Abstract
Sphingosine-1-phosphate (S1P) is a bioactive sphingolipid whose actions are essential for many physiological processes including angiogenesis, lymphocyte trafficking and development. In addition, S1P serves asamuscle trophic factor that enables efficient muscle regeneration. This is due in part to S1P's ability to activate quiescent muscle stem cells called satellite cells (SCs) that are needed for muscle repair. However, the molecular mechanism by which S1P activates SCs has not been well understood. Further, strategies for harnessing S1P signaling to recruit SCs for therapeutic benefit have been lacking. S1P is irreversibly catabolized by S1P lyase (SPL), a highly conserved enzyme that catalyzes the cleavage of S1P at carbon bond C2–3, resulting in formation of hexadecenal and ethanolamine-phosphate. SPL enhances apoptosis through substrate- and product-dependent events, thereby regulating cellular responses to chemotherapy, radiation and ischemia. SPL is undetectable in resting murine skeletal muscle. However, we recently found that SPL is dynamically upregulated in skeletal muscle after injury. SPL upregulation occurred in the context of a tightly orchestrated genetic program that resulted in a transient S1P signal in response to muscle injury. S1P activated quiescent SCs via a sphingosine-1-phosphate receptor 2 (S1P2)/signal transducer and activator of transcription 3 (STAT3)-dependent pathway, thereby facilitating skeletal muscle regeneration. Mdx mice, which serve as a model for muscular dystrophy (MD), exhibited skeletal muscle SPL upregulation and S1P deficiency. Pharmacological SPL inhibition raised skeletal muscle S1P levels, enhanced SC recruitment and improved mdx skeletal muscle regeneration. These findings reveal how S1P can activate SCs and indicate that SPL suppression may provide a therapeutic strategy for myopathies. This article is part of a Special Issue entitled Advances in Lysophospholipid Research.
Keywords: Satellite cell, Skeletal muscle, Sphingosine phosphate lyase, Sphingosine-1-phosphate, Sphingolipid, STAT3
1. Introduction
Sphingosine-1-phosphate (S1P) is a bioactive lipid that binds to a family of five G protein coupled receptors [1]. Through activation of S1P receptors and their G protein partners, S1P modulates the activities of adenylyl cyclase, the Ras-related C3 botulinum toxin substrate (Ras)/mitogen activated protein kinase cascade, AKT and STAT3 signaling, phospholipase C and small Ras-homologous (Rho) GTPases. Through these actions, S1P signaling affects cell survival, proliferation, migration and cell–cell interactions [2]. Recent studies have revealed the additional capability of S1P to modulate the activity of intracellular targets including NFκB, histone deacetylases and mitochondrial proteins through molecular mechanisms that appear to be independent of S1P receptors [3]. S1P signaling is essential for many physiological processes including angiogenesis [4,5], hematopoietic cell trafficking [6–8], embryonic development [9,10] and skeletal muscle homeostasis [11].
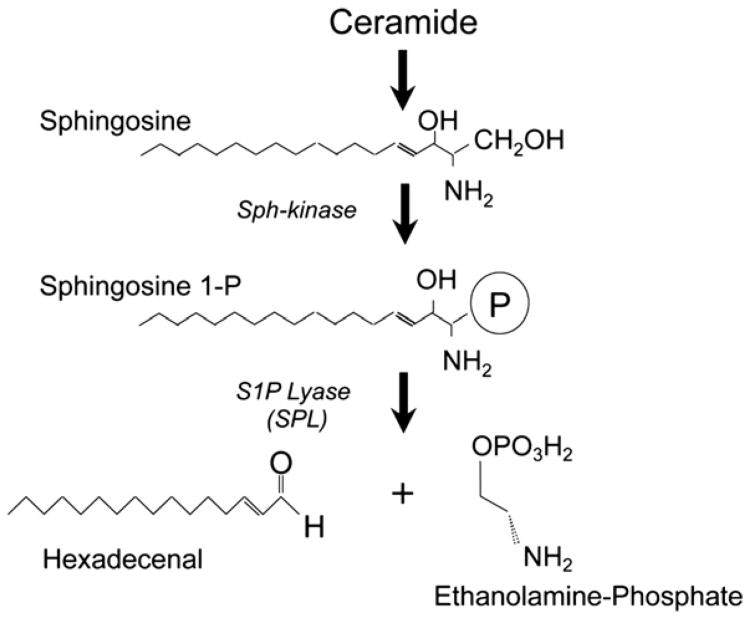
S1P is generated from sphingosine by a phosphorylation reaction catalyzed by the sphingosine kinases, SphK1 and SphK2 [12]. Sphingosine can be regenerated from S1P through the actions of S1P-specific phosphatases (Sgpp1 and Sgpp2) [13] and nonspecific lipid phosphatases (LPPs) [14]. However, only one enzyme, S1P lyase (SPL), is responsible for the irreversible catabolism of S1P (Fig. 1) [15,16]. SPL is an integral membrane protein located in the endoplasmic reticulum. SPL degrades S1P in the cytosol by catalyzing its cleavage at the C2–3 carbon bond, resulting in the formation of ethanolamine phosphate and the long chain aldehyde, hexadecenal. S1P activity has a major impact on intracellular S1P stores as well as tissue and circulating S1P levels, illustrated by the marked accumulation of S1P observed in cells, model organisms and genetically modified mice in which the gene encoding SPL has been knocked down or knocked out [17–19]. By removing S1P in cellular, vascular and tissue compartments, SPL regulates the S1P pools available for interactions with intracellular targets and for autocrine and paracrine receptor-mediated signaling [20].
Fig. 1.
Formation of sphingosine 1-phosphate. Diagram of the formation of sphingosine 1-phosphate starting from ceramide which is a central molecule of sphingolipid metabolism. Sphingosine kinases (Sph-kinase) catalyze the phosphorylation of sphingosine, generating S1P. S1P lyase (SPL) catalyzes the cleavage of S1P into the long chain aldehyde hexadecenal and ethanolamine-phosphate.
Based on the ability of SPL to generate S1P gradients required for lymphocyte trafficking, SPL inhibition via treatment with the drug LX2931 (Lexicon Pharmaceuticals) is now being tested as a strategy for immunomodulation in the treatment of autoimmune disease such as rheumatoid arthritis [21]. SPL induces apoptosis in response to stressful conditions including DNA damage and ischemia through mechanisms involving both substrate depletion and generation of reactive aldehydes [22,23]. Delivery of a soluble bacterial SPL has also been shown to reduce circulating S1P levels and block the proangiogenic effects of S1P receptor signaling [24]. These findings suggest that modulation of SPL may be useful in the prevention and treatment of a wide variety of human diseases including autoimmunity, cancer, ischemic tissue damage, fibrosis, inflammation and pathological angiogenesis.
