SUMMARY
The protocols currently used for protein structure determination by NMR depend on the determination of a large number of upper distance limits for proton-proton pairs. Typically, this task is performed manually by an experienced researcher rather than automatically by using a specific computer program. To assess whether it is indeed possible to generate in a fully automated manner NMR structures adequate for deposition in the Protein Data Bank, we gathered ten experimental datasets with unassigned NOESY peak lists for various proteins of unknown structure, computed structures for each of them using different, fully automatic programs, and compared the results to each other and to the manually solved reference structures that were not available at the time the data were provided. This constitutes a stringent “blind” assessment similar to the CASP and CAPRI initiatives. This study demonstrates the feasibility of routine, fully automated protein structure determination by NMR.
INTRODUCTION
The typical protocol for protein structure determination by NMR spectroscopy involves a number of sequential steps (Wüthrich, 1986). First, the chemical shifts (CS) observed in multidimensional NMR spectra are assigned sequence-specifically to their corresponding protein atoms (the resonance assignment step). Second, thousands of through-space dipolar coupling effects, known as nuclear Overhauser effects (NOEs), are identified in multidimensional NOESY spectra (peak picking), assigned and converted into inter-atomic distance restraints (NOESY assignment step). Additional conformational restraints can result from e.g. measurements of residual dipolar couplings (RDCs), scalar couplings, and CS data. Third, software programs are used to generate a set of protein conformations (called a bundle of conformers) that should satisfy these experimental restraints (structure generation step). The bundle of conformers is often energetically refined through restrained molecular dynamics simulations (structure refinement step). Alternative protocols have been proposed that do not involve the use of distance restraints, thus skipping the NOESY assignment step and exploiting instead RDCs (Hus et al., 2001; Zweckstetter and Bax, 2001) and/or CS data (Cavalli et al., 2007; Shen et al., 2008; Wishart et al., 2008; Raman et al., 2010b).
The NOESY assignment and structure generation steps are performed in an integrated manner over several iterations in order to maximize the number of conformational restraints obtained while guaranteeing the self-consistency of all distance restraints (measured a posteriori from the absence of significant distance restraint violations). Many of the tasks in the NOESY assignment step are repetitive, although non-trivial, yet typically they must be performed by a skilled researcher. A considerable bookkeeping effort is also needed in order to converge to a self-consistent set of conformational restraints from which the final bundle of low pseudo-energy conformers is calculated. For these reasons, and to enhance reproducibility, automation of the aforementioned steps has been actively pursued (Markwick et al., 2008; Güntert, 2009; Guerry and Herrmann, 2011). Protocols aimed at the integration of all steps of the protocol for protein structure determination by NMR have also appeared (López-Mendez and Güntert, 2006).
In 2009 we launched the community-wide initiative called “Critical assessment of Automated Structure Determination of proteins by NMR (CASD-NMR)” (Rosato et al., 2009) (http://www.wenmr.eu/wenmr/casd-nmr), with the aim to assess whether automated methods addressing the NOESY assignment (if needed), structure generation and structure refinement steps can, in a fully automated manner, produce protein structures that closely match the structures manually determined by experts using the same experimental data (“reference structures”). To this end, we have released regularly over one year NMR data sets consisting of assigned chemical shift lists and unassigned NOESY peak lists, while the reference structures determined from the same data were kept “on hold” by the Protein Data Bank (PDB) (Berman et al., 2000), and were thus unavailable to the participants. Each of these data sets is referred to as a “masked” (or “blind”) data set. The protocols used to determine the reference structures are summarized in the Supplemental information, and typically involved manual refinement (such as fixing assignments or removing artifacts) of initial, partly automated NOESY assignments performed with various tools. The final, iteratively obtained lists of resonance assignments and NOESY peak positions were subsequently provided to the CASD initiative.
Here we report the results obtained in the first round of CASD-NMR (CASD-NMR2010) for a total of ten masked data sets, provided by the NIH Protein Structure Initiative, for monomeric proteins of 60 to 150 amino acids. All the input data as well as the structures generated in this study can be freely downloaded from http://www.wenmr.eu/wenmr/casd-nmr. CASD-NMR2010 did not address automation methods for determining resonance assignments and for NOESY peak picking. We chose to postpone the assessment of these parts of the process until the NOE assignment and structure calculation steps will have been demonstrated to be truly robust.
The present results demonstrate that routine application of NMR structure calculation methods integrating NOE cross peak assignment and structure generation is both feasible and reliable. Furthermore, the recently developed approaches based on the use of NMR chemical shift data to generate structural models were found to benefit significantly when supplemented with information from unassigned NOESY peak lists.
RESULTS
Accuracy and convergence of structure calculations
CASD-NMR2010 involved three groups of automated methods (Table 1): those using NOESY data to obtain distance restraints for structure calculations (CYANA (Herrmann et al., 2002a), UNIO (Herrmann et al., 2002b), ASDP (Huang et al., 2006) and ARIA (Rieping et al., 2007)), those using chemical shift data augmented by NOESY data (CS-DP-Rosetta (Raman et al., 2010a), which uses NOESY information to re-rank its CS-based results, and Cheshire-YAPP, which uses CS-generated structures to perform NOESY assignments and extract distance restraints), and those relying exclusively on CS data as experimental information (Cheshire (Cavalli et al., 2007) and CS-Rosetta (Shen et al., 2008)). The NOESY-based methods include a structure refinement step after structure generation with the aforementioned programs. Both steps exploit all automatically assigned restraints. A variety of programs has been used for the refinement (also in the case of the reference structures).
Table 1. Features of the programs used in CASD-NMR2010.
Y indicates this type of information is directly used in structure calculations, s indicates that it is used as a support to derive additional restraints and/or to improve scoring. Details are given in the Methods section.
| Software | NOEs | Chemical shifts* |
Comments |
|---|---|---|---|
| CYANA | Y | s | Includes torsion angle restraints generated on the basis of the chemical shift values |
| UNIO | Y | s | Includes torsion angle restraints generated on the basis of the chemical shift values |
| ARIA | Y | s | Includes torsion angle restraints generated on the basis of the chemical shift values |
| ASDP | Y | s | Includes torsion angle restraints generated on the basis of the chemical shift values; uses the DP-score measure (Huang et al., 2005) to re-rank the structural models |
| Cheshire-Yapp | s | Y | Uses structural models initially generated using only CS data to assign NOEs, derive distance restraints and refine the best-scoring initial 100 models |
| CS-DP-Rosetta | s | Y | Uses the unassigned NOESY peak lists and the DP-score measure (Huang et al., 2005) to re-rank the structural models |
| Cheshire | Y | ||
| CS-Rosetta | Y |
Used as direct structural restraints, rather than to derive secondary structure information or torsion angle restraints
For each data set, we used the deviation of the backbone coordinates (RMSD) to quantify the degree of convergence (i.e. the similarity) among the automatically generated structures as well as their closeness to the reference structure determined under manual supervision. Assuming that the reference structure is correct, the RMSD to it becomes a measure of accuracy. We computed the RMSD to the reference for the structures generated by all the methods (Tables 1 and 2, Figure 1A). As the RMSD calculations require the a priori definition of residue ranges to be superimposed, a consensus RMSD range comprising the well-ordered residues in the reference structure was chosen for each dataset (Supplemental Table S1). In order to avoid a possible bias from this selection when evaluating the similarity to the reference structure, we computed also the Global Distance Test Total Score (GDT_TS, Figure 1B), which does not require residue ranges to be predefined and is independent of protein size. The GDT_TS score has been developed in the frame of the Local-Global alignment method (Zemla, 2003) for structure comparison and has been extensively used in CASP assessments (Clarke et al., 2007). It is defined by GDT_TS = (P1 + P2 + P4 + P8)/4, where Pd is the percentage of residues that can be superimposed under a distance cutoff of d Å. This definition reduces the dependence on the choice of the distance cutoff by averaging over four different distance cutoff values. GDT_TS and backbone RMSD to the reference are anticorrelated, i.e. high structural similarity corresponds to low RMSD and high GDT_TS values. Another structure similarity score that equally does not require the definition of residue ranges, the TM-Score (Zhang and Skolnick, 2004), was found to be strongly linearly correlated with GDT_TS for our datasets (not shown).
