply with adequate neurogenic potential for scale-up CNS regeneration as stem cell therapy to be translated to patients in clinical trials.
Materials and Methods
Culture of undifferentiated hESCs under the defined culture
The hESC lines WA01 and WA09 (WiCell Research Institute) and newly-derived biologics-free hESCs (Xcel-hESCs) [29] were used in this study. The defined culture systems consist of DMEM/F-12 or KO-DMEM (knockout-DMEM) (80%), Knockout Serum Replacement (KO) (20%), L-alanyl-L-gln or L-gln (2 mM), MEM nonessential amino acids (MNAA, 1X), β-Mercaptoethanol (β-ME, 100 μM) (all from Invitrogen), human purified laminin (Sigma) or laminin/collagen (growth factor reduced Matrigel, BD Bioscience) as the matrix protein, and bFGF (basic fibroblast growth factor, 20 ng/ml) (PeproTech Inc). The KO can be replaced with defined essential factors containing MEM essential amino acids (MEAA, 1X), human insulin (20 mg/ml) (Sigma), and ascorbic acid (50 mg/ml) (Sigma), in which activin A (50 ng/ml, Sigma), human albumin (10 mg/ml, Sigma), and human transferrin (8 mg/ml, Sigma) were added in order to increase cell survival and maintain normal shape and healthy colonies. Long-term stable expansion of pluripotent hESCs maintained under the defined culture (~50–150 passages, ~12–36 months) has been published in our previous reports [26,29]. These hESCs were stored by cryopreservation over their long-term expansion periods.
Neuronal lineage-specific differentiation direct from the pluripotent state of hESCs
Pluripotent hESCs maintained under the defined culture (~50–150 passages, ~ 2–36 months) [26,29] were used to generate hESC derivatives in this study. Undifferentiated hESCs maintained under the defined culture conditions were treated with RA (10μM) 3 days after seeding for 4–5 days. These neuroectoderm-differentiated hESCs were transferred to a serum-free suspension culture to allow floating neuroblasts (hESC-I hNuPs) to form in the hESC media lacking bFGF for 4–5 days. To limit the effects of long-term culture on those hESC-I hNuPs, only cells within the first 6 passages were used for the analyses. For further differentiating into a neuronal phenotype, the neuroblasts were then permitted to attach to a tissue culture plate in a defined medium containing DMEM/F-12, N-2 supplement (1%), heparin (8 mg/ml), VEGF (20 ng/ml), NT-3 (10 ng/ml), and BDNF (10 ng/ml). β-III-tubulin- and Map-2-expressing, extensively neurite-bearing cells (hESC-I hNus) and pigmented cells, typical of those in the ventral mesencephalon, were observed within 2 weeks of continuous cultivation, increased in numbers with time. The generated hESC neuronal derivatives were stored by cryopreservation over passaging periods.
Generation of hESC-D hNSCs and CNS-D hNSCs
A homogenous population of hESC-D hNSCs was generated by standard well-established procedures described previously [22–25]. Briefly, differentiation of hESCs was initiated by growing hESCs in a suspension culture as embryoid bodies (EBs). The differentiating EBs were then allowed to attach to tissue culture plates and develop into clusters of neural rosettes. A homogeneous population of Nestin, Vimentin, and Musashi-expressing hNSCs was isolated, characterized, and maintained in a defined neural stem cell media containing bFGF (20 ng/ ml). To limit the effects of long-term culture on those hESC-D hNSCs, only cells within the first 6 passages were used for the analyses. The CNS-D hNSCs were isolated directly from the ventricular zone of the telencephelon (a neurectoderm-derived structure) of two 11–13 week human fetal cadavers (HFB2030 and HFT13) [4,20,21]. These hNSCs were propagated without genetic manipulation following a serial growth factor and engraftment selection process, characterized, and maintained in a karyotypically normal state in a defined NSC media containing bFGF (20 ng/ml) and leukemia inhibitory factor (LIF) (10 ng/ml) as previously described [4,20,21]. Fresh vials of low passage cells from the initial isolation and derivation of these hNSCs were used for these studies. The homogeneity and comparability of hNSCs was verified by insuring that >95% of the cells expressed standard neural stem/progenitor markers, including Nestin, Musashi, and Sox-2, and no longer expressed markers associated with pluripotency, including Oct-4, SSEA-4, Tra-1-60, Tra-1-80, or markers of non-neural lineages. Neural differentiation of hNSCs was initiated by removal of bFGF followed by culture in a defined medium containing neurotrophin-3 (NT-3, 10 ng/ml) and brain-derived neurotrophic factor (BDNF, 10 ng/ml) for 2–3 weeks before further analysis. These hNSCs, either derived from hESCs or CNS, were stored by cryopreservation over passaging periods.
Immunofluorescence and Deconvolution Microscopy
Cells were grown in 12- or 24-well plates, fixed with 4% paraformaldehyde, and blocked in phosphate buffered saline (PBS) containing 0.1% Triton X-100 and 2% bovine serum albumin (BSA). Cells were further incubated with the primary antibody (Santa Cruz Biotech.; Upstate Biotech.; and Millipore) in wash buffer (0.1% Triton X-100 in PBS) at 4°C overnight, and then with the secondary antibody (Invitrogen) in wash buffer for 45 min at room temperature. After staining with DAPI, cells were mounted onto a microscope slide and visualized under an immunofluorescence and deconvolution microscope. The specificity of all antibodies were independently verified on known positive and negative control cells before being used in these studies.
High-Resolution Three-Dimensional Quantitative Intracellular Imaging
The cell-image-based assays for protein intensity and intracellular localization pattern (e.g., cytoplasmic-to-nuclear translocation, nuclear foci formation) were carried out using a Beckman Coulter Q3DM IC-100/GE-Amersham IN Cell Analyzer 1000 and Cytoshop software.
Analysis of Nucleoprotein Complexes
Chromosomal proteins were cross-linked to DNA by adding 1% formaldehyde directly to culture medium and incubating at 37°C for 10 min. Cells were isolated and resuspended in PBS. This cell mixture was loaded into a double-chamber cytofunnel and centrifuged in a Cytospin 3 (Shandon) at 900 rpm for 5 min onto clean glass microscope slides. Immediately after centrifugation, slides were placed in lysis buffer (25 mM Tris [pH 7.5], 500 mM NaCl, and 1% Triton X-100) for 15 min, then processed for immunofluorescence and deconvolution microscopic analysis.