The first SPL gene to be identified was the Saccharomyces cerevisiae dihydrosphingosine phosphate lyase 1 (Dpl1) gene [25]. Over the past 15 years, the SPL genes of various bacterial, fungal, nematode, plant and mammalian genomes as well as murine and human Sgpl1 genes encoding SPL have been identified and characterized. In many cases, knockout models have been generated, and the mutant phenotypes in these organisms have revealed critical roles for SPL in cellular function, development and physiology. Interestingly, Drosophila Sply mutants lacking SPL expression exhibit a myopathic phenotype affecting the muscles of the thorax that power the wings and enable flight [26]. Based on this observation, we hypothesized that SPL has an essential and conserved role in skeletal muscle homeostasis. Using a murine model of skeletal muscle injury, we have demonstrated that an S1P signal is generated in response to muscle injury and, through activation of S1P2, leads to downstream events that involve the transcription factor STAT3 and the recruitment of skeletal muscle stem cells called satellite cells (SCs) that are needed for efficient skeletal muscle regeneration [27]. Further, we found that dystrophic mdx mice, which serve as a model of Duchenne MD, are S1P-deficient due to chronic injury-induced upregulation of SPL. Pharmacological inhibition of SPL improved SC recruitment and muscle regeneration in a STAT3-dependent manner in mdx mice, thereby illustrating the potential utility of targeting SPL for therapeutic benefit in MD.
This review will highlight our recent findings on the role and mechanism of action of S1P signaling in SC recruitment and muscle regeneration. These observations will be related to the known functions of S1P as a skeletal muscle trophic factor and SC activator. We present some new findings regarding the potential cellular sources of SPL in injured muscle and demonstrate the presence of SPL expression in SC-derived myoblasts. We will discuss remaining questions and propose potential next steps toward further elucidating the biology and clinical potential of modulating S1P metabolism and signaling for therapeutic purposes in human diseases affecting skeletal muscle. We refer readers interested in learning more about SPL structure, function and regulation to numerous recent reviews describing the biochemical characterization of SPL, its subcellular localization, tissue distribution, regulation, role in development, function in apoptosis, development of SPL inhibitors, and structure/function relationships predicted by recent crystallization of a bacterial SPL [16,20,28–31].
2. The muscular dystrophies
MDs are a heterogeneous group of genetic diseases characterized by the progressive loss of skeletal muscle strength associated with pathological features including pseudohypertrophy, muscle necrosis and fiber splitting, regeneration and centralized nuclei, variation in fiber size, and eventual muscle replacement by adipose and fibrotic tissues [32]. Together, these effects compromise patient mobility and quality of life, and in the most severe cases lead to premature death. In 1987, Eric Hoffman identified mutations in the dystrophin gene as the cause of the most common and severe form of MD, Duchenne MD (DMD), which affects 1 in 4000 newborn males [33]. The dystrophin protein links the plasma membrane to the internal cytoskeleton through interactions with γ-actin and with a plasma membrane complex called the dystroglycan-associated protein complex (DGC). The DGC also interacts with the extracellular matrix (ECM). These connections anchor the plasma membrane internally and externally, thereby facilitating the distribution of forces generated upon muscle contraction. The lateral distribution of force stabilizes the myofiber and prevents membrane disruption with each contraction. Subsequent to the cloning of dystrophin, over 30 genes have been linked to hereditary MDs [32,34,35]. MD mutations are found in the components of the DGC, DGC-interacting proteins, and enzymes that regulate the expression, modification, and function of the DGC. In addition, mutations in the ECM proteins laminin and collagen VI also cause MD [35]. Disruption of the normal bridge between ECM, DGC and γ-actin in patients with MD leads to high stress on fragile membranes, resulting in membrane lesions that overwhelm the membrane repair and muscle regeneration systems, ending finally in calcium influx and muscle cell death [34,36,37]. In addition, there is evidence that modifying factors independent of the DGC may influence the severity of dystrophy in MD [32,38]. Novel treatments for MD focus on reducing muscle atrophy, inhibiting cell death pathways, and restoring membrane protein scaffolding and repair through the use of skeletal muscle stem cells [39]. Characterizing the signaling pathways that control these processes may reveal new targets for therapeutic intervention in MD and muscle wasting disease. While it remains controversial whether or not SC reserves and/or functions are reduced/impaired in MD, finding ways to expand endogenous stem cell reserves and/or enhance their myogenic potential may be of therapeutic benefit in MD.
3. S1P signaling, SCs and skeletal muscle regeneration
Adult skeletal muscle is a unique tissue that has the capacity to regenerate after injury [40]. Skeletal muscle regeneration involves the activation of SCs found between the basal lamina and sarcolemma of each myofiber (Fig. 2). After muscle injury, SCs are stimulated to leave their quiescent state and reenter the cell cycle. SCs proliferate, and their progeny (myoblasts) adheretoand fuse with the damaged muscle cell (multinucleated syncytia or myofiber), stimulating a program of differentiation that leads to regeneration of healthy tissue [41–43].
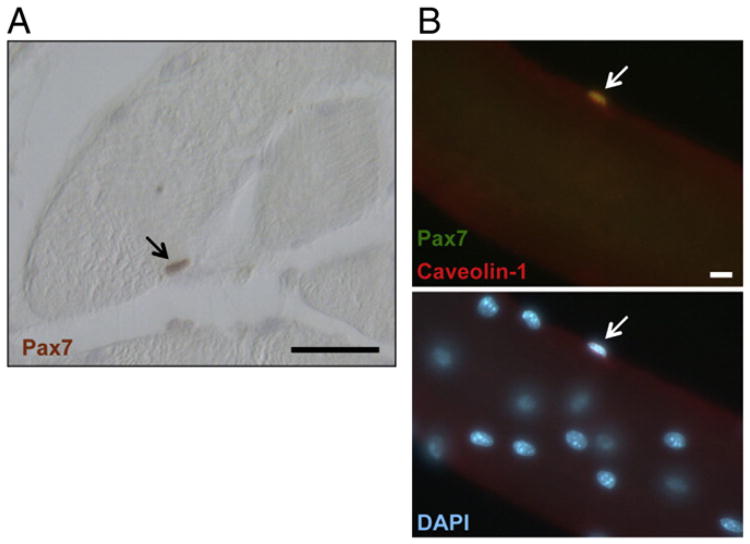
Fig. 2.
SC localization. (A) SC stained with Pax7 antibody (red, marked by arrow) in a cross-section of human skeletal muscle. Scale bar 50 μm. (B) Upper panel, co-immunostaining (merged yellow) of SCs with Pax7 (green) and Caveolin-1 (red) antibodies in isolated fibers of adult mouse muscle; lower panel, DAPI (blue) counterstained for nuclei. Scale bar 20 μm arrow points to SC. Taken from A.S. de la Garza-Rodea [85].
S1P generated from catabolism of muscle membrane sphingolipids was recently identified as the signal that causes SCs to enter the cell cycle, whereas chemical inhibition of S1P formation prevented skeletal muscle regeneration [44–46]. This suggests a central role for S1P in skeletal muscle homeostasis. However, the mechanism responsible for this was not known. Rodent muscles express three of the five known S1P receptors, and S1P exerts a trophic action on denervated muscle fibers [46]. Sphingolipids including S1P have also been implicated in the regulation of muscle cell signaling, contractile properties, insulin resistance and muscle fiber trophism [47]. These findings suggest that S1Pmetabolism and signaling contribute to muscle homeostasis and repair, although the fundamental mechanisms responsible for the actions of S1P in muscle remain poorly understood. The chronic/degenerative phase of MD is caused by a failure of skeletal muscle regeneration to keep up with the ongoing injury and destruction of muscle fibers [48]. This may be accounted for in part by a reduction in SC reserves or SC myogenic potential. Elucidating the specific roles that S1P signaling plays inSCbiology, muscle regeneration, aging and MD may help devise new approaches to maintaining muscle function and reducing loss of muscle mass in these diseases.