Table 2. Targets for CASD-NMR and overview of the accuracy of the various approaches.
The targets, defined by NESG target id’s, are ordered by the time of release, from the oldest (VpR247) to the most recent (CtR69A). See also Supplemental Tables S1 and S3−S4.
| Target name |
PDB Code |
Sequen ce length |
Average pairwise RMSD within the reference (Å)* |
Backbone RMSD (Å) *,+ / GDT_TS * score (%) to the reference structure | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CYANA | UNIO | ARIA | ASDP | Cheshire- Yapp |
CS-DP- Rosetta |
Cheshire | CS- Rosetta |
||||
| VpR247 | 2KIF | 106 | 0.7 | 0.8 / 91 | 0.9 / 92 | 2.7 / 71§ | 1.8 / 81 | n.a. | 1.4 / 78 | 1.7 / 78 | 14.6 / 43 |
| AR3436A | 2KJ6 | 97 | 1.4 | 2.0 / 65 | 2.2 / 61 | n.a. | 1.4 / 66 | n.a. | 3.3 / 55 | 4.5 / 56 | 3.3 / 47 |
| HR5537A | 2KK1 | 135 | 1.0 | 1.3 / 89 | 1.6 / 83 | 2.4 / 76§ | 1.7 / 84 | n.a. | 1.6 / 86 | 2.1 / 77 | 2.2 / 76 |
|
ET109A (reduced) |
2KKX | 102 | 0.6 | 1.2 / 90 | 1.7 / 85 | 1.5 / 87§ | 1.4 / 90 | 1.5 / 86 | 2.0 / 82 | n.a. | 4.2 / 58 |
|
ET109A (oxidized) |
2KKY | 102 | 0.6 | 0.9/ 92 | 1.1 / 90 | 1.2 / 89§ | 1.0 / 91 | n.a. | 1.6 / 84 | n.a. | 14.3 / 30 |
| AtT13 | 2KNR | 121 | 0.6 | 1.9 / 85 | 1.7 / 91 | 2.5 / 84§ | 2.1 / 84 | n.a. | 6.8 / 65 | n.a. | 11.2 / 32 |
| PgR122A | 2KMM | 73 | 0.7 | 1.1 / 85 | 1.0 / 87 | 1.6 / 74§ | 1.0 / 86 | n.a. | 0.9 / 88 | 1.1 / 87 | 1.3 / 83 |
| NeR103A | 2KPM | 105 | 1.7 | 1.0 / 86 | 0.9 / 89 | 1.0 / 86# | 1.6 / 80 | 1.5 / 78 | 1.4 / 81 | n.a. | 2.8 / 62 |
| CgR26A | 2KPT | 148 | 1.6 | 0.8 / 94 | 0.8 / 94 | 0.5 / 87# | 1.0 / 93 | 0.8 / 97 | 2.6 / 78 | n.a. | 4.0 / 62 |
| CtR69A | 2KRU | 63 | 0.4 | 0.6 / 92 | 0.9 / 86 | 0.6 / 90# | 0.7 / 89 | n.a. | 0.6 / 90 | 1.2 / 79 | 1.0 / 83 |
| Number of submitted targets | 10 | 10 | 9 | 10 | 3 | 10 | 5 | 10 | |||
|
Number of successful targets (RMSD ≤ 2.0 Å or GDT_TS ≥ 80%) |
10 | 9 | 7 | 10 | 3 | 7 | 3 | 2 | |||
For the backbone atoms of ordered residues, as defined by PSVS using dihedral angle order parameters(Bhattacharya et al., 2007) (Supplemental Table S1)
Backbone RMSD between the average conformer of each structure and the average conformer of the reference structure
Determined with the ARIA-Soft protocol
Determined with the ARIA-BayW protocol
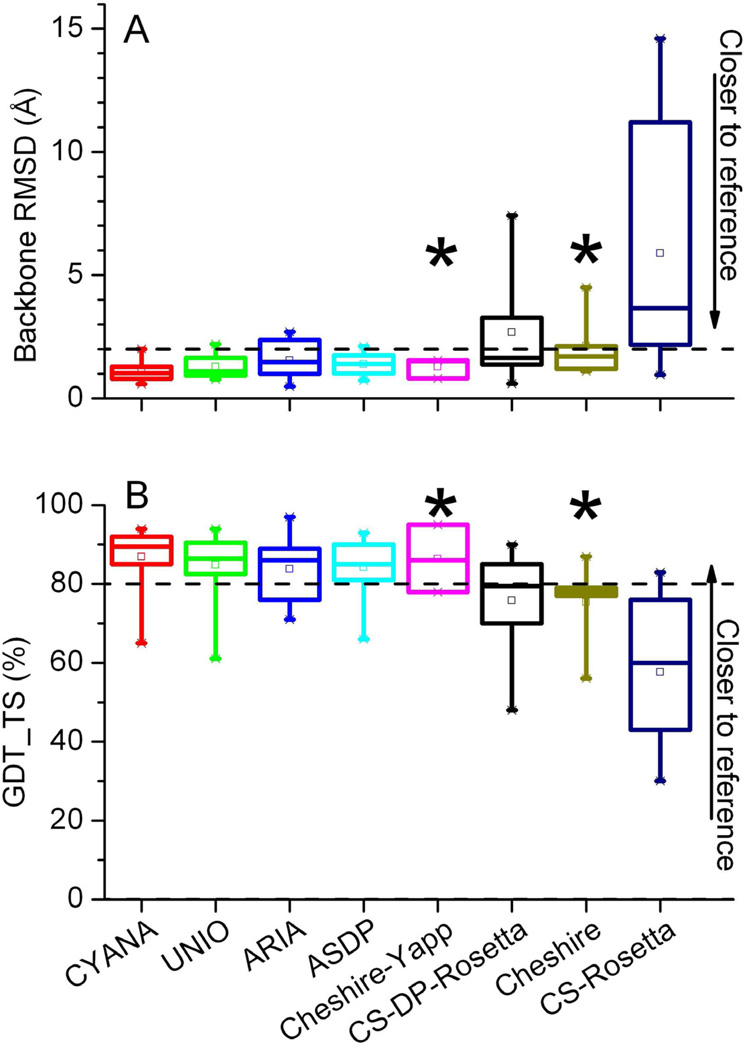
Figure 1. Structural similarity between reference and CASD-NMR2010 structures.