Transplantation into mouse brain
Human neuronal progenitors were generated from undifferentiated hESCs maintained under the defined culture by retinoic acid induction. The human neuronal progenitor cells in suspension culture were pulled together and dissociated with a brief treatment (~1min.) of Accutase (Invitrogen) followed by gently triturated into single cell suspension, and resuspended in PBS (Mg2+ & Ca2+ free) (Invitrogen) at a density of 50,000/ml. A total of 10×6 human donor cells (20 ml at 50,000/ml) were injected into the cerebral ventricles of immunocompetent wild type and SOD1 mutant newborn mice (n = 54) (The Jackson Laboratory). Controls were similarly transplanted with the same amount of human fibroblast cells (Hs27, ATCC). After at least 3 months post-grafting, the mice were sacrificed and processed for histological and immunocytochemical analysis of the transplanted brain. No graft overgrowth, formation of teratomas or neoplasms, or appearance of non-neuronal cell types was observed in at least 54 transplanted animals and at least 1 year after transplantation (the latest time point examined). Transplanted wild type mice developed hyper-active behavior, such as fast movement and fast spin (see videos at http://www.sdrmi.org). The animal experiments were reviewed and approved by the relevant institutional animal care and use committees.
Results and Discussion
The in vitro neuroectoderm-derived nuclear-Nurr1-positive hESC-I hNuPs do not express the canonical hNSC markers
Previously, we have identified the minimal essential components for sustaining the epiblast pluripotence of hESCs in a defined culture system, serving as a platform for de novo derivation of clinically-suitable hESCs and effectively directing such hESCs uniformly towards functional lineages by small molecule induction [26–30]. In order to achieve uniformly conversion of pluripotent hESCs to a lineage-specific fate, we have used the defined culture system to screen the differentiation inducing effect of a variety of small molecules and growth factors on the pluripotent state of hESCs. We found that such defined conditions rendered small molecule RA sufficient to induce the specification of neuroectoderm direct from the pluripotent state of hESCs and trigger neuronal lineage-specific progression to human neuronal progenitors (hESC-I hNuPs) and neurons (hESC-I hNus) in the developing CNS with high efficiency [26,27,30]. RA induces a cascade of neuronal lineage-specific differentiation events direct from the pluripotent state of hESCs by promoting nuclear translocation of the neuronal specific transcription factor Nurr1, a member of the orphan nuclear hormone receptor super-family implicated in ventral neuronal development [26,27]. Genome-scale profiling of microRNA differential expression showed that the expression of pluripotence-associated hsa-miR-302 family was silenced and the expression of Hox miRNA hsa-miR-10 family that regulates gene expression predominantly in neuroectoderm was induced to high levels in those hESC neuronal lineage-specific derivatives, suggesting that hESC-I hNuPs have acquired a neuroectodermal identity through RA induction of pluripotent hESCs in vitro [30].
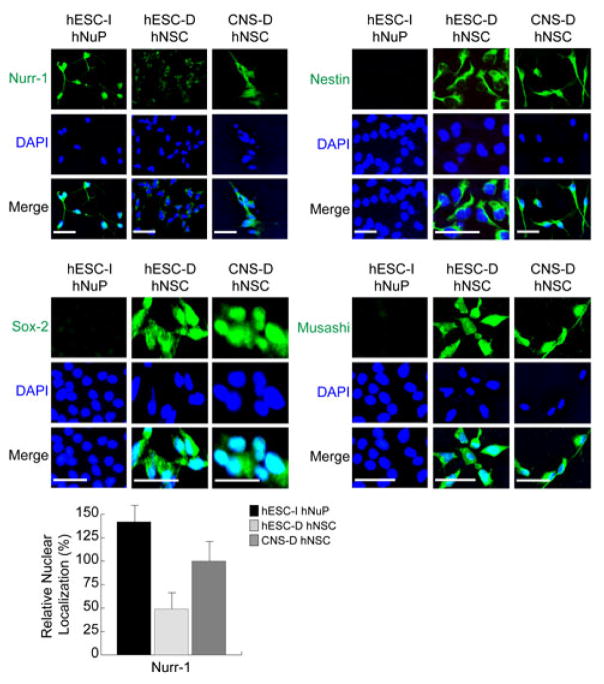
Compared to the two prototypical neuroepithelial-like nestin-positive hNSCs either derived from hESCs or CNS, hESC-I hNuPs did not express the canonical early neural lineage stem/progenitor cell markers of hNSCs, including Nestin, Musashi, and Sox-2 [4,5,18–26], but assumed uniformly strong expression and nuclear localization of the neuronal-specific transcription factor Nurr-1 (Figure 1). Although CNS-derived hNSCs (CNS-D hNSCs), which have acquired their neurectodermal identity through in vivo developmental processes [4,5,18–21], show moderate expression and nuclear localization of Nurr-1, in hESC-derived hNSCs (hESC-D hNSCs) [5,22–26], Nurr-1 localizes to the cell-surface and cytoplasm, suggesting its being inactive (Figure 1). The relative nuclear localization of Nurr1 in these human stem cell derivatives was further verified by quantitative analysis with high-resolution 3-dimensional intracellular microscopic imaging system [34] (Figure 1). These observations suggest that, unlike the prototypical neuroepithelial-like nestin-positive hNSCs, these nuclear-Nurr1-positive hESC-I hNuPs are an hESC neuronal lineage-specific derivative in the developing CNS during human embryonic neurogenesis.
Figure 1. The in vitro neuroectoderm-derived nuclear-Nurr1-positive hESC-I hNuPs do not express the canonical hNSC markers.
hESC-I hNuPs display strong expression and nuclear localization of Nurr-1, compared to fetal CNS-derived hNSCs (CNS-D hNSCs) that show moderate expression and nuclear localization of Nurr-1 and hESC-derived hNSCs (hESC-D hNSCs) that show cell-surface and cytoplasm localization of Nurr-1. hESC-I hNuPs did not express the canonical early neural lineage stem/progenitor cell markers of hNSCs, including Nestin, Musashi, and Sox-2, compared to the two neuroepithelial-like nestin-positive hNSCs either derived from hESCs or CNS. The relative nuclear localization of Nurr1 in these human stem cell derivatives was further verified by quantitative intracellular imaging analysis. DAPI stains nuclei. The nuclear localization of Nurr1 in CNS-D hNSCs is set at 100%. Scale bars: 5 μm.