4. The Sply mutantasan invertebrate model for muscular dystrophy
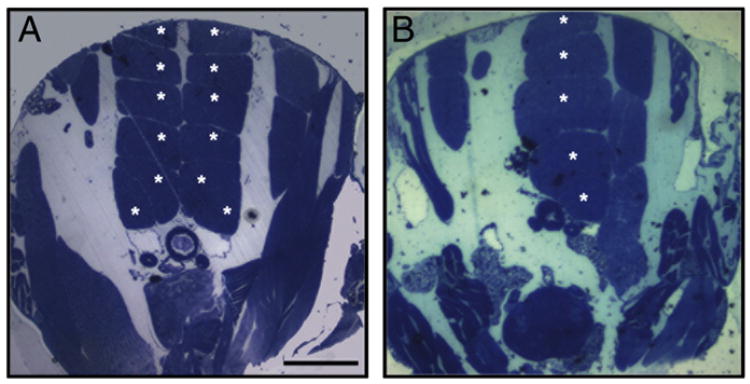
Sply mutants harboring a transposon insertion mutation in the gene encoding the Drosophila homolog of SPL cannot catalyze the degradation of S1P and accumulate sphingolipid intermediates. Sply mutants are flightless due to degeneration of the thoracic flight muscles that power the wings [26]. The thoraces of wild type fruit flies exhibit an invariant pattern of six pairs of dorsal longitudinal muscles (DLMs) as shown in Fig. 3A. In the Sply mutants, one or more fibers of the DLM are typically absent, and the severity of this myopathy was observed to increase with age (Fig. 3B). The Sply flightless phenotype can be recapitulated by knockdown of Sply in myoblast precursor cells using RNA interference, indicating that the defect is intrinsic to muscle (our unpublished observations). Interestingly, we have observed that RNAi-mediated knockdown of Sply expression in Drosophila S2 cells leads to reduction of dystrophin expression (our unpublished observations). These cumulative findings provide dramatic evidence that sphingolipid metabolism plays a crucial and intrinsic role in muscle stability and that tight control over sphingolipid metabolism is necessary to maintain muscle homeostasis. They also specifically implicate SPL in skeletal muscle homeostasis and suggest that the Sply mutant may potentially serve as an invertebrate model of DMD.
Fig. 3.
The Drosophila Sply mutant myopathy. Methylene blue and Azure II stain of transversally sectioned thoraces of a normal fruit fly (A) and Sply mutant (B). The 6 pairs of dorsal longitudinal skeletal muscles (asterisks) present in wild type Drosophila (A). Degenerating muscles in a Sply mutant (B). Scale bar 100 μm.
5. Murine muscle injury elicits a metabolic response that produces an S1P signal
The genetic and molecular pathways that govern muscle development are largely conserved between Drosophila and humans [49,50]. To explore whether the Sply phenotype accurately predicts a role for S1P signaling in mammalian skeletal muscle homeostasis, we first examined the expression of the murine SPL gene Sgpl1 in skeletal muscle at rest. Our findings indicate that SPL is expressed at low or undetectable levels in resting skeletal muscle [27]. However, we have observed that SPL expression is induced in response to various stressful conditions, such as radiation and ischemia [22,51]. This raised the possibility that expression of the lyase, while hidden at rest, may be “exposed” in response to injury. Therefore, we examined the effect of muscle injury on Sgpl1 as well as other genes involved in S1P biosynthesis, catabolism and signaling. To accomplish this, we employed a commonly used system for analyzing murine skeletal muscle injury, the notexin (NTX) injury model. NTX is a snake venom phospholipase A2 that when injected into muscle causes small lesions in the sarcolemma, resulting in muscle fiber degeneration. Due to the capacity of skeletal muscle for regeneration, the muscle recovers from this injury over the course of two to four weeks, [37,52,53]. Within 6 h of NTX injection into the gastrocnemius muscles of wild type mice, we observed induction of Sphk1 gene expression 100-fold over baseline levels as determined by quantitative RT-PCR. The expression of Sgpl1 was induced 20-fold over baseline several days after injury, concomitant with upregulation of the muscle transcription factor, Myf5. Other genes involved in S1P signaling including Sphk2, Sgpp1 and genes encoding S1P receptors 1-3 were upregulated after injury, but the changes were more modest than those of Sphk1 and Sgpl1. Using additional detection methods, we confirmed that SPL gene and protein expression are induced in muscle fibers after treatment with NTX. Not surprisingly, when we measured S1P plasma levels, we found that they are increased by 50% compared to baseline levels within the first 24 h after injury [27]. Thus, a dynamic and tightly orchestrated genetic program modulates S1P metabolism and receptor expression in response to muscle injury. These changes lead to the generation of a plasma S1P “signal.” Based on these findings, it became important to determine the significance of that signal for skeletal muscle regeneration.
6. SphK1 is essential for efficient skeletal muscle regeneration and SC functions
Two sphingosine kinases (SphK1 and SphK2) contribute to S1P biosynthesis in mammals. In mice, the disruption of one SphK gene is not lethal, provided the other is functional. Although SphK1 and SphK2 knockout mice appear healthy under routine conditions, some phenotypes have been revealed in response to specific stressful stimuli. For example, SphK1 knockout mice fail to achieve cardiac preconditioning and are more susceptible than control mice to anaphylactic shock, whereas SphK2 knockout mice are more susceptible to kidney ischemic injury [54–57]. These findings suggest that the developmental functions of SphKs are redundant, whereas individual SphKs may be responsible for specific functions in postnatal life.
Therefore, to address whether the profound upregulation of SphK1 and corresponding peak in plasma S1P levels we observed within hours of intramuscular NTX injection signified a specific function for SphK1 in the injured muscle, we compared the biochemical and functional consequences of NTX muscle injury in SphK1 global knockout mice and wild type controls. We found that the skeletal muscles of wild type mice were largely repaired by 10 days after injury. The point of maximal injury exhibited mostly newly regenerated fibers containing a centralized nucleus, the hallmark of a regenerated muscle fiber. In contrast, at the same time point the SphK1 knockout mouse muscles exhibited many residual necrotic fibers and only partial regeneration at the site of maximal injury. Importantly, when we measured SCs at the injury site, SphK1 knockout mice lacked a robust recruitment of SCs compared to wild type mice. In addition, plasma S1P levels in SphK1 knockout mice were approximately half the levels found in the circulation of wild type mice throughout the course of the experiment [27].
To study the requirement for S1P signaling in SC functions in more detail, we isolated SCs from the injured muscle fibers of wild type and SphK1 knockout mice and characterized them by measuring their capacity for proliferation and differentiation using in vitro assays. In comparison to wild type SCs incubated in media supplemented with wild type serum, we observed that SphK1 knockout SCs incubated in media supplemented with SphK1 knockout serum did not perform as well, exhibiting reduced proliferation and differentiation. The functions of the SphK1 knockout SCs were enhanced by incubation with wild type serum, although not to the level of wild type SCs. Conversely, wild type SCs incubated in medium containing serum from SphK1 knockout mice were also functionally compromised, although not as severely as SphK1 knockout SCs [27]. These experiments revealed that SphK1 induction and S1P signaling contribute to efficient SC recruitment and muscle regeneration in response to acute injury. They also suggest that after muscle injury SCs may respond to S1P signals derived from multiple sources. S1P production by SphK1 within the SC itself may be required for autocrine stimulation. In addition, when the sequestered niche of the SC is compromised by injury, SCs are likely exposed to S1P derived from the injured fiber, the vasculature or from other source within the muscle fiber microenvironment. It seems likely that maximal SC activation requires S1P signals generated from both sources. However, it is also possible that SphK1 knockout mice fail to mobilize other circulating factors independent of or complementary to S1P that are needed for maximal SC activation and recruitment. Further investigation of the cytokine responses in SphK1 knockout mice may reveal more about the contribution of SphK1 and S1P signaling to the global inflammatory response to skeletal muscle injury.