RMSD (A) and GDT_TS score (B) deviation of the backbone coordinates (for ordered residues only; see Supplemental Table S1) with respect to the reference structure for the various algorithms. GDT_TS is the average fraction of residues that can be superimposed to within four different distance cutoffs (1, 2, 4 and 8 Å) and ranges between 0 and 100%. For each structure, the automatically generated average conformer has been used for calculations. The dashed lines are at 2 Å for RMSD and at 80% of superimposable residues for GDT_TS, corresponding to our thresholds for acceptable performance. See also Supplemental Table S2.
The box parameters are as follows: the box range goes from the first to the third quartile; box whiskers identify the minimum and maximum values; the square within the box identifies the mean; the thick line in the box identifies the median.
The starred boxes correspond to algorithms for which less than 60% of the targets were submitted.
The backbone RMSD values to the reference for the structures generated by NOESY restraint-based methods were in the range 0.6–2.7 Å whereas the range for GDT_TS scores were 61–94% (Table 2 and Supplemental Table S2). Setting thresholds for an acceptable structural accuracy (here assumed to be quantified by similarity to the reference structure) at an RMSD from the reference structure ≤ 2 Å (Nederveen et al., 2005; Andrec et al., 2007) and GDT_TS ≥ 80% (Clarke et al., 2007), three of the four NOESY-based programs (CYANA, UNIO and ASDP) automatically and consistently generated acceptable structures, based on one (90–100% of the instances) or simultaneously both (80–90% of the instances) parameters (Table 2). The RMSD was always ≤ 2.2 Å, whereas the lowest GDT_TS was 61% (78% upon exclusion of target AR3436). The fourth program, ARIA, performed acceptably for nearly 80% of the targets, with the best results obtained with a recently developed logharmonic potential combined with a Bayesian determination of restraint weights (protocol ARIA-BayW) (Bernard et al., 2011), which produced structures with excellent GDT_TS and RMSD values for the three most recent targets.
Regarding CS-based methods augmented with NOESY data, Cheshire-YAPP, which was developed during CASD-NMR2010 and run on three randomly selected targets, featured a similarity to the corresponding reference structures in-line with NOESY restraint-driven methods. Cheshire-YAPP uses initial (pure CS) Cheshire models to assign NOESY distance restraints used to refine the models. For CS-DP-Rosetta, which uses NOESY information only to re-rank the CS-based models, the deviation from the manual reference structures was close to that of the NOESY restraint methods, with a range of RMSD and GDT_TS values of, respectively, 0.3–3.3 Å and 55–90% and 70% of targets falling within the thresholds described above. Finally, pure CS-based methods had the poorest performance in terms of closeness to the reference structures, as it is apparent from Table 2 and Figure 1. Note that the poorer appearance of the CS-Rosetta server, which was run via the web server developed in the e-NMR project (Bonvin et al., 2010), is partly due to inclusion of non-converged solutions in the comparison. It can be concluded that NOESY-based methods delivered more consistent and robust performances than CS-based methods (resulting in smaller boxes in Fig. 1A–B), yielding structures on average closer to the reference. NOESY-filtering as in CS-DP-Rosetta could recover some but not all of the consistency and reliability of the restraint-driven methods (see also below). Notably, the CS-methods (regardless of whether augmented with NOESY information) are computationally much more demanding than NOESY-based methods.
Regarding individual targets, the one with the lowest performance across all methods was AR3436A (Table 2), a 97-amino acid protein. Our target selection included three proteins with more than 100 residues (HR5536A, AtT13 and CgR26A), for all of which NOESY-based methods were able to automatically generate accurate structures. Instead, purely CS-based methods failed for all of them, whereas CS-based methods augmented with NOESY data were successful in nearly all cases.
All the results examined in the preceding paragraphs address the degree of similarity to the manually solved reference structure. Additional insight can be obtained by the evaluation of the degree of convergence among the different programs. This has been measured as the mean RMSD among the average conformers obtained with the automatically generated methods (Supplemental Table S3). For the NOESY-based algorithms, the mean RMSD for each target was in the range 0.9–3.0 Å, with four targets featuring a mean RMSD lower than 1.0 Å and eight targets being within 2.0 Å. If CS-based methods augmented with NOE cross peak information are also included, the mean RMSD range widens slightly up to 3.3 Å, still with eight targets having a mean RMSD lower than the 2.0 Å threshold. Instead, inclusion of all methods yielded values as large as 6.2 Å (Supplemental Table S3). The present evaluation of convergence is much more stringent than the standard re-calculation with different random number seeds, because in each calculation the NOE assignments have been determined independently and with different methods.
A further measure of accuracy would be the comparison with a completely independent structure determination. This is at present possible for only two targets (VpR247 and PgR122A), for which the PDB contains X-ray structures of relatively close homologues (40–50% sequence identity). These allowed us to build reliable structural models that can be used as the structural reference for comparisons (Supplemental Table S4). For PgR122A, the relevant structure is 3HVZ (Forouhar et al., 2009). The homology model of PgR122A built on this structure shows a backbone RMSD of 0.77 Å to the average coordinates of the reference structure. All methods yielded structures within 1.5 Å from the homology model, with the majority being actually within 1 Å. For VpR247 there are several related crystal structures of the S. pombe homologue, in the free or ligand-bound form. The model built on the DNA-complexed protein (3GX4 (Tubbs et al., 2009)) is closer to the reference VpR247 structure than the model built on the free protein (3GVA), with backbone RMSD values of 1.4 Å and 2.1 Å, respectively. Similarly, nearly all the automatically generated structures are more similar to the former than the latter model. With the exception of the ARIA and CS-Rosetta server structures (Supplemental Table S4), all structures are within 2.0 Å from the 3GX4-based model, whereas they are in the range 1.7–2.2 Å from the 3GVA model. These results may suggest that the free VpR247 protein in solution populates a different conformational state than its S. pombe homologue in the crystal structure. This state would be relatively similar to the DNA-bound conformation.
Geometric and stereochemical quality
The geometric and stereochemical quality is another important property of a structure that must be checked prior to deposition in the PDB. We evaluated this aspect using the PSVS (Bhattacharya et al., 2007) (http://psvs-1_4-dev.nesg.org/) and CING (http://nmr.cmbi.ru.nl/cing/) validation suites (Supplemental Table S2), which assess several quality measures. The Verify3D (Eisenberg et al., 1997) and ProsaII (Sippl, 1993) scores, which evaluate the global fold likelihood, were not significantly different for the CASD-NMR or the reference structures and featured relatively wide ranges for all the algorithms. Instead, the Procheck-all (Laskowski et al., 1996) score, which assesses the distribution of all the protein dihedral angles, and the MolProbity clashscore (Davis et al., 2007), which assesses the occurrence of high-energy interatomic contacts, differed among the CASD-NMR structures, even though their ranges over all targets overlapped with the reference structures (Fig. 2). The ranges of Procheck-all values for the structures generated by the Rosetta-based algorithms are narrow and on average significantly better than for the other structures (Figure 2B). Also the MolProbity clashscore tends to be better for the Rosetta-based structures (Figure 2A). Given the fact that the latter structures tend to be the most dissimilar from the reference, it appears that the geometric and stereochemical quality of the structures is not a good indicator of their accuracy, as defined above (Fig. 2 and Table 2). The geometric and stereochemical quality of the structures is largely determined by the algorithm and the force field used in the structure refinement step. This can be appreciated also by comparing the scores of the various NOESY-based results, which can vary appreciably even for structures closely similar to the reference. The importance of force fields is due partly to the fact that NMR data cannot define parameters such as bond lengths or bond angles, which however are often restrained also during X-ray structure determinations. Studies affording a deeper understanding of the effects of structure refinement as a function of the quantity and quality of the NMR data available would be quite useful. Nonetheless, it can be stated that accurate structures should satisfy both stereochemical requirements and the available experimental information.