The in vitro neuroectoderm-derived hESC-I hNuPs retain an embryonic acetylated globally active chromatin
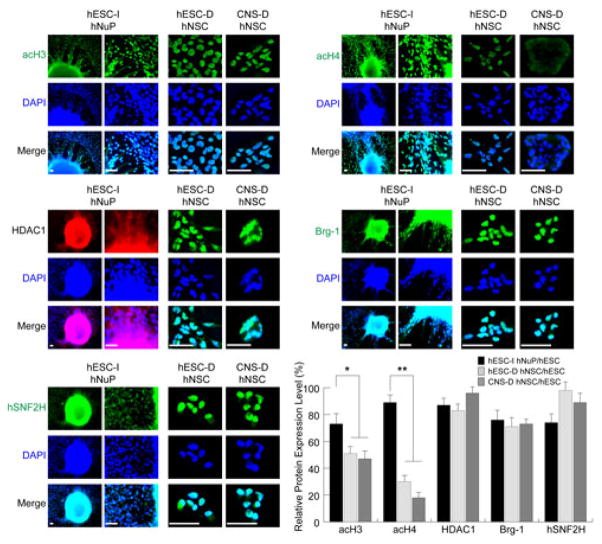
In previous report, we examined the global chromatin dynamics in the pluripotent hESCs maintained under the defined culture and found that the pluripotency of undifferentiated hESCs that display normal stable expansion is associated with high levels of expression and nuclear localization of active chromatin remodeling factors that include acetylated histone H3 and H4, Brg-1, hSNF2H, and HDAC1; weak expression or cytoplasmic localization of repressive chromatin remodeling factors that are implicated in transcriptional silencing; and residual H3 K9 methylation [11]. Having achieved uniformly conversion of pluripotent hESCs to a neuronal lineage, in this study, we further examined the expression and intracellular distribution patterns of a set of chromatin modifiers in hESC-I hNuPs, compared to hESC- and CNS-derived hNSCs (Figures 2 and 3). We observed that these in vitro neuroectoderm-derived Nurr1-positive hESC-I hNuPs expressed high levels of active chromatin modifiers that have been associated with the chromatin states of embryonic stem cells and their derivatives [11–17], including acetylated histones H3 and H4 (acH3 and acH4), histone deacetylase 1 (HDAC1), ATP-dependent active chromatin-remodeling factors (Brg-1 and hSNF2H) (Figure 2), suggesting that hESC-I hNuPs retain an embryonic acetylated globally active chromatin state. In contrast, the prototypical neuroepithelial-like nestin-positive hNSCs either derived from hESCs or the fetal CNS tissue displayed decreasing histone H3 acetylation (~ 2 fold) and decreasing histone H4 acetylation (~ 3–6 fold), despite strong expression of active chromatin-remodeling factors HDAC1, Brg-1, and hSNF2H (Figure 2). In either hESC-derived hNSCs or CNS-derived hNSCs, significantly higher degrees of decreasing in histone H4 acetylation, in comparison with histone H3 acetylation, were observed (Figure 2). Chromatin remodeling factors are ATP-utilizing motor proteins that mediate the interaction of proteins with nucleosomal DNA by DNA/nucleosome-translocation [11]. Brg-1 is a subunit of the Swi/Snf chromatin remodeling complex implicated in the regulation of cellular proliferation and as a tumor suppressor; while hSNF2H is a human homolog of the ISWI family of chromatin remodeling proteins [11]. HDAC1 is a general maintenance histone deacetylase that sustains global transcription at a basal level [11]. Quantitative analysis of the expression levels of the active chromatin-modifying molecules in these human stem cell derivatives, compared to their expression levels in pluripotent hESCs, with intracellular imaging analysis further confirmed their expression patterns (Figure 2).
Figure 2. The in vitro neuroectoderm-derived hESC-I hNuPs retain an embryonic acetylated globally active chromatin.
The strong expression and nuclear localization of a set of active chromatin modifiers in hESC-I hNuPs, compared to hESC-D and CNS-D hNSCs, including acetylated histones H3 and H4 (acH3 and acH4), HDAC1, chromatin-remodeling factors Brg-1 and hSN-F2H. DAPI stains nuclei. Quantitative intracellular imaging analysis shows that the prototypical neuroepithelial-like hNSCs either derived from hESCs or the fetal CNS tissue display decreasing histone H3 acetylation (~ 2 fold) and decreasing histone H4 acetylation (~ 3–6 fold), despite strong expression of active chromatin-remodeling factors HDAC1, Brg-1, and hSNF2H. The expression levels of these active chromatin-modifying molecules in these human stem cell derivatives were quantified and plotted as relative levels in comparison to their expression levels in pluripotent hESCs. The expression levels of these active chromosomal proteins in pluripotent hESCs are set at 100%. Scale bars: 5 μm. Quantification data are presented as Mean ± SD; Statistical significance was defined as *P < 0.05, **P < 0.01.