7. Dystrophic mice exist in a state of S1P deficiency
Considering the dramatic changes in S1P metabolism and signaling that we observed in response to acute muscle injury, we predicted that chronic, global skeletal muscle injury that occurs in patients suffering from MD might also have an impact on S1P levels and metabolism. If so, this could have potential consequences for muscle regeneration and SC recruitment in these conditions and, conversely, could present opportunities for therapeutic intervention. Male C57BL/10ScSn-Dmdmdx/J (mdx) mice harbor a single point mutation in the mouse dystrophin gene located on the X chromosome [58]. This mutation is equivalent to the mutation that causes DMD in humans, making mdx mice a useful model of the disease, although the murine version is milder (due to the presence of revertant fibers and utrophin expression) and has greater capacity for regeneration due to other genetic factors [59]. When we compared the expression levels of the genes by RT-PCR involved in S1P metabolism in the muscles of mdx mice and control mice of the same background, we found no changes in Sphk1 or Sphk2 levels but a significant increase in Sgpl1 expression. The upregulation of Sgpl1 was confirmed by immunoblotting, which revealed a very high level of SPL expression in the dystrophic muscles of mdx mice compared to controls. Skeletal muscle comprises the largest single organ of the body making up approximately half of the body's mass. Thus, it was perhaps not too surprising that the upregulation of SPL in dystrophic skeletal muscle correlated with a significant reduction of plasma S1P in mdx mice compared to controls. The S1P levels in mdx mouse plasma were as low as those observed in SphK1 knockout mice [27]. This suggested that muscle regeneration and SC recruitment might be compromised by progressive S1P deficiency in dystrophic muscles. Further, it suggests that inhibiting SPL might provide a useful way to boost S1P levels and improve efficiency of skeletal muscle regeneration as therapeutic.
8. SPL expression in SCs, myoblasts, myofibers and other cells of skeletal muscle
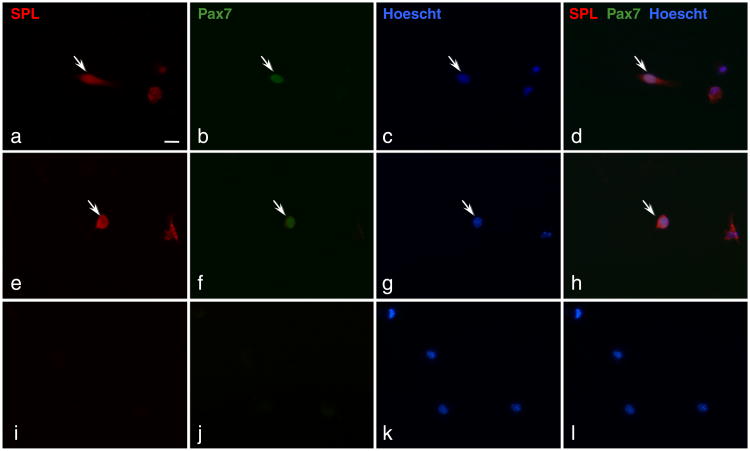
The findings above point toward an important role for S1P in SC proliferation and a potential role in muscular dystrophy. However, the role in the muscle regeneration process of intracellular S1P and of SPL, the enzyme responsible for degrading S1P and controlling its intracellular levels, have not been fully explored. Specifically, our study did not investigate the cellular source of SPL responsible for increasing SPL levels in skeletal muscle in the aftermath of injury. In our laboratory, we have conducted a pilot study in which SC-derived myoblasts were isolated from mdx mouse skeletal muscles, cultured for 24 h, fixed and stained with specific antibodies for SPL and paired box protein-7 (Pax7), the latter which serves as a marker for SCs. Fig. 4 shows that majority of cells we isolated express SPL. Further, some of these cells were positive for both Pax7 and SPL. This suggests that SPL expression is present in activated SCs and/or in SC progeny that have differentiated to myoblasts. Our observation is consistent with other results from our laboratory indicating that murine C2C12 cells express high levels of SPL and that knockdown of SPL in this cell line has profound effects on the ability of the cells to differentiate into myoblasts (our unpublished observations).
Fig. 4.
Mdx SC-derived myoblasts express SPL. SC-derived myoblasts were isolated from skeletal muscle of mdx mice. Cells were in culture for 24 h before being fixed and stained with specific antibodies for SPL and Pax7. Nuclei were counterstained with Hoechst 33342. Two areas (a to d) and (e to h) show a Pax7 positive cell among several SPL positive cells. (i to l) cells stained with secondary antibodies (Alexa 488 and Alexa 546) and Hoechst only. Scale bar 100 μm.
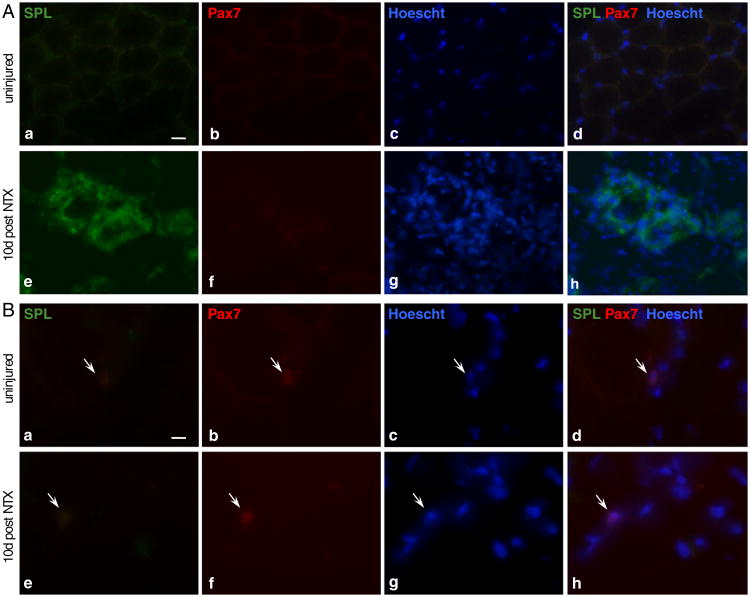
Skeletal muscle regeneration involves the complex interaction of many cell types, not only the injured fiber and SCs, but also vascular cells, vascular support cells and inflammatory cells that enter the skeletal muscle interstitium in response to injury. In other studies, we have observed that bone marrow-derived blood cells amply express SPL (Borowsky et al., submitted). Thus, inflammatory cells that infiltrate injured skeletal muscle likely contribute to the total amount of skeletal muscle SPL pool after injury. To directly address this possibility, as well as to further explore the presence of SPL in cells within the SC–myoblast–myofiber continuum, cryosections of wild type skeletal muscle (uninjured and 10 days post NTX injection) were stained for SPL and Pax7. We observed that quiescent SC and muscle fibers at rest do not express detectible amounts of SPL (Fig. 5). In response to injury, many mononucleated infiltrating cells were found to express SPL, confirming that some of the SPL “upregulation” we observed in injured muscle can be explained by inflammatory changes. However, we did not observe SPL expression in the regenerating muscle fibers or SCs in situ at 10 days after injury. Taken together with our finding of SPL expression in isolated SCs and SC-derived myoblasts in culture, our histological results suggest that SPL upregulation may largely represent an inflammatory response to muscle injury, but that SPL may also be transiently expressedin activated SCs and/or in the progenyof activated SCs (Fig. 5). Further imaging studies that follow SPL and Pax7 expression in situ throughout the time course of the first week after injury will be required to define whether SPL expression within SC-derived myoblasts may contribute to localized S1P gradients and affect the S1P levels of regenerating fibers in the aftermath of muscle injury. It is interesting to note that acute stress responses have been characterized by rapid and significant lymphopenia. Further, snake venoms, including notexin, have been shown to induce lymphopenia in rats [60,61]. The kinetics of induced lymphopenia after notexin injection, which occurs within 24 h, is similar to the kinetics of SphK1 upregulation and S1P increase that we have observed after notexin-induced injury in mice. Thus, there may be a causal connection between the increased S1P release and retention of cells in peripheral tissues as a mechanism leading to notexin-induced lymphopenia.