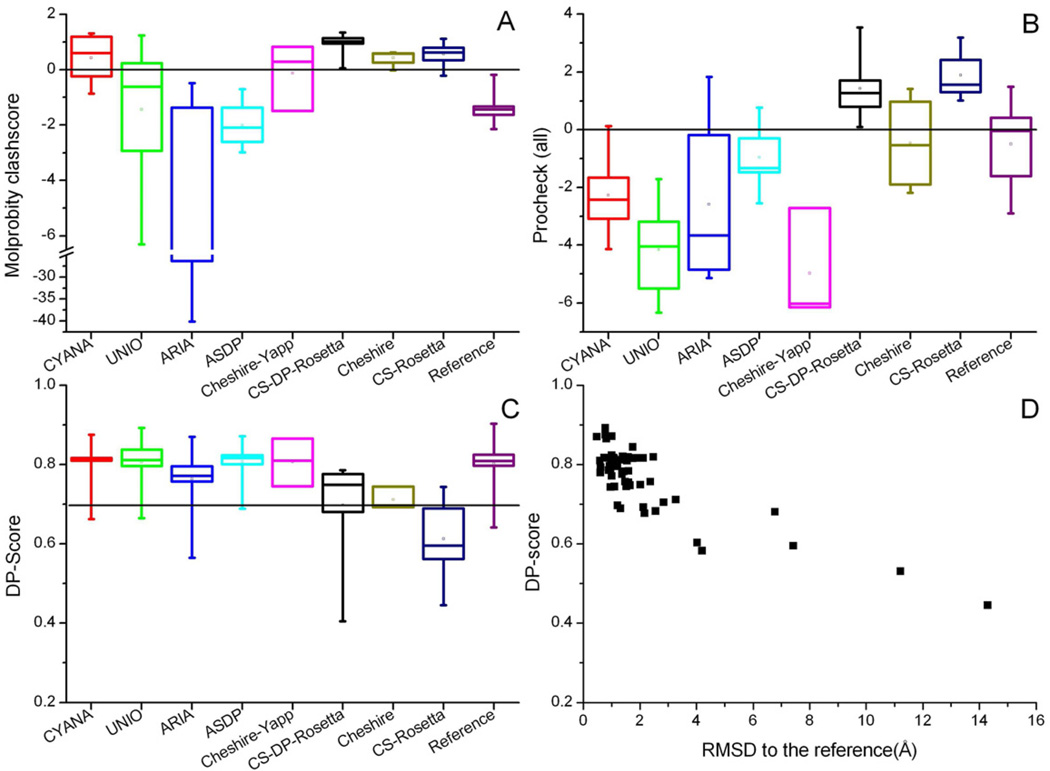
Figure 2. Quality of CASD-NMR2010 structures.
Molprobity (A) and Procheck-all (B) Z-score values describe the distribution of, respectively, all protein dihedral angles and high energy interatomic contacts for the automatically generated and the reference structures. The Z-score is the deviation of the value calculated for a given structure from the average calculated for a set of 150 high-resolution X-ray structures (Bhattacharya et al., 2007), expressed in units of the standard deviation. A positive Z-score indicates that the corresponding structure quality score is better than the average, whereas a negative value indicates that the structure analyzed is worse than the average. DP-Scores (C) describe the agreement between the structures and the unassigned NOESY peak lists, and range from 0 (worst) to 1 (best) (Huang et al., 2005). The dashed line corresponds to the 0.7 threshold described in the main text. The box parameters are as in Figure 1. Panel (D) reports DP-scores as a function of the backbone RMSD to the reference structure, for all CASD-NMR2010 structures. See also Supplemental Figure S1 and Supplemental Tables S5 and S6.
Quality measured by agreement with the data
A different kind of structure validation assesses the completeness of experimental data and its agreement with the structure. Because it is difficult to compare structures directly to the raw experimental NMR data, these analyses were performed with respect to partially interpreted experimental data, e.g. after peak picking and CS assignment. The DP-score (Figure 2C) is a measure of the goodness-of-fit of the unassigned NOESY peak lists to a structure, ranging from 0 to 1 (Huang et al., 2005). This data-based quality measure featured a significant correlation to structure accuracy (Figure 2D; Supplemental Table S5). A DP-score cutoff of ≥0.7 allowed the identification of acceptable CASD-NMR structures with a reliability of 94% (Supplemental Table S6), based on the available refined peak lists. On the other hand, all structures with an RMSD to the reference larger than 3.0 Å or a GDT_TS score lower than 60% had DP-scores lower than 0.6, except for a single CS-DP-Rosetta structure. For comparison, the DP-score values for the reference structures were in the 0.64–0.90 range. It is important to note that the 0.7 DP-score threshold value was determined using refined peak lists, which might facilitate the discrimination, e.g. by reducing the number of artifact peaks that cannot be accounted for. If automatically peak-picked NOESY lists, which potentially contain a significant amount of artifacts that however cannot be excluded at the outset of a NMR structure determination, were used, presumably the DP-score threshold would be shifted toward lower values. It is interesting to observe that for the AR3436A target, which was previously mentioned as the one for which we observed the poorest overall performance, the average DP-score was as low as 0.60; for the other targets the range of average DP-scores was 0.72–0.81.
The different approaches extracted significantly varying numbers of NOESY-based distance restraints for a given target. The information contained within these restraint sets, as determined by the QUEEN procedure (Nabuurs et al., 2003), is highly variable and, after excluding outliers, did not correlate significantly with the RMSD between automatically generated and reference structures nor with the DP-score (Supplemental Table S2). The reference structures spanned a range of information content essentially as wide as that of all the NOESY-based structures. On average, automatically generated structures had a non-significant tendency towards higher information content than the reference structures. Nevertheless, even structures with information content as low as about 0.1 bits/atom are found within 2.0 Å RMSD from the reference structure; the occurrence of these very low information content values is due to loose (5.5 Å) upper bounds constituting >90% of the restraints.
DISCUSSION
In summary, the CASD-NMR 2010 initiative has successfully proven, without the possible bias inherent in test calculations of targets with previously known structure, that, given almost complete CS assignments, the automated calculation of NMR structures of small proteins from “clean”, unassigned NOESY peak lists is routinely feasible. NOESY-based methods yield structures that are typically within 2.0 Å of the corresponding manually solved structures and within 2.5 Å in all but one of the 49 cases reported here. This conclusion is also supported by the good convergence of these algorithms, which is within 3.0 Å for all targets and within 2.0 Å for eight targets out of ten. Comparison with the crystal structures of homologous proteins, limited to the Pgr122A and VpR247 targets, provided similar conclusions.
Another notable result of the present investigation is that whereas the performance of methods for NMR structure determination based only on CS data is not yet fully reliable, augmenting these methods with different schemes to exploit unassigned (refined) NOESY peak lists recovers to a significant extent the robustness of the NOESY-based methods, as judged both by similarity to the manually solved structures and by looking at the convergence of the various methods. For the size range addressed by our target selection (up to 150 amino acids), the protein size does not impact significantly on the success rate of the approaches that include NOESY data.