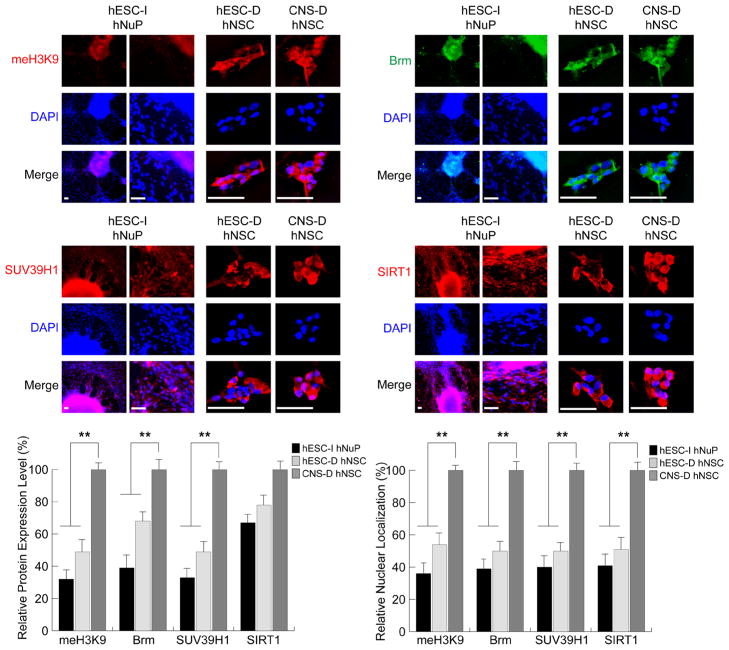
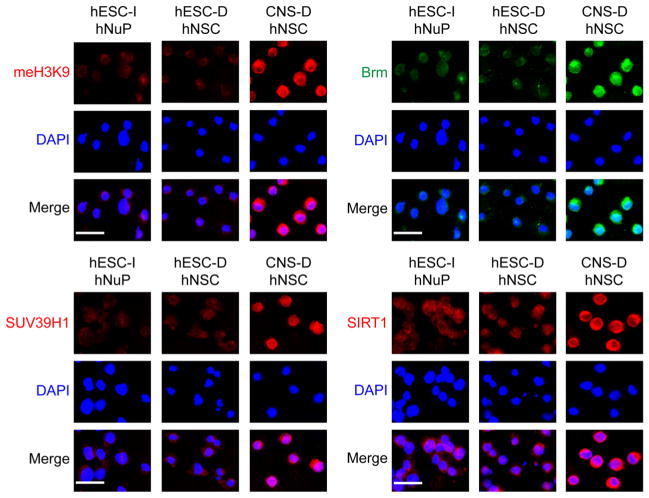
Consistent with this observation, several repressive chromatin remodeling factors regulating histone H3K9 methylation and mediating chromatin-silencing in development [31–33], including the class III NAD+-dependent histone deacetylase SIRT1, the histone methyltransferase (HMT) SUV39H1, the chromatin remodeling factors Brm, and the K9 methylated histone H3 (meH3K9) were localized to the cytoplasm (SIRT1, SUV39H1, Brm, meH3K9) and/or expressed at low levels (SUV39H1, Brm, meH3K9), suggesting that they were inactive in hESC-I hNuPs, similar to the expression patterns in hESC-derived hNSCs (Figure 3). In contrast, the fetal CNS tissue derived hNSCs displayed strong expression and nuclear location of these repressive chromatin modifiers, including SIRT1, SUV39H1, Brm, meH3K9 (Figure 3). Quantitative analysis of expression levels and nuclear localization in these human stem cell derivatives with intracellular imaging analysis further confirmed the spatial and temporal patterns of these chromatin-modifying molecules (Figure 3). These observations suggest that, in spite of neuronal lineage specification from pluripotent hESCs, these in vitro neuroectoderm-derived Nurr1-positive hESC-I hNuPs retain an embryonic acetylated globally active chromatin.
Figure 3. Repressive chromatin remodeling factors regulating histone H3K9 methylation and mediating chromatin-silencing are inactive in hESC-I hNuPs.
The weak expression and cytoplasmic localization of repressive chromatin remodeling factors in hESC-I hNuPs, including H3K9 methylated histones, Brm, SUV39H1, and SIRT1, similar to the expression patterns in hESC-derived hNSCs. In contrast, the CNS-D hNSCs display strong expression and nuclear location of these repressive chromatin modifiers. DAPI stains nuclei. Quantitative intracellular imaging analysis of expression levels and nuclear localization in these human stem cell derivatives confirmed the spatial and temporal patterns of these chromatin-modifying molecules. The expression levels and nuclear localization of these repressive chromatin-modifying molecules in these human stem cell derivatives were quantified and plotted as relative levels in comparison to their expression levels and nuclear localization in CNS-D hNSCs. The expression levels and nuclear localization of these repressive chromosomal proteins in CNS-D hNSCs are set at 100%. Scale bars: 5 μm. Quantification data are presented as Mean ± SD; Statistical significance was defined as *P < 0.05, **P < 0.01.
Epigenomic landscape profiles of nucleoprotein complexes reveal embryonic chromatin plasticity of the hESC neuronal lineage-specific derivative
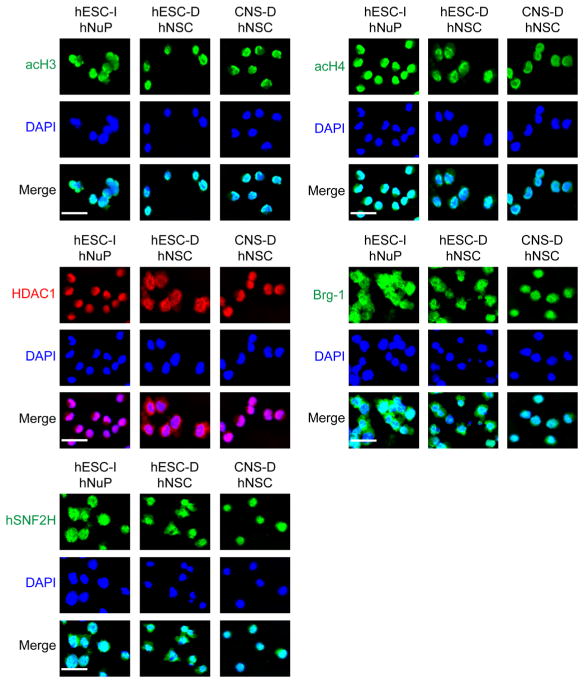
To better view the genome-wide localization patterns of these chromosomal proteins, immunofluorescence and deconvolution microscopic analysis of the formaldehyde cross-linked nucleoprotein complexes in the nuclei of these human stem cell derivatives was performed [35,36]. Analysis of the cross-linked nucleoprotein complexes revealed that the basal and active chromatin remodeling factors acH3, acH4, Brg-1, hSNF2H, and HDAC1 formed large chromatin-localized nucleoprotein complexes and remained strongly expressed in these human stem cell derivatives (Figure 4), suggesting that they might be critical for maintaining the undifferentiated state of human stem cell derivatives in general. Interestingly, acH4 chromatin deposition displayed a pattern of global distribution on the genome of hESC-I hNuPs, as compared to the patterns of discrete distribution of acH4 chromatin deposition on the genomes of hNSCs and the patterns of discrete distribution of acH3 chromatin deposition on the multipotent genomes of all the human stem cell derivatives examined (Figure 4). A considerable amount of evidence suggests that acetylation of histone H3 and H4 has distinct functional and temporal patterns [33]. The H3 modifications seem to be connected to proper control of gene expression, whereas acetylation of H4 seems to be most important in histone deposition and chromatin structure [31,33]. Of the four lysine residues in the N-terminal tail of H4 (K5, 8, 12, 16), lysine 16 (K16) is the specific target of SIRT1 and plays a unique role in regulating chromatin structure [31,33]. Histone H4 K16 acetylation is important in epigenetic regulation as substantiated by its being the only lysine residue among the N-terminal tails of all histones that is targeted by an exclusive category of histone acetyltransferases (HATs) as well as HDACs, such as the MYST family of HATs and the class III NAD-dependent HADCs to mediate silencing of chromatin locus during phenotype switch in human development [31,33].