Fig. 5.
SPL is expressed in inflammatory cells of degenerating/regenerating skeletal muscle. Cross-sections of cryopreserved skeletal muscle of an uninjured wild type (C57BL/6) mouse at rest (upper panels) and 10 days post-NTX-injury (bottom panels). Muscles were excised, sectioned, stained with antibodies specific for SPL and Pax7 and counterstained with Hoechst 33342. A: (a to d) resting myofibers do not express SPL; (e to h) strong stain is observed in degenerated/necrotic myofibers undergoing regeneration. Scale bar 100 μm. B: SCs (arrows) that were detected in both injured and uninjured muscles did not co-stain for SPL. Scale bar 10 μm.
9. SPL inhibition improves the response to acute muscle injury in mdx mice
Tetrahydroxybutylimidazole (THI) is a byproduct of sugar caramelization that is found in many food substances containing caramel food coloring type III. THI was recently shown to serve as an immunomodulatory agent by virtue of its ability to inhibit SPL [62]. Chemical derivatives of THI have subsequently been developed and are undergoing clinical evaluation for therapeutic efficacy in patients with rheumatoid arthritis [21]. To test the effect of SPL inhibition on muscle regeneration in a dystrophic model, we compared the short-term effects of THI administration compared to vehicle in mdx mice after acute muscle injury with NTX. We observed a significant improvement in the number of regenerating fibers after injury when mice received THI. Quantification of SCs by both flow cytometry and immunostaining revealed that THI treatment improved SC activation as well [27]. These important findings demonstrate that SPL inhibition may have potential as a strategy to improve SC function and recruitment in the context of muscular dystrophy. Interestingly, T-cells have been shown to promote pathology of mdx mice [63]. Further, the immunosuppressant rapamycin ameliorates the dystrophic phenotype of mdx mice [64]. One of the main effects of THI and genetic knockout of SPL is retention of lymphocytes in lymphoid organs and preventing them from reaching sites of injury and inflammation. Therefore, it is conceivable that such immunosuppressive activity may add to the potential therapeutic efficacy of SPL inhibition in disease states that involve muscle injury.
10. THI promotes muscle regeneration and SC recruitment through STAT3 signaling
Zammit and colleagues showed that S1P contributes to the activation of SCs [44], but the molecular mechanism underlying this effect was not well understood. The ability of THI to promote muscle regeneration and SC recruitment gave us a tool with which to investigate this question in vivo. Recent studies by the laboratory of Hua Yu demonstrated the existence of cross-talk between signaling pathways involving S1P and the transcriptional regulator STAT3 [65]. The interactions between S1P and STAT3 were found to be responsible for the chronic STAT3 activation in some types of cancer. Additional reports provided further evidence for S1P and STAT3 signaling interactions in the context of prostate cancer and cardiomyocyte function [66,67]. When we measured STAT3 activation in the skeletal muscles of mdx mice 5 days after NTX injury, THI treatment appeared to have a positive effect, as shown by increased phosphorylation on STAT3 Tyrosine 703 [27]. Importantly, inhibition of STAT3 signaling using the small molecule WP1066 reversed the ability of THI to promote muscle regeneration and increase the number of SCs several days after injury, indicating the importance of STAT3 activation in this process.
11. S1P activates SCs via S1P2/STAT3-dependent repression of cell cycle inhibitors
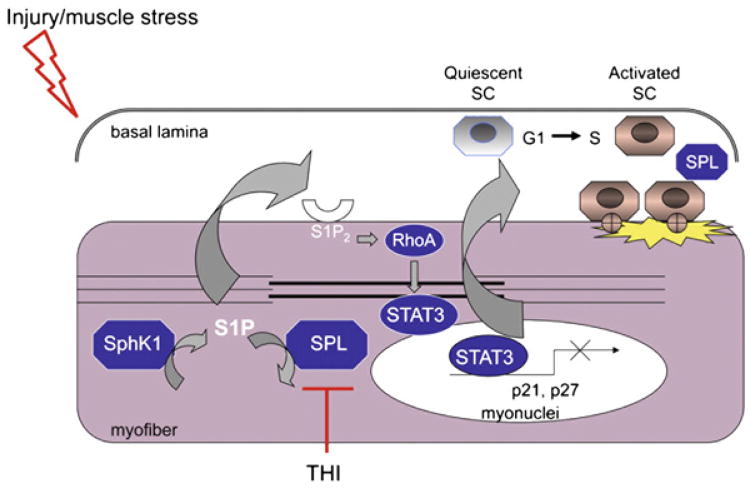
Since S1P can act via receptor-dependent as well as intracellular, receptor-independent mechanisms, we next measured the effect of inhibitors of the three major S1P receptors in muscle, S1P1-3, on STAT3 activation in isolated SCs. Since the isolation of SCs from injured muscle fibers readily activates them, we were not surprised to find a significant level of phosphorylated, active STAT3 at baseline. We found that treatment of SCs with S1P2 inhibitor prevented STAT3 activation in SC-derived myoblasts, whereas inhibitors of S1P1 and S1P3 did not alter STAT3 activation. We found that siRNA-mediated knockdown of S1P2 also reduced STAT3 activation in murine SC-derived myoblasts [27]. Importantly, the inhibition of S1P2 led to the upregulation of two cell cycle inhibitor proteins, p21 and p27. These proteins are known to be under repressive control by STAT3. We interpreted these findings as revealing a plausible mechanism by which S1P recruits SCs into the cell cycle, i.e. by activating STAT3, leading to repression of cell cycle inhibitors, thereby allowing the SC to re-enter the cell cycle and begin proliferating in response to injury. A schematic diagram of this mechanism is illustrated in Fig. 6. There remained one additional question we hoped to answer, i.e., by what mechanism did S1P ligation to S1P2 induce the activation of STAT3? We suspected that this process might involve members of the Rho family of small GTPases, including Ras-related C3 botulinum toxin substrate (Rac) 1, cell division control protein 42 homolog, and RhoA. These proteins function downstream of G protein coupled receptors including S1P2 and have been shown to promote tyrosine phosphorylation of STAT3 and its translocation to the nucleus in various cellular contexts including carcinogenesis. Our results using a combination of Rac1 and RhoA inhibitors and plasmid constructs with which we modulated Rac1 and RhoA expression and activity demonstrated that S1P2 activation inhibits Rac1 and activates RhoA, thereby promoting STAT3 phosphorylation on Tyrosine 703 in myoblasts (Fig. 6) [27].