On average, the automatically generated and the reference structures are of comparable geometric and stereochemical quality. These quality measures do not correlate with the similarity to the reference structure, as measured by either the backbone RMSD or the GDT_TS score. Indeed, even structures with a significantly wrong fold can feature excellent geometric and stereochemical quality measures. Our findings thus reinforce previous indications that the structure refinement protocol is a major determinant of these parameters (Nabuurs et al., 2006; Saccenti and Rosato, 2008). The use of an indicator, the DP-score, quantifying the agreement between the structures and the unassigned NOESY data was useful to discriminate good or problematic structures. The DP-score featured a good correlation with both the backbone RMSD and the GDT_TS score; with the present refined peak lists, a DP-score threshold of 0.7 could be applied to identify accurate structures with a 94% precision. Conversely, all structures further than 3.0 Å from the reference had a DP-score lower than 0.6. For the AR3436A target the automated methods obtained the lowest accuracy (Table 2) and the poorest convergence (Supplemental Table S3). AR3436A is also the target with the lowest DP-score for the reference structure as well as on average over all CASD-NMR2010 structures. It is possible that the available data did not permit capturing some features of the protein, e.g. related to its dynamics.
For a given target, the various automated NOESY-based methods could yield varying levels of NOESY assignments and, consequently, quite different numbers of structural restraints. Interestingly, this factor did not correlate appreciably with the DP-score (which refers to the unassigned lists) of the calculated structure nor with its geometric and stereochemical quality, as mentioned above. Overall, we can thus conclude that indicators of agreement with non-interpreted experimental data are useful to validate NMR structures. Geometric and stereochemical parameters are not sufficient to guarantee accuracy; nevertheless they should be taken into account as necessary features of high-quality protein structures; i.e. good structures should have both good agreement with non-interpreted experimental data (e.g. DP-score) and good geometric and stereochemical parameters.
The automated structure calculations addressed in this contribution are non-supervised, with the exclusively NOESY-based methods being typically fast (with calculation times on a single CPU of the order of hours, including refinement) and routine and CS-based methods being relatively CPU-intensive (with estimated calculation times on a single CPU of the order of 103–104 hours, making it mandatory to employ large clusters or distributed computing for these calculations) and less dependable. A fair criticism to the setup of CASD-NMR2010 is that the NOESY peak lists provided had been refined against initial structural models during the determination of the reference structure and were therefore almost devoid of artifacts. This simplifies the task for NOESY-based approaches and for CS-methods augmented by NOESY data. However, considering their highly satisfactory performance observed here, the peak list refinement may not be necessary if the quality of the NOESY spectra and the completeness of the chemical shift assignments are high. To investigate this, we have initiated a second round of CASD-NMR using new masked NOESY data sets that have been generated using exclusively automated peak-picking procedures. This second round will further consolidate the methodological improvements fostered by the 2010 round.
EXPERIMENTAL PROCEDURES
Data distribution
Masked data sets for CASD-NMR 2010 (whose amino acidic sequences are given in Supplemental Table S1) comprised chemical shift assignments in BMRB format and unassigned NOESY peak lists in SPARKY and/or XEASY/CARA format. The data were made available both via the CASD-NMR website (www.wenmr.eu/wenmr/casd-nmr) and a dedicated page at the Protein Structure Initiative (PSI Knowledge Base (http://kb.psi-structuralgenomics.org/). For two targets raw NOESY spectra were also made available. At the time of release, all participants were notified of the availability of a new data set as well as of the date of release of the corresponding structure from the PDB (about eight weeks later). The automatically calculated structures and all restraints were deposited directly by the participants into a password-protected database again via the CASD-NMR website.
Residual dipolar coupling data and hydrogen bond restraints were not used in the CASD-NMR 2010 project.
Calculation Protocols
Each method developer team carried out calculations with their own program, as detailed below.
CYANA
Structure calculations by the CYANA method (Güntert, 2009) used as input data from the blind data sets the protein sequence, the list of assigned chemical shifts, and the unassigned NOESY peak lists. Torsion angle restraints were generated on the basis of the chemical shift values with the program TALOS+ (Shen et al., 2009) for the backbone torsion angles ϕ and ψ of non-proline residues with a prediction classified as “Good” by TALOS+. The torsion angle restraints were centered at the predicted average value and their full width was set to four times the predicted standard deviation or 20°, whichever was larger. Th e program CYANA was used for seven cycles of combined automated NOE assignment(Herrmann et al., 2002a) and structure calculation by torsion angle dynamics (TAD) (Güntert et al., 1997). The tolerance for the matching of chemical shifts and NOESY peak positions was set to 0.03 ppm for 1H and 0.5 ppm for 13C and 15N. Peak intensities were converted into upper distance bounds according to a 1/r6-relationship. The standard CYANA simulated annealing schedule was applied to 100 randomly generated conformers with 15000 TAD steps. NOE distance restraints involving 1H atoms with degenerate chemical shifts, e.g., methyl groups, were treated as ambiguous distance restraints using 1/r6-summation over the distances to the individual 1H atoms. Non-stereospecifically assigned methyls and methylene protons were treated by automatic swapping of restraints between diastereotopic partners (Folmer et al., 1997) during the seven cycles of automated NOE assignment and by pseudoatom correction and symmetrization (Güntert et al., 1991; Güntert, 1998) for the final structure calculation. The 20 conformers with the lowest final CYANA target function values were embedded in an 8 Å shell of explicit water molecules and subjected to restrained energy refinement using the program OPALp (Koradi et al., 2000; Luginbühl et al., 1996). A maximum of 3000 steps of restrained conjugate gradient minimization were applied, using the standard AMBER force field (Ponder and Case, 2003) and pseudo-potentials proportional to the sixth power of the NOE upper distance bound violations and the square of the torsion angle restraint violations, respectively. The entire procedure was driven by the program CYANA, which was also used for parallelization of all time-consuming steps on 10–100 processors of a Linux cluster system with Intel quad-core 2.4 GHz processors.
UNIO
For all blind data sets NOE assignment were performed using the modules ATNOS/CANDID and/or the CANDID module alone incorporated into the software UNIO (Herrmann et al., 2002a; Herrmann et al., 2002b), depending if NOE peak lists or NOESY spectra were provided for a given CASD-target. The standard UNIO protocol with seven cycles of peak picking with ATNOS, if NOESY spectra were provided, and NOE assignment with CANDID was used. During the first six UNIO-ATNOS/CANDID cycles, ambiguous distance restraints were used (Nilges, 1997). At the outset of the spectral analysis, UNIO-ATNOS/CANDID used highly permissive criteria to identify and assign a comprehensive set of peaks in the NOESY spectra or the unassigned peak lists provided. Only the knowledge of the covalent polypeptide structure and the chemical shifts were exploited to guide NOE cross peak identification and NOE assignment. In the second and subsequent cycles, the intermediate protein 3D structures were used as an additional guide for the interpretation of the NOESY spectra or unassigned peak lists. The output in each ATNOS/CANDID cycle consisted of assigned NOE peak lists for each input spectrum and a final set of meaningful upper limit distance restraints which constituted the input for the TAD algorithm of CYANA for structure calculation (Güntert et al., 1997). In addition, torsion angle restraints for the backbone dihedral angles ϕ and ψ derived from Cα chemical shifts were automatically generated in UNIO and added to the input for each cycle of structure calculation (Spera and Bax, 1991; Luginbühl et al., 1995). For the final structure calculation in cycle 7, only distance restraints that could be unambiguously assigned based on the protein 3D structure from cycle 6 were retained.