Figure 4.
Epigenomic landscape profiles of active nucleoprotein complexes reveal embryonic chromatin plasticity of the hESC neuronal lineage-specific derivative. Images of the cross-linked nucleoprotein complexes show the extent of chromatin-localization and the distribution patterns of acH3, acH4, HDAC1, Brg-1, and hSNF2H, in these human stem cell derivatives. Scale bars: 2 μm.
Analysis of the cross-linked nucleoprotein complexes revealed that the repressive chromatin remodeling factors SIRT1, SUV39H1, Brm, and, consequently, H3K9 methylated histones (meH3K9) remained weakly expressed in hESC lineage-specific derivatives (Figure 5), suggesting that hESC-I hNuPs retain hESC globally active open chromatin state [11]. The chromatin depositions of SIRT1, SUV39H1, Brm, and meH3K9 were significantly increased in CNS-derived hNSCs, compared to those expressed at low levels in hESC-I hNuPs and hESC-derived hNSCs (Figure 5), suggesting that CNS-derived tissue-resident hNSCs had acquired a more silenced chromatin.
Figure 5. Repressive nucleoprotein complexes remain weakly expressed in hESC lineage-specific derivatives.
The chromatin depositions of SIRT1, SUV39H1, Brm, and me-H3K9 were significantly increased in CNS-D hNSCs, compared to those expressed at low levels in hESC-I hNuPs and hESC-D hNSCs, suggesting that CNS-derived tissue-resident hNSCs had acquired a more silenced chromatin. Scale bars: 2 μm.
The neuroectoderm-derived hESC neuronal lineage-specific derivative is safely engraftable and highly neurogenic in vitro and in vivo
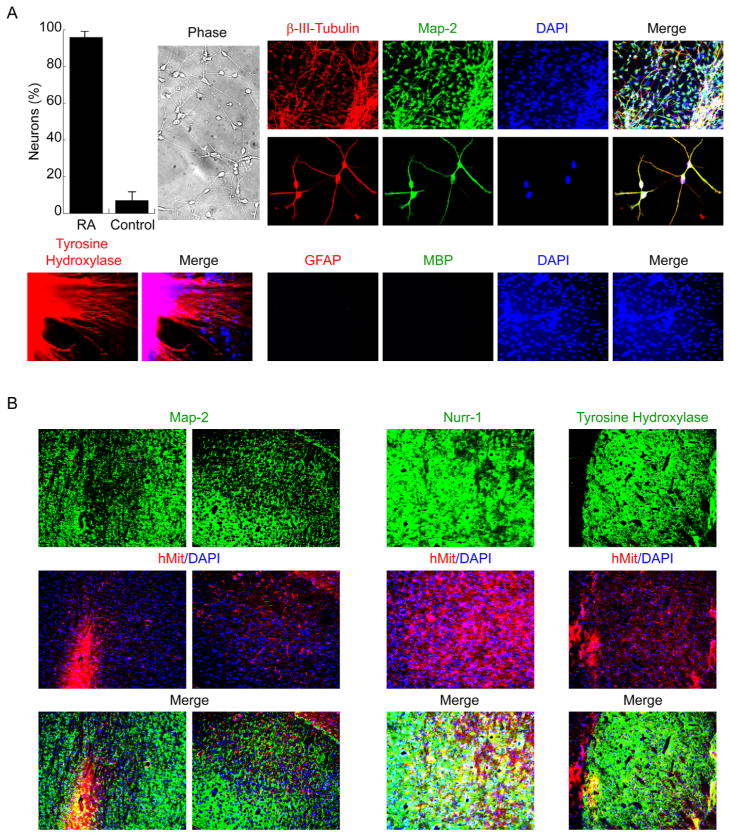
To confirm these in vitro neuroectoderm-derived Nurr-1-positive hESC-I hNuPs commit to a neuronal fate, hESC-I hNuPs were permitted to attach in a defined media containing neurotrophic factors without bFGF to further differentiate into neurons [26,27]. Under these neuronal differentiation conditions, hESC-I hNuPs yielded exclusively neurons that expressed neuronal marker β-III-tubulin and co-expressed Map-2 with a drastic increase in efficiency (~95%) when compared to similarly cultured cells derived from untreated embryoid bodies (EBs) as the control (Figure 6A). That these in vitro neuroectoderm-derived hESC-I hNuPs differentiate exclusively to a neuronal fate was confirmed by the absence of markers associated with other neural cells such as glial lineage, as indicated by no cell expressing GFAP and MBP (Figure 6A). Accordingly, a large proportion of these hESC-derived neuronal cells began to express tyrosine hydroxylase (TH) (Figure 6A), consistent with the early stages of acquiring catecholaminergic or dopaminergic potential [26,27]. By contrast, the prototypical neuroepithelial-like nestin-positive hNSCs derived either from hESCs or CNS can spontaneously differentiate into a mixed population of cells containing undifferentiated hNSCs, neurons (10–30%), astrocytes, and oligodendrocytes in vitro and in vivo [4,5,18–26]. These observations suggest that, unlike the prototypical neuroepithelial-like nestin-positive hNSCs, these in vitro neuroectoderm-derived Nurr1-positive hESC-I hNuPs are a more neuronal lineage-specific human stem cell derivative in the developing CNS, yielding neurons exclusively, as they do not differentiate into other neural cell types such as oligodendrocytes and astrocytes (Figure 6A).