Fig. 6.
Role of S1P in response to skeletal muscle repair. After skeletal muscle injury, SphK1 generates S1P, which activates S1PR2, leading to RhoA and STAT3 activation, thereby repressing small cell cycle inhibitor proteins. SCs reenter the cell cycle and contribute to muscle regeneration. Inhibition of SPL using THI leads to a boost in S1P signaling, thereby promoting SC activation and efficient muscle regeneration. Note that the localization of SPL is assigned to the myofiber for convenience. However, SPL in SCs and inflammatory cells may also regulate S1P levels in and around the injured muscle.
12. Other recent findings regarding S1P, SCs and skeletal muscle regeneration
Extensive research has been conducted to better understand the role of S1P in the biology of SCs and the process of skeletal muscle regeneration. S1P has been shown to stimulate the proliferation of quiescent SCs in vivo after cardiotoxin-induced damage [44]. Further, in vitro studies have demonstrated that exogenous S1P increases the intake of labeled thymidine incorporation in a PI3K-dependent manner and that the proliferative action of the sphingolipid depends on S1P2 and S1P3 [68]. The involvement of S1P in the migration of SCs has been also explored. Employing S1P receptor agonists and antagonists as well as specific siRNAs, Calise et al. found that S1P1 and S1P4 are involved in mediating the pro-migratory effects in SCs [68].
The C2C12 murine SC line is widely used to study in vitro skeletal muscle growth and differentiation. Early studies by Bruni and colleagues demonstrated that addition of S1P to cultured C2C12 cells led to the activation of phospholipase D [69], monomeric GTPase Rho [70] and increased cytosolic calcium [71]. C2C12 cells do not express S1P5 but do have S1P1–S1P4. The most highly expressed S1P receptors in myoblasts and myotubes are S1P1 and S1P3, respectively. During myogenic differentiation, S1P2 is down-regulated and almost absent in myotubes [72]. The dynamic modulation and critical role of S1P2 in the myogenic process has been observed in other studies, as well. Danieli-Betto et al. [73] observed an increase in expression of S1P1 after muscle damage induced by bupivacaine. S1P2 was noted to be absent in quiescent cells but transiently expressed in the early phase of regeneration. The expression patterns of the S1P receptors during myogenic differentiation in vitro and in vivo suggest that they exert complex and possibly counteracting signals in this process. Future studies should elucidate how these receptor signals are synchronized to coordinately regulate the sequential phases of SC and skeletal muscle response to injury required for efficient muscle regeneration.
13. S1P and cell therapy for skeletal muscle regeneration
The main goal of cell therapy in patients with muscle injury, disease or sarcopenia (frailty of aging) is to repair tissue damage and replenish lost myofibers through systemic or local delivery of cells with myoregenerative properties and thereby restore skeletal muscle function. SCs, recognized as true muscle progenitor cells, have been extensively explored for this purpose in laboratory animals and in patients. However, one of the major problems encountered in these studies was the limited cell survival of the transplanted SCs [74,75]. In the last few years, in addition to SCs and SC-derived myoblasts, other somatic cell types exhibiting myogenic potential have been identified and characterized. Some of these cell types (most of them multipotent stem cells) have proven capable of contributing to myogenic regeneration (e.g. endothelial cells, mesoangioblasts) [76–78], and a few [e.g. pericytes, endothelial cells, mesenchymal stem cells (MSCs)] have demonstrated the ability to incorporate directly into the SC niche [79–82]. Pericytes and mesoangioblast are blood vessel-associated stem cells that originate from embryonic and postnatal tissue respectively. Both of these cell types hold promise for clinical application, because a significant proportion is capable of reaching skeletal muscles following systemic administration. In mesoangioblasts, S1P strongly stimulates proliferation and protects from apoptosis elicited by different stimuli [83]. Treatment of mesoangioblast ex vivo with S1P enhanced their survival after delivery intramuscular into mice [78]. This exciting finding supports the view that S1P-preconditioned allogeneic mesoangioblast cells might enhance their survival, overcoming a critical barrier to cell therapy.
In order to explore the potential impact of S1P/S1PR2 signaling on STAT3 activation, we applied exogenous S1P to selected cell types: human umbilical vein endothelial cells (HUVECs), the pericyte cell line C3H10T1/2 and M25.2 cells, a clonally-derived population of mesoangioblasts generated by Suzanne Berry-Miller (University of Illinois). Of all the cells we tested, SC-derived myoblasts responded most robustly to S1P with regard to the activation of STAT3 [27]. In contrast, HUVECs and M25.2 mesoangioblast cells exhibited minimal STAT3 phosphorylation in response to S1P, and STAT3 phosphorylation was undetectable in C3H101/2 cells under the conditions we employed. However, additional experiments will be required to establish whether endogenous or exogenous S1P signaling plays a role in the biology of these cell types or on their potential for myogenic differentiation and in vivo incorporation of these cells into muscle.
MSCs are characterized by their capacity for extensive proliferation ex vivo, relative ease of isolation and ability to differentiate into adipogenic, osteogenic, chondrogenic and myogenic lineages. S1P has been shown to stimulate MSCs isolated from adipose tissue to differentiate into smooth muscle in a dose-dependent manner [84]. S1P2 was shown to be a mediator of the differentiation toward smooth muscle, whereas S1P3 was shown to be less important in this process. Although these studies are intriguing, the potential of MSCs to be used for cell therapy in skeletal muscle disease, and the extent to which S1P signaling and metabolism may be used to enhance cell therapy using this source of stem cells remains to be further explored.
14. Summary
The involvement of sphingolipids in many aspects of skeletal muscle biology is becoming increasingly clear (see Donati and Bruni this issue). Further, the evidence supporting a central role for S1P signaling and metabolism in SC biology and the coordinated control of skeletal muscle regeneration is now irrefutable in light of results from investigations of S1P signaling in myoregeneration by many research groups. These cumulative findings have revealed at least the theoretical potential for harnessing S1P's actions to modulate the activation of SCs and enhance muscle repair for therapeutic purposes in a variety of clinical contexts, including muscle wasting diseases, trauma and sarcopenia, wherein reduction of SC numbers contributes to the frailty of aging. Considering the availability of small molecule S1P receptor agonists and antagonists and SPL inhibitors already in clinical trials for other diseases, it seems likely that these basic research findings will be translated into more clinically relevant large animal model systems in the near future.
Acknowledgments
This work was funded by NIH grant GM66954, Muscular Dystrophy Association grant MDA217712 and CIRM fellowship TG2-01164.Weare grateful to Derron Herr and Greg Harris for photomicrographs of Sply flight muscles, Nelle Cronen for expert administrative assistance and Eric Hoffman and Suzanne Berry-Miller for many helpful discussions.