The 20 conformers with the lowest residual CYANA target function values obtained from cycle 7 were energy-refined in a water shell with the program OPALp (Koradi et al., 2000; Luginbühl et al., 1996) using the AMBER force field (Ponder and Case, 2003).
ASDP
13C chemical shift was first referenced based on the LACS method(Wang et al., 2005). AutoStructure’s topology-constrained distance network algorithm (Huang et al., 2006) was used to assign NOE peaks, using the list of resonance assignments, and the unassigned NOESY peak lists. The tolerance to match chemical shifts with NOE peak positions was set to 0.05 ppm for 1H and 0.5 ppm for 13C and 15N. Distance constraints were generated based on these NOE assignments. Dihedral angle constraints were generated using TALOS+ (Shen et al., 2009), using only sites with TALOS+ scores = 10 and constraining the dihedrals to the defined range ± 20° or twice the standard deviation, whichever was larger. One hundred structures were generated using CYANA standard structural calculation module (Güntert et al., 1997) and DP-scores (Huang et al., 2005) were calculated for all 100 structures. We then computed a new score: (target function/100)-DP for each model, and the 20 models with highest scores were selected for additional iterative 5 cycles of NOE analysis with AutoStructure and structure generation with CYANA (Güntert et al., 1997). After six cycles of ASDP analysis, the resulting structures were energy-refined using CNS (Brunger, 2007) with explicit water. If any TALOS+ dihedral angle constraints were observed to be violated in all 20 models, they were removed and the ASDP / CNS refinement process was repeated.
ARIA
Two protocols were used: one (ARIA-Soft) based on the standard soft-square distance restraint potential, the other (ARIA-BayW) based on a log-harmonic potential shape (Rieping et al., 2005) and iterative determination of the optimal data weight (Habeck et al., 2006; Nilges et al., 2008). ARIA 2.2 (Rieping et al., 2007) was used with the ARIA-Soft protocol, and ARIA 2.3 with the more recent ARIA-BayW protocol. ARIA-Soft was applied to targets VpR247, HR5537A, ET109A, AtT13, PgR122A, whereas ARIA-BayW was applied to targets NeR103A, CgR26A and CtR69A. Dihedral angle restraints were generated from chemical shifts with the program TALOS+ (Shen et al., 2009) for the backbone torsion angles ϕ and ψ. The predictions classified as “good” by TALOS+ were converted into restraints with the script talos2xplor.tcl. For analyzing NOESY crosspeaks, the tolerance for matching chemical shifts and peak positions was set to 0.04 and 0.02 ppm for indirect and direct 1H dimensions and to 0.5 ppm for 13C and 15N.
For each calculation, we ran eight ARIA iterations in a simplified, geometric force field, and one refinement iteration in water with full electrostatics (Linge et al., 2003). Structures were calculated with CNS (Brunger, 2007), recompiled with specific ARIA subroutines. The standard four-phase ARIA simulated annealing protocol was used, with 2200 TAD steps at 20000 K, 2200 TAD steps cooling from 20000K down to 0K, 10000 Cartesian cooling steps from 2000K to 1000K, and 8000 cooling steps from 1000K to 50K. Molecular dynamics was followed by 200 steps of conjugate gradient minimization. For the water refinement, we used heating from 100 to 500 K in steps of 100 K with 750 dynamics steps at each temperature, during which positional restraints on the heavy atom positions were progressively relaxed; 2000 steps of refinement at 500K; cooling to 25K in steps of 25K, with 1000 integration steps at each temperature, followed by 200 steps conjugate gradient minimization. The log-harmonic potential and the Bayesian weight determination were only used in the final cooling phase, minimization and water refinement. 50 conformers were randomly generated and annealed; the 15 conformers with lowest (extended) hybrid energy were analyzed to refine the restraint list. After the eighth iteration, the 10 conformers with the lowest energy were refined in water.
CHESHIRE
In the structure calculations two protocols were used, CHESHIRE and CHESHIRE-YAPP. CHESHIRE uses only chemical shifts, while CHESHIRE-YAPP uses a combination of chemical shifts and unassigned NOESY peak lists.
CHESHIRE consists of a three-phase computational procedure (Cavalli et al., 2007). In the first phase, the chemical shifts and the intrinsic secondary structure propensities of amino acid triplets are used to predict the protein secondary structure. In the second phase, the secondary structure predictions and the chemical shifts are used to predict backbone torsion angles. These angles are screened against a database to create a library of trial conformations of three and nine residue fragments spanning the sequence of the protein. In the third phase, a molecular fragment replacement strategy is used to assemble low-resolution structural models. The information provided by chemical shifts is used in this phase to guide the assembly of the fragments. The resulting structures are refined with a hybrid molecular dynamics and Monte Carlo conformational search using a scoring function defined by: (1) the agreement between experimental and calculated chemical shifts, and (2) the energy of a molecular mechanics force field. This scoring function ensures that a structure is associated with a low CHESHIRE score only if it has a low value of the molecular mechanics energy and is highly consistent with experimental chemical shifts. Typically 50,000 structures were generated for each target and the best scoring one was submitted. This protocol was used for five targets (VpR247, AR3436A, HR5537A, PGR122A and CtR69A).
The CHESHIRE-YAPP protocol uses the best scoring 500–1000 high-resolution structures generated by CHESHIRE to select compatible NOEs from the unassigned NOESY peak lists. NOEs are selected using an iterative protocol. In the first step, atoms are assigned to each spectral dimension using a chemical shift tolerance of 0.03 ppm for 1H and 0.3 ppm for 13C and 15N. Then, chemical shift-based assignments that are violated by more than 2Å in 50 or more of the best 500 CHESHIRE structures are removed. The remaining restraints are used to refine the best scoring 100 CHESHIRE structures. The last two steps are repeated 4 times with a threshold for violations of 1.5, 1.0, 0.5 and 0.2Å. This protocol was used for three targets (ET109A, NeR103A and CGR103A).
CS-DP-ROSETTA
Fragments were picked using the original CS-Rosetta fragment picker (Shen et al., 2008). Decoys were generated on Rosetta@home using 50,000 boinc work units (ca. 200,000 CPU hours). This resulted in 105–106 decoys, depending on the target. Decoys were generated with the standard CS-Rosetta protocol(Shen et al., 2008) and relaxed in full-atom resolution, as described by Raman et al.(Raman et al., 2010b). The best 1000 decoys were selected by score and their DP-score was calculated with AutoStructure (version 2.2.1)(Huang et al., 2005). To finally rank the models, we computed the final score S = R + 1000(1-DP)(Raman et al., 2010a), with R for the Rosetta full-atom score and DP for the DP-score, and selected the 10–20 best models for submission to the CASD website.