Figure 6. The neuroectoderm-derived hESC neuronal lineage-specific derivative is highly neurogenic in vitro and in vivo.
(A). Under neuronal differentiation conditions, hESC-I hNuPs yielded exclusively neurons that expressed neuronal marker β-III-tubulin and co-expressed Map-2 with a drastic increase in efficiency when compared to similarly cultured cells derived from un treated embryoid bodies (EBs) as the control. No cell expressed glial lineage marker GFAP and MBP. Accordingly, a large proportion of these hESC-derived neuronal cells began to express tyrosine hydroxylase (TH).
(B). hESC-I hNuPs were injected into the cerebral ventricles of newborn mice. Histological analysis of transplanted mice showed well-dispersed and well-integrated human neurons at a high prevalence, indicated by anti-human mitochondrial antibody (hMit) and their immunoreactivity to Map-2, including Nurr1-positve and TH-positive DA neurons, within neurogenic regions of the brain. DAPI nuclear marker stains all cells in the field.
To address whether these neuroectoderm-derived Nurr1-positive hESC-I hNuPs could be safely engrafted in the brain and could migrate and retain their neurogenic ability in vivo, hESC-I hNuPs were transplanted into the cerebral ventricles of newborn mice of both wild type and SOD1, the transgenic mouse model expressing mutated forms of the enzyme copper-zinc superoxide dismutase 1 (SOD1) that replicate with fidelity the onset and progression of the disease of Amyotrophic Lateral Sclerosis (ALS) and have been largely used to test therapies to be translated to patients in clinical trials [37]. This route allows excellent access to the subventricular zone (SVZ), a secondary germinal zone from which cells widely migrate and respond to appropriate regional developmental cues [4]. The animal experimental protocols were reviewed and approved by the relevant institutional animal care and use committees. After at least 3 months post-grafting, the mice were sacrificed and processed for histological analysis. Transplanted hESC-I hNuPs survived, engrafted, and migrated widely and yielded well-dispersed and well-integrated human neurons at a high prevalence, including Nurr1-positve dopaminergic (DA) neurons, within neurogenic regions of the brain (Figure 6B), demonstrating their potential for neuron replacement therapy. No graft overgrowth, formation of teratomas or neoplasms, or appearance of non-neuronal cell types was observed following engraftment. Transplanted wild type mice developed hyper-active behavior, such as fast movement and fast spin (see videos at http://www.sdrmi.org), which also suggested that transplanted human neuronal cells had survived and integrated into the mouse brain to function and control mouse behavior. SOD1 mouse is an animal model of ALS, a fatal neurodegenerative disease characterized by progressive degeneration of motor neurons due to toxic astrocytes [37–39]. No human neuron from engrafted human cells was found in the spinal cord of transplanted SOD1 mice to mediate behavior improvement. It is unclear whether it is because the transplanted human cells could not migrate out of brain into the spinal cord or could not survive the toxic astrocytes of SOD1 mice or both. These findings suggest that this novel engraftable hESC neuronal lineage-specific derivative will dramatically increase the clinical efficacy of graft-dependent neuron replacement and safety of hESC-derived cellular products for scale-up CNS regeneration.
Conclusion
Using chromatin marks to judge the intrinsic plasticity of human stem cell lineage-specific derivatives for cell-based therapies
We must bear in mind that the pluripotent hESC itself cannot be used for therapeutic applications. It has been recognized that pluripotent hESCs must be transformed into fate-restricted derivatives before use for cell therapy. Realizing the therapeutic potential of pluripotent hESC derivatives will demand a better understanding of how a pluripotent cell becomes progressively constrained in its fate options to the lineages of tissue or organ in need of repair. It is obvious that we do not want the hESC so uncommitted that it retains its ability to yield teratoma or to give rise to cell types inappropriate to the engrafted organ. Neither, however, do we want the hESC so terminally-differentiated that it cannot integrate in vivo, respond to microenvironmental cues, and yield the diversity of cross-talking cell types that might be necessary to reconstitute an organ’s function [4,5,18,19]. We are coming to recognize – particularly in the CNS -- that an ideal cell source for derivation of therapeutic stem cells should have the ability for long-term stable large-scale expansion in vitro as well as the capacity to yield, following engraftment, an array of appropriate lineage-specific cell types necessary for reconstituting the CNS structure and circuitry [4,5,18,19,26,29]. Based on the clinical indication, we will need to choose the human stem cell that has sufficient plasticity to yield a range of requisite interacting components yet enough neural lineage specificity not to give rise to lineages and cell types inappropriate or undesirable in the CNS.
Turning stem cells into the desired cell types necessary for restitution of damaged host structure and circuitry depends on interactions between extrinsic signals in the host and autonomous programs intrinsic to the grafted cell [5,18,19]. Transplantation studies suggest that stem cells from different sources vary greatly in their capacity to reconstruct the damaged structures. To date, the most promising results have been obtained using somatic stem cells isolated directly from the tissue or organ system of interest, that is, employing cells restricted to the lineage in need of repair [4,5,18–21]. However, as tissue-resident stem cells become more restricted in their developmental potential with aging, their capacity to be directed by host cues in a region-specific manner is downregulated or lost. The pluripotent hESCs, on the other hand, start with the capacity to respond to an exceptionally wide spectrum of developmental signals and to generate a variety of cell types, some of which may be desired for structural and functional recovery while others may not [2,3,5,22–26]. The pluripotent cell-derived grafts, even when used successfully in therapeutic paradigms, tend to display phenotypic instability and pose oncogenic risk [2,3,5]. Safe and effective clinical translation of stem cell biology requires a better knowledge of the inherent cellular mechanisms maintaining plasticity and then stabilizing lineage-specific commitment. Our studies address a heretofore under-appreciated mechanism whereby the global remodeling of chromatin structure may be fundamental to defining the intrinsic potential of human stem cell populations or derivatives.