Abbreviations
- Dpl1
dihydrosphingosine phosphate lyase 1
- DLMs
dorsal longitudinal muscles
- DMD
Duchenne muscular dystrophy
- DGC
dystroglycan-associated protein complex
- ECM
extracellular matrix
- HUVECs
human umbilical vein endothelial cells
- LPPs
lipid phosphatases
- MSCs
mesenchymal stem cells
- MD
muscular dystrophy
- NTX
notexin
- Pax7
paired box protein-7
- Rho
Ras-homologous
- Rac
Ras-related C3 botulinum toxin substrate
- SPL
S1P lyase
- SCs
satellite cells
- STAT3
signal transducer and activator of transcription 3
- S1P
sphingosine-1-phosphate
- S1P2
sphingosine-1-phosphate receptor 2
- THI
tetrahydroxybutylimidazole
Footnotes
This article is part of a Special Issue entitled Advances in Lysophospholipid Research.
References
- 1.Maceyka M, Milstien S, Spiegel S. Sphingosine-1-phosphate: the Swiss army knife of sphingolipid signaling. J Lipid Res. 2008;50:S272–S276. doi: 10.1194/jlr.R800065-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Young N, Van Brocklyn JR. Signal transduction of sphingosine-1-phosphate G protein-coupled receptors. Sci World J. 2006;6:946–966. doi: 10.1100/tsw.2006.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maceyka M, Harikumar KB, Milstien S, Spiegel S. Sphingosine-1-phosphate signaling and its role in disease. Trends Cell Biol. 2012;22:50–60. doi: 10.1016/j.tcb.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Su SC, Bayless KJ. Utilizing sphingosine-1-phosphate to stimulate sprouting angiogenesis. Methods Mol Biol. 2012;874:201–213. doi: 10.1007/978-1-61779-800-9_16. [DOI] [PubMed] [Google Scholar]
- 5.Argraves KM, Wilkerson BA, Argraves WS. Sphingosine-1-phosphate signaling in vasculogenesis and angiogenesis. World J Biol Chem. 2010;1:291–297. doi: 10.4331/wjbc.v1.i10.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bréart B, Ramos-Perez WD, Mendoza A, Salous AK, Gobert M, Huang Y, Adams RH, Lafaille JJ, Escalante-Alcalde D, Morris AJ, Schwab SR. Lipid phosphate phosphatase 3 enables efficient thymic egress. J Exp Med. 2011;208:1267–1278. doi: 10.1084/jem.20102551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jenne CN, Enders A, Rivera R, Watson SR, Bankovich AJ, Pereira JP, Xu Y, Roots CM, Beilke JN, Banerjee A, Reiner SL, Miller SA, Weinmann AS, Goodnow CC, Lanier LL, Cyster JG, Chun J. T-bet-dependent S1P5 expression in NK cells promotes egress from lymph nodes and bone marrow. J Exp Med. 2009;206:2469–2481. doi: 10.1084/jem.20090525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keul P, Lucke S, von Wnuck Lipinski K, Bode C, Graler M, Heusch G, Levkau B. Sphingosine-1-phosphate receptor 3 promotes recruitment of monocyte/macrophages in inflammation and atherosclerosis. Circ Res. 2011;108:314–323. doi: 10.1161/CIRCRESAHA.110.235028. [DOI] [PubMed] [Google Scholar]
- 9.Kupperman E, An S, Osborne N, Waldron S, Stainier DY. A sphingosine-1-phosphate receptor regulates cell migration during vertebrate heart development. Nature. 2000;406:192–195. doi: 10.1038/35018092. [DOI] [PubMed] [Google Scholar]
- 10.Mizugishi K, Yamashita T, Olivera A, Miller G, Spiegel S, Proia R. Essential role for sphingosine kinases in neural and vascular development. Mol Cell Biol. 2005;25:11113–11121. doi: 10.1128/MCB.25.24.11113-11121.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donati C, Meacci E, Nuti F, Becciolini L, Farnararo M, Bruni P. Sphingosine 1-phosphate regulates myogenic differentiation: a major role for S1P2 receptor. FASEB J. 2005;19:449–451. doi: 10.1096/fj.04-1780fje. [DOI] [PubMed] [Google Scholar]
- 12.Spiegel S, Milstien S. Functions of the multifaceted family of sphingosine kinases and some close relatives. J Biol Chem. 2006;282:2125–2129. doi: 10.1074/jbc.R600028200. [DOI] [PubMed] [Google Scholar]
- 13.Pyne S, Lee SC, Long J, Pyne NJ. Role of sphingosine kinases and lipid phosphate phosphatases in regulating spatial sphingosine 1-phosphate signalling in health and disease. Cell Signal. 2009;21:14–21. doi: 10.1016/j.cellsig.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 14.Pyne S, Long J, Ktistakis N, Pyne N. Lipid phosphate phosphatases and lipid phosphate signalling. Biochem Soc Trans. 2005;33:1370–1374. doi: 10.1042/BST0331370. [DOI] [PubMed] [Google Scholar]
- 15.Kumar A, Saba JD. Lyase to live by: sphingosine phosphate lyase as a therapeutic target. Expert Opin Ther Targets. 2009;13:1013–1025. doi: 10.1517/14728220903039722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bourquin F, Capitani G, Grutter MG. PLP-dependent enzymes as entry and exit gates of sphingolipid metabolism. Protein Sci. 2011;20:1492–1508. doi: 10.1002/pro.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siow D, Anderson C, Berdyshev E, Skobeleva A, Pitson S, Wattenberg B. Intra-cellular localization of sphingosine kinase 1 alters access to substrate pools but does not affect the degradative fate of sphingosine-1-phosphate. J Lipid Res. 2010;51:2546–2559. doi: 10.1194/jlr.M004374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vogel P, Donoviel MS, Read R, Hansen GM, Hazlewood J, Anderson SJ, Sun W, Swaffield J, Oravecz T. Incomplete inhibition of sphingosine 1-phosphate lyase modulates immune system function yet prevents early lethality and non-lymphoid lesions. PLoS One. 2009;4:e4112. doi: 10.1371/journal.pone.0004112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bektas M, Allende ML, Lee BG, Chen W, Amar MJ, Remaley AT, Saba JD, Proia RL. Sphingosine 1-phosphate lyase deficiency disrupts lipid homeostasis in liver. J Biol Chem. 2010;285:10880–10889. doi: 10.1074/jbc.M109.081489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Serra M, Saba JD. Sphingosine 1-phosphate lyase, a key regulator of sphingosine 1-phosphate signaling and function. Adv Enzyme Regul. 2010;50:349–362. doi: 10.1016/j.advenzreg.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bagdanoff JT, Donoviel MS, Nouraldeen A, Carlsen M, Jessop TC, Tarver J, Aleem S, Dong L, Zhang H, Boteju L, Hazelwood J, Yan J, Bednarz M, Layek S, Owusu IB, Gopinathan S, Moran L, Lai Z, Kramer J, Kimball SD, Yalamanchili P, Heydorn WE, Frazier KS, Brooks B, Brown P, Wilson A, Sonnenburg WK, Main A, Carson KG, Oravecz T, Augeri DJ. Inhibition of sphingosine 1-phosphate lyase for the treatment of rheumatoid arthritis: discovery of (E)-1-(4-((1R,2S,3R)-1,2,3,4-tetrahydroxybutyl)-1H-imidazol-2-yl)ethanone oxime (LX2931) and (1R,2S,3R)-1-(2-(isoxazol-3-yl)-1H-imidazol-4-yl)butane-1,2,3,4-tetraol (LX2932) J Med Chem. 2010;53:8650–8662. doi: 10.1021/jm101183p. [DOI] [PubMed] [Google Scholar]