CS-ROSETTA (Web Server)
The CS-Rosetta webserver developed under the eNMR project (Bonvin et al., 2010) was used. Firstly, the supplied NMR chemical shift data were pre-checked on chemical shift referencing and possible errors, using the standard pre-check option of the TALOS+ program (Shen et al., 2009). TALOS+ was then used to identify flexible residues at the termini of the protein (those classified as either “Dynamic” or “Not classified” by TALOS+). These and any histidine tags were removed. The resulting cleaned TALOS+ file was submitted to the server. For each target 50000 models were generated on the Grid following the standard CS-ROSETTA protocol (Shen et al., 2008) using the original CS-Rosetta fragment picker and Rosetta version 2.3.0. The 1000 best ROSETTA score models were rescored using chemical shift rescoring as in the CS-ROSETTA protocol. After rescoring, if convergence was observed in the top five models (backbone RMSD below 2Å), these were submitted as prediction for CASD-NMR, otherwise only the top scoring model was submitted. For the last two targets, we implemented a novel smoothing procedure on the Rosetta raw score: for each model, a smoothed score was calculated as a Gaussian-weighted average score calculated over all structural neighbors within a 4.5Å Cα RMSD cutoff. The smoothing was performed on the top 5000 models. The top 1000 models after smoothing were then rescored using the regular CS-scoring in CS-ROSETTA. This smoothing procedure removes some of the noise in the raw score and strengthens any weak correlation that might be present in the data set.
Highlights.
Automated assignment and structure calculation from NMR NOESY spectra were assessed
Routine, fully automated determination of protein structures is feasible
Good stereochemical and geometric quality alone does not indicate structure accuracy
Supplementary Material
Acknowledgements
Financial support to CASD-NMR was provided by the European Community FP7 e-Infrastructure “e-NMR” and "WeNMR" projects (grants 213010 and 261572). We acknowledge financial support from the CNRS and the Institut Pasteur (M.N. and T.M.), from the Lichtenberg program of the Volkswagen Foundation, the Deutsche Forschungsgemeinschaft (DFG) and the Japan Society for the Promotion of Science (P.G.), from the National Institutes of General Medical Science Protein Structure Initiative program (grants U54 GM074958 and U54 GM094597, G.T.M., C.A., M.K., and T.S), from the Brussels Institute for Research and Innovation (Innoviris, grant BB2B 2010-1-12, W.V.). The support of the national GRID Initiatives of Belgium, Italy, Germany, the Netherlands (via the Dutch BiG Grid project), Portugal, UK, South Africa, Taiwan and the Latin America GRID infrastructure via the Gisela project is acknowledged for the use of web portals, computing and storage facilities.
Abbreviations
- RMSD
root mean square deviation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Andrec M, Snyder DA, Zhou Z, Young J, Montelione GT, Levy RM. A large data set comparison of protein structures determined by crystallography and NMR: statistical test for structural differences and the effect of crystal packing. Proteins. 2007;69:449–465. doi: 10.1002/prot.21507. [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard A, Vranken WF, Bardiaux B, Nilges M, Malliavin TE. Bayesian estimation of NMR restraint potential and weight: A validation on a representative set of protein structures. Proteins. 2011;79:1525–1537. doi: 10.1002/prot.22980. [DOI] [PubMed] [Google Scholar]
- Bhattacharya A, Tejero R, Montelione GT. Evaluating protein structures determined by structural genomics consortia. Proteins. 2007;66:778–795. doi: 10.1002/prot.21165. [DOI] [PubMed] [Google Scholar]
- Bonvin AMJJ, Rosato A, Wassenaar T. The eNMR platform for structural biology. J. Struct. Funct. Genomics. 2010;11:1–8. doi: 10.1007/s10969-010-9084-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat. Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- Cavalli A, Salvatella X, Dobson CM, Vendruscolo M. Protein structure determination from NMR chemical shifts. Proc. Natl. Acad. Sci. USA. 2007;104:9615–9620. doi: 10.1073/pnas.0610313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke ND, Ezkurdia I, Kopp J, Read RJ, Schwede T, Tress M. Domain definition and target classification for CASP7. Proteins. 2007;69(Suppl 8):10–18. doi: 10.1002/prot.21686. [DOI] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, III, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg D, Luthy R, Bowie JU. VERIFY3D: assessment of protein models with three-dimensional profiles. Methods Enzymol. 1997;277:396–404. doi: 10.1016/s0076-6879(97)77022-8. [DOI] [PubMed] [Google Scholar]
- Folmer RHA, Hilbers CW, Konings RNH, Nilges M. Floating stereospecific assignment revisited: Application to an 18 kDa protein and comparison with J-coupling data. Journal of Biomolecular NMR. 1997;9:245–258. doi: 10.1023/a:1018670623695. [DOI] [PubMed] [Google Scholar]
- Forouhar F, Lew S, Seetharaman J, Sahdev S, Xiao R, Ciccosanti C, Lee D, Everett JK, Nair R, Acton TB, et al. Crystal Structure of the TGS domain of the CLOLEP_03100 protein from Clostridium leptum, Northeast Structural Genomics Consortium Target QlR13A. 2009 [Google Scholar]
- Guerry P, Herrmann T. Advances in automated NMR protein structure determination. Q. Rev. Biophys. 2011;44:257–309. doi: 10.1017/S0033583510000326. [DOI] [PubMed] [Google Scholar]
- Güntert P. Structure calculation of biological macromolecules from NMR data. Q Rev Biophys. 1998;31:145–237. doi: 10.1017/s0033583598003436. [DOI] [PubMed] [Google Scholar]
- Güntert P. Automated structure determination from NMR spectra. Eur. Biophys. J. 2009;38:129–143. doi: 10.1007/s00249-008-0367-z. [DOI] [PubMed] [Google Scholar]
- Güntert P, Braun W, Wüthrich K. Efficient computation of three-dimensional protein structures in solution from nuclear magnetic resonance data using the program DIANA and the supporting programs CALIBA, HABAS and GLOMSA. J. Mol. Biol. 1991;217:517–530. doi: 10.1016/0022-2836(91)90754-t. [DOI] [PubMed] [Google Scholar]
- Güntert P, Mumenthaler C, Wüthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- Habeck M, Rieping W, Nilges M. Weighting of experimental evidence in macromolecular structure determination. Proc. Natl. Acad. Sci. U. S. A. 2006;103:1756–1761. doi: 10.1073/pnas.0506412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann T, Güntert P, Wüthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 2002a;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- Herrmann T, Güntert P, Wüthrich K. Protein NMR structure determination with automated NOE-identification in the NOESY spectra using the new software ATNOS. J. Biomol. NMR. 2002b;24:171–189. doi: 10.1023/a:1021614115432. [DOI] [PubMed] [Google Scholar]
- Huang YJ, Powers R, Montelione GT. Protein NMR recall, precision, and F-measure scores (RPF scores): structure quality assessment measures based on information retrieval statistics. J. Am. Chem. Soc. 2005;127:1665–1674. doi: 10.1021/ja047109h. [DOI] [PubMed] [Google Scholar]