In summary, this study shows that the intrinsic plasticity of human stem cell derivatives can be differentiated by their epigenomic features. These in vitro neuroectoderm-derived hESC neuronal lineage-specific derivative expressed high levels of active chromatin modifiers that have been associated with the chromatin states of embryonic stem cells and their derivatives [11–17], including acH3 and acH4, HDAC1, Brg-1, and hSNF2H, suggesting that hESC-I hNuPs retain an embryonic acetylated globally active chromatin state. Consistent with this observation, several repressive chromatin remodeling factors regulating histone H3K9 methylation and mediating chromatin-silencing [31–33], including SIRT1, SUV39H1, and Brm, were localized to the cytoplasm, suggesting that they were inactive in hESC-I hNuPs. These chromatin marks described here might help us to select the human stem cell derivative prospectively that strikes the proper balance desired for future therapeutic tasks to insure the safety and efficacy of stem cell engraftment when translated to patients in clinical trials. These Nurr1-positive hESC-I hNuPs, which did not express the canonical hNSC markers, yielded neurons efficiently (> 90%) and exclusively, as they did not differentiate into glial cells. Following engraftment in the brain, hESC-I hNuPs yielded well-dispersed and well-integrated human neurons at a high prevalence. By contrast, the prototypical neuroepithelial-like nestin-positive hNSCs derived either from hESCs or CNS can spontaneously differentiate into a mixed population of cells containing undifferentiated hNSCs, neurons (10–30%), astrocytes, and oligodendrocytes in vitro and in vivo [4,5,18–26]. These observations suggest that, unlike the prototypical neuroepithelial-like nestin-positive hNSCs, these in vitro neuroectoderm-derived Nurr1-positive hESC-I hNuPs are a more neuronal lineage-specific and plastic human stem cell derivative in the developing CNS, providing an engraftable human embryonic neuronal progenitor in high purity and large supply with adequate neurogenic potential for scale-up CNS regeneration as stem cell therapy to be translated to patients in clinical trials.
Acknowledgments
Acknowledgements and funding
XHP has been supported by National Institute of Health (NIH) grants from National Institute on Aging (NIHK01AG024496) and The Eunice Kennedy Shriver National Institute of Child Health and Human Development (NIHR21HD056530).
Footnotes
This is an open access article distributed under the terms of Creative Commons Attribution License (http://creativecommons.org/licenses/by/3.0), This permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Competing interests
The authors declare competing interests. XHP is the founder of Xcelthera and has intellectual properties related to hESCs.
References
- 1.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Roy NS, Cleren C, Singh SK, Yang L, Beal MF, Goldman SA. Functional engraftment of human ES cell-derived dopaminergic neurons enriched by coculture with telomerase-immortalized midbrain astrocytes. Nat Med. 2006;12:1259–1268. doi: 10.1038/nm1495. [DOI] [PubMed] [Google Scholar]
- 3.Wernig M, Zhao JP, Pruszak J, Hedlund E, Fu D, Soldner F, Broccoli V, Constantine-Paton M, Isacson O, Jaenisch R. Neurons derived from reprogrammed fibroblasts functionally integrate into the fetal brain and improve symptoms of rats with Parkinson’s disease. Proc Natl Acad Sci U S A. 2008;105:5856–5861. doi: 10.1073/pnas.0801677105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Redmond DE, Jr, Bjugstad KB, Teng YD, Ourednik V, Ourednik J, Wakeman DR, Parsons XH, Gonzalez R, Blanchard BC, Kim SU, Gu Z, Lipton SA, Markakis EA, Roth RH, Elsworth JD, Sladek JR, Jr, Sidman RL, Snyder EY. Behavioral improvement in a primate Parkinson’s model is associated with multiple homeostatic effects of human neural stem cells. Proc Natl Acad Sci U S A. 2007;104:12175–12180. doi: 10.1073/pnas.0704091104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parsons XH, Teng YD, Snyder EY. Important precautions when deriving patient-specific neural elements from pluripotent cells. Cytotherapy. 2009;11:815–824. doi: 10.3109/14653240903180092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shamblott MJ, Axelman J, Littlefield JW, Blumenthal PD, Huggins GR, Cui Y, Cheng L, Gearhart JD. Human embryonic germ cell derivatives express a broad range of developmentally distinct markers and proliferate extensively in vitro. Proc Natl Acad Sci U S A. 2001;98:113–118. doi: 10.1073/pnas.021537998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 8.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 9.Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 10.Kim JB, Sebastiano V, Wu G, Arauzo-Bravo MJ, Sasse P, Gentile L, Ko K, Ruau D, Ehrich M, van den Boom D, Meyer J, Hubner K, Bernemann C, Ortmeier C, Zenke M, Fleischmann BK, Zaehres H, Scholer HR. Oct4-induced pluripotency in adult neural stem cells. Cell. 2009;136:411–419. doi: 10.1016/j.cell.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 11.Parsons XH. The dynamics of global chromatin remodeling are pivotal for tracking the normal pluripotency of human embryonic stem cells. Anatom Physiol. S3:002. [PMC free article] [PubMed] [Google Scholar]
- 12.Spivakov M, Fisher AG. Epigenetic signatures of stem-cell identity. Nat Rev Genet. 2007;8:263–271. doi: 10.1038/nrg2046. [DOI] [PubMed] [Google Scholar]
- 13.Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, Fisher AG. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 14.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 15.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O’Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao XD, Han X, Chew JL, Liu J, Chiu KP, Choo A, Orlov YL, Sung WK, Shahab A, Kuznetsov VA, Bourque G, Oh S, Ruan Y, Ng HH, Wei CL. Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell. 2007;1:286–298. doi: 10.1016/j.stem.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 17.Pan G, Tian S, Nie J, Yang C, Ruotti V, Wei H, Jonsdottir GA, Stewart R, Thomson JA. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007;1:299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 18.Martino G, Pluchino S. The therapeutic potential of neural stem cells. Nat Rev Neurosci. 2006;7:395–406. doi: 10.1038/nrn1908. [DOI] [PubMed] [Google Scholar]