- 22.Kumar A, Oskouian B, Fyrst H, Zhang M, Paris F, Saba JD. S1P lyase regulates DNA damage responses through a novel sphingolipid feedback mechanism. Cell Death Dis. 2011;2:e119. doi: 10.1038/cddis.2011.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar A, Byun HS, Bittman R, Saba JD. The sphingolipid degradation product trans-2-hexadecenal induces cytoskeletal reorganization and apoptosis in a JNK-dependent manner. Cell Signal. 2011;23:11346–11353. doi: 10.1016/j.cellsig.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huwiler A, Bourquin F, Kotelevets N, Pastukhov O, Capitani G, Grutter MG, Zangemeister-Wittke U. A prokaryotic S1P lyase degrades extracellular S1P in vitro and in vivo: implication for treating hyperproliferative disorders. PLoS One. 2011;6:e22436. doi: 10.1371/journal.pone.0022436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saba JD, Nara F, Bielawska A, Garrett S, Hannun YA. The BST1 gene of Saccharomyces cerevisiae is the sphingosine-1-phosphate lyase. J Biol Chem. 1997;272:26087–26090. doi: 10.1074/jbc.272.42.26087. [DOI] [PubMed] [Google Scholar]
- 26.Herr DR, Fyrst H, Phan V, Heinecke K, Georges R, Harris GL, Saba JD. Sply regulation of sphingolipid signaling molecules is essential for Drosophila development. Development. 2003;130:2443–2453. doi: 10.1242/dev.00456. [DOI] [PubMed] [Google Scholar]
- 27.Loh K, Leong W, Carlson M, Oskouian B, Kumar A, Fyrst H, Zhang M, Proia R, Hoffman E, Saba J. Sphingosine-1-phosphate enhances satellite cell activation in dystrophic muscles through a S1PR2/STAT3 signaling pathway. PLoS One. 2012;7:e37218. doi: 10.1371/journal.pone.0037218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Veldhoven PP. Sphingosine-1-phosphate lyase. In: Merrill AH Jr, Hannun YA, editors. Sphingolipid Metabolism and Cell Signaling Part A. Vol. 311. Academic Press; New York: 2000. pp. 244–254. [Google Scholar]
- 29.Graler MH. Targeting sphingosine 1-phosphate (S1P) levels and S1P receptor functions for therapeutic immune interventions. Cell Physiol Biochem. 2010;26:79–86. doi: 10.1159/000315108. [DOI] [PubMed] [Google Scholar]
- 30.Alexander S, Alexander H. Lead genetic studies in Dictyostelium discoideum and translational studies in human cells demonstrate that sphingolipids are key regulators of sensitivity to cisplatin and other anticancer drugs. Semin Cell Dev Biol. 2010;22:97–104. doi: 10.1016/j.semcdb.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 31.Aguilar A, Saba JD. Truth and consequences of sphingosine-1-phosphate lyase. Adv Enzyme Regul. 2012;52:17–30. doi: 10.1016/j.advenzreg.2011.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deconinck N, Dan B. Pathophysiology of Duchenne muscular dystrophy: current hypotheses. Pediatr Neurol. 2007;36:1–7. doi: 10.1016/j.pediatrneurol.2006.09.016. [DOI] [PubMed] [Google Scholar]
- 33.Hoffman EP, Brown RH, Jr, Kunkel LM. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- 34.Kanagawa M, Toda T. The genetic and molecular basis of muscular dystrophy: roles of cell-matrix linkage in the pathogenesis. J Hum Genet. 2006;51:915–926. doi: 10.1007/s10038-006-0056-7. [DOI] [PubMed] [Google Scholar]
- 35.Schessl J, Zou Y, Bonnemann CG. Congenital muscular dystrophies and the extracellular matrix. Semin Pediatr Neurol. 2006;13:80–89. doi: 10.1016/j.spen.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 36.Bansal D, Miyake K, Vogel SS, Groh S, Chen CC, Williamson R, McNeil PL, Campbell KP. Defective membrane repair in dysferlin-deficient muscular dystrophy. Nature. 2003;423:168–172. doi: 10.1038/nature01573. [DOI] [PubMed] [Google Scholar]
- 37.Heslop L, Morgan JE, Partridge TA. Evidence for a myogenic stem cell that is exhausted in dystrophic muscle. J Cell Sci. 2000;113(Pt 12):2299–2308. doi: 10.1242/jcs.113.12.2299. [DOI] [PubMed] [Google Scholar]
- 38.Evans NP, Misyak SA, Robertson JL, Bassaganya-Riera J, Grange RW. Immune-mediated mechanisms potentially regulate the disease time-course of duchenne muscular dystrophy and provide targets for therapeutic intervention. PM R. 2009;1:755–768. doi: 10.1016/j.pmrj.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoffman EP, Escolar D. Translating mighty mice into neuromuscular therapeutics: is bigger muscle better? Am J Pathol. 2006;168:1775–1778. doi: 10.2353/ajpath.2006.060270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Figeac N, Daczewska M, Marcelle C, Jagla K. Muscle stem cells and model systems for their investigation. Dev Dyn. 2007;236:3332–3342. doi: 10.1002/dvdy.21345. [DOI] [PubMed] [Google Scholar]
- 41.Ehrhardt J, Morgan J. Regenerative capacity of skeletal muscle. Curr Opin Neurol. 2005;18:548–553. doi: 10.1097/01.wco.0000177382.62156.82. [DOI] [PubMed] [Google Scholar]
- 42.Grefte S, Kuijpers-Jagtman AM, Torensma R, Von den Hoff JW. Skeletal muscle development and regeneration. Stem Cells Dev. 2007;16:857–868. doi: 10.1089/scd.2007.0058. [DOI] [PubMed] [Google Scholar]
- 43.Kuang S, Kuroda K, Le Grand F, Rudnicki MA. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell. 2007;129:999–1010. doi: 10.1016/j.cell.2007.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagata Y, Partridge T, Matsuda R, Zammit P. Entry of muscle satellite cells into the cell cycle requires sphingolipid signaling. J Cell Biol. 2006;174:245–253. doi: 10.1083/jcb.200605028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Danieli-Betto D, Germinario E, Esposito A, Megighian A, Midrio M, Ravara B, Damiani E, Libera LD, Sabbadini RA, Betto R. Sphingosine 1-phosphate protects mouse extensor digitorum longus skeletal muscle during fatigue. Am J Physiol Cell Physiol. 2005;288:C1367–C1373. doi: 10.1152/ajpcell.00246.2004. [DOI] [PubMed] [Google Scholar]
- 46.Zanin M, Germinario E, Dalla Libera L, Sandona D, Sabbadini RA, Betto R, Danieli-Betto D. Trophic action of sphingosine 1-phosphate in denervated rat soleus muscle. Am J Physiol Cell Physiol. 2008;294:C36–C46. doi: 10.1152/ajpcell.00164.2007. [DOI] [PubMed] [Google Scholar]
- 47.Bruni P, Donati C. Pleiotropic effects of sphingolipids in skeletal muscle. Cell