- Huang YJ, Tejero R, Powers R, Montelione GT. A topology-constrained distance network algorithm for protein structure determination from NOESY data. Proteins. 2006;62:587–603. doi: 10.1002/prot.20820. [DOI] [PubMed] [Google Scholar]
- Hus JC, Marion D, Blackledge M. Determination of Protein Backbone Structure Using Only Residual Dipolar Couplings. J. Am. Chem. Soc. 2001;123:1541–1542. doi: 10.1021/ja005590f. [DOI] [PubMed] [Google Scholar]
- Koradi R, Billeter M, Güntert P. Point-centered domain decomposition for parallel molecular dynamics simulation. Computer Physics Communications. 2000;124:139–147. [Google Scholar]
- Laskowski RA, Rullmann JAC, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- Linge JP, Williams MA, Spronk CAEM, Bonvin AMJJ, Nilges M. Refinement of protein structures in explicit solvent. Proteins. 2003;50:496–506. doi: 10.1002/prot.10299. [DOI] [PubMed] [Google Scholar]
- López-Mendez B, Güntert P. Automated protein structure determination from NMR spectra. J. Am. Chem. Soc. 2006;128:13112–13122. doi: 10.1021/ja061136l. [DOI] [PubMed] [Google Scholar]
- Luginbühl P, Güntert P, Billeter M, Wüthrich K. The new program OPAL for molecular dynamics simulations and energy refinements of biological macromolecules. J. Biomol. NMR. 1996;8:136–146. doi: 10.1007/BF00211160. [DOI] [PubMed] [Google Scholar]
- Luginbühl P, Szyperski T, Wüthrich K. Statistical Basis for the Use of 13Cα Chemical-Shifts in Protein-Structure Determination. J. Magn. Reson. Ser. B. 1995;109:229–233. [Google Scholar]
- Markwick PR, Malliavin T, Nilges M. Structural biology by NMR: structure, dynamics, and interactions. PLoS. Comput. Biol. 2008;4:e1000168. doi: 10.1371/journal.pcbi.1000168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabuurs SB, Spronk CA, Krieger E, Maassen H, Vriend G, Vuister GW. Quantitative evaluation of experimental NMR restraints. J. Am. Chem. Soc. 2003;125:12026–12034. doi: 10.1021/ja035440f. [DOI] [PubMed] [Google Scholar]
- Nabuurs SB, Spronk CA, Vuister GW, Vriend G. Traditional biomolecular structure determination by NMR spectroscopy allows for major errors. PLoS. Comput. Biol. 2006;2:e9. doi: 10.1371/journal.pcbi.0020009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nederveen AJ, Doreleijers JF, Vranken W, Miller Z, Spronk CA, Nabuurs SB, Güntert P, Livny M, Markley JL, Nilges M, et al. RECOORD: a recalculated coordinate database of 500+ proteins from the PDB using restraints from the BioMagResBank. Proteins. 2005;59:662–672. doi: 10.1002/prot.20408. [DOI] [PubMed] [Google Scholar]
- Nilges M. Ambiguous distance data in the calculation of NMR structures. Fold. Des. 1997;2:S53–S57. doi: 10.1016/s1359-0278(97)00064-3. [DOI] [PubMed] [Google Scholar]
- Nilges M, Bernard A, Bardiaux B, Malliavin T, Habeck M, Rieping W. Accurate NMR structures through minimization of an extended hybrid energy. Structure. 2008;16:1305–1312. doi: 10.1016/j.str.2008.07.008. [DOI] [PubMed] [Google Scholar]
- Ponder JW, Case DA. Force fields for protein simulations. Adv. Prot. Chem. 2003;66:27–85. doi: 10.1016/s0065-3233(03)66002-x. [DOI] [PubMed] [Google Scholar]
- Raman S, Huang YJ, Mao B, Rossi P, Aramini JM, Liu G, Montelione GT, Baker D. Accurate automated protein NMR structure determination using unassigned NOESY data. J. Am. Chem. Soc. 2010a;132:202–207. doi: 10.1021/ja905934c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman S, Lange OF, Rossi P, Tyka M, Wang X, Aramini J, Liu G, Ramelot TA, Eletsky A, Szyperski T, et al. NMR structure determination for larger proteins using backbone-only data. Science. 2010b;327:1014–1018. doi: 10.1126/science.1183649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieping W, Habeck M, Bardiaux B, Bernard A, Malliavin TE, Nilges M. ARIA2: Automated NOE assignment and data integration in NMR structure calculation. Bioinformatics. 2007;23:381–382. doi: 10.1093/bioinformatics/btl589. [DOI] [PubMed] [Google Scholar]
- Rieping W, Habeck M, Nilges M. Modeling errors in NOE data with a log-normal distribution improves the quality of NMR structures. J. Am. Chem. Soc. 2005;127:16026–16027. doi: 10.1021/ja055092c. [DOI] [PubMed] [Google Scholar]
- Rosato A, Bagaria A, Baker D, Bardiaux B, Cavalli A, Doreleijers JF, Giachetti A, Guerry P, Güntert P, Herrmann T, et al. CASD-NMR: critical assessment of automated structure determination by NMR. Nat. Methods. 2009;6:625–626. doi: 10.1038/nmeth0909-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccenti E, Rosato A. The war of tools: how can NMR spectroscopists detect errors in their structures? J. Biomol. NMR. 2008;40:251–261. doi: 10.1007/s10858-008-9228-4. [DOI] [PubMed] [Google Scholar]
- Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS plus : a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. Journal of Biomolecular NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Lange O, Delaglio F, Rossi P, Aramini JM, Liu G, Eletsky A, Wu Y, Singarapu KK, Lemak A, et al. Consistent blind protein structure generation from NMR chemical shift data. Proc. Natl. Acad. Sci. U. S. A. 2008;105:4685–4690. doi: 10.1073/pnas.0800256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sippl MJ. Recognition of Errors in the Three-Dimensional Structures. Proteins Struct. Funct. Genet. 1993;17:355–362. doi: 10.1002/prot.340170404. [DOI] [PubMed] [Google Scholar]
- Spera S, Bax A. Empirical Correlation between Protein Backbone Conformation and Cα and Cβ 13C Nuclear Magnetic Resonance Chemical Shifts. J. Am. Chem. Soc. 1991;113:5490–5492. [Google Scholar]
- Tubbs JL, Latypov V, Kanugula S, Butt A, Melikishvili M, Kraehenbuehl R, Fleck O, Marriott A, Watson AJ, Verbeek B, et al. Flipping of alkylated DNA damage bridges base and nucleotide excision repair. Nature. 2009;459:808–813. doi: 10.1038/nature08076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Eghbalnia HR, Bahrami A, Markley JL. Linear analysis of carbon-13 chemical shift differences and its application to the detection and correction of errors in referencing and spin system identifications. J. Biomol. NMR. 2005;32:13–22. doi: 10.1007/s10858-005-1717-0. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Arndt D, Berjanskii M, Tang P, Zhou J, Lin G. CS23D: a web server for rapid protein structure generation using NMR chemical shifts and sequence data. Nucleic Acids Res. 2008;36:W496–W502. doi: 10.1093/nar/gkn305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wüthrich K. NMR of Proteins and Nucleic Acids. New York: Wiley; 1986. [Google Scholar]
- Zemla A. LGA: A method for finding 3D similarities in protein structures. Nucleic Acids Res. 2003;31:3370–3374. doi: 10.1093/nar/gkg571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Skolnick J. Scoring function for automated assessment of protein structure template quality. Proteins. 2004;57:702–710. doi: 10.1002/prot.20264. [DOI] [PubMed] [Google Scholar]
- Zweckstetter M, Bax A. Single-step determination of protein substructures using dipolar couplings: aid to structural genomics. J. Am. Chem. Soc. 2001;123:9490–9491. doi: 10.1021/ja016496h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.