- 19.Bjorklund A, Lindvall O. Cell replacement therapies for central nervous system disorders. Nat Neurosci. 2000;3:537–544. doi: 10.1038/75705. [DOI] [PubMed] [Google Scholar]
- 20.Ourednik J, Ourednik V, Lynch WP, Schachner M, Snyder EY. Neural stem cells display an inherent mechanism for rescuing dysfunctional neurons. Nat Biotechnol. 2002;20:1103–1110. doi: 10.1038/nbt750. [DOI] [PubMed] [Google Scholar]
- 21.Kelly S, Bliss TM, Shah AK, Sun GH, Ma M, Foo WC, Masel J, Yenari MA, Weissman IL, Uchida N, Palmer T, Steinberg GK. Transplanted human fetal neural stem cells survive, migrate, and differentiate in ischemic rat cerebral cortex. Proc Natl Acad Sci U S A. 2004;101:11839–11844. doi: 10.1073/pnas.0404474101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang SC, Wernig M, Duncan ID, Brustle O, Thomson JA. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat Biotechnol. 2001;19:1129–1133. doi: 10.1038/nbt1201-1129. [DOI] [PubMed] [Google Scholar]
- 23.Elkabetz Y, Panagiotakos G, Al Shamy G, Socci ND, Tabar V, Studer L. Human ES cell-derived neural rosettes reveal a functionally distinct early neural stem cell stage. Genes Dev. 2008;22:152–165. doi: 10.1101/gad.1616208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koch P, Opitz T, Steinbeck JA, Ladewig J, Brustle O. A rosette-type, self-renewing human ES cell-derived neural stem cell with potential for in vitro instruction and synaptic integration. Proc Natl Acad Sci U S A. 2009;106:3225–3230. doi: 10.1073/pnas.0808387106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Daadi MM, Li Z, Arac A, Grueter BA, Sofilos M, Malenka RC, Wu JC, Steinberg GK. Molecular and magnetic resonance imaging of human embryonic stem cell-derived neural stem cell grafts in ischemic rat brain. Mol Ther. 2009;17:1282–1291. doi: 10.1038/mt.2009.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parsons XH, Teng YD, Moore DA, Snyder EY. Patents on technologies of human tissue and organ regeneration from pluripotent human embryonic stem cells. Recent Patents on Regenerative Medicine. 2011;1:142–163. doi: 10.2174/2210297311101020142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parsons XH, Teng YD, Parsons JF, Snyder EY, Smotrich DB, Moore DA. Efficient derivation of human neuronal progenitors and neurons from pluripotent human embryonic stem cells with small molecule induction. J Vis Exp. 2011:e3273. doi: 10.3791/3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parsons XH, Teng YD, Parsons JF, Snyder EY, Smotrich DB, Moore DA. Efficient derivation of human cardiac precursors and cardiomyocytes from pluripotent human embryonic stem cells with small molecule induction. J Vis Exp. 2011:e3274. doi: 10.3791/3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parsons JF, Smotrich DB, Gonzalez R, Snyder EY, Moore DA, Parsons XH. Defining Conditions for Sustaining Epiblast Pluripotence Enables Direct Induction of Clinically-Suitable Human Myocardial Grafts from Biologics-Free Human Embryonic Stem Cells. J Clin Exp Cardiolog. 2012:S9. doi: 10.4172/2155-9880.s9-001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parsons XH. MicroRNA profiling reveals distinct mechanisms governing cardiac and neural lineage-specification of pluripotent human embryonic stem cells. J Stem Cell Res Ther. 2012;2:124. doi: 10.4172/2157-7633.1000124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parsons XH, Garcia SN, Pillus L, Kadonaga JT. Histone deacetylation by Sir2 generates a transcriptionally repressed nucleoprotein complex. Proc Natl Acad Sci U S A. 2003;100:1609–1614. doi: 10.1073/pnas.0434064100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harikrishnan KN, Chow MZ, Baker EK, Pal S, Bassal S, Brasacchio D, Wang L, Craig JM, Jones PL, Sif S, El-Osta A. Brahma links the SWI/SNF chromatin-remodeling complex with MeCP2-dependent transcriptional silencing. Nat Genet. 2005;37:254–264. doi: 10.1038/ng1516. [DOI] [PubMed] [Google Scholar]
- 33.Vaquero A, Scher M, Erdjument-Bromage H, Tempst P, Serrano L, Reinberg D. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature. 2007;450:440–444. doi: 10.1038/nature06268. [DOI] [PubMed] [Google Scholar]
- 34.Shen F, Hodgson L, Rabinovich A, Pertz O, Hahn K, Price JH. Functional proteometrics for cell migration. Cytometry A. 2006;69:563–572. doi: 10.1002/cyto.a.20283. [DOI] [PubMed] [Google Scholar]
- 35.Sullivan BA, Karpen GH. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Struct Mol Biol. 2004;11:1076–1083. doi: 10.1038/nsmb845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schones DE, Zhao K. Genome-wide approaches to studying chromatin modifications. Nat Rev Genet. 2008;9:179–191. doi: 10.1038/nrg2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Contestabile A. Amyotrophic lateral sclerosis: from research to therapeutic attempts and therapeutic perspectives. Curr Med Chem. 2011;18:5655–5665. doi: 10.2174/092986711798347289. [DOI] [PubMed] [Google Scholar]
- 38.Papadeas ST, Kraig SE, O’Banion C, Lepore AC, Maragakis NJ. Astrocytes carrying the superoxide dismutase 1 (SOD1G93A) mutation induce wild-type motor neuron degeneration in vivo. Proc Natl Acad Sci U S A. 2011;108:17803–17808. doi: 10.1073/pnas.1103141108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haidet-Phillips AM, Hester ME, Miranda CJ, Meyer K, Braun L, Frakes A, Song S, Likhite S, Murtha MJ, Foust KD, Rao M, Eagle A, Kammesheidt A, Christensen A, Mendell JR, Burghes AH, Kaspar BK. Astrocytes from familial and sporadic ALS patients are toxic to motor neurons. Nat Biotechnol. 2011;29:824–828. doi: 10.1038/nbt.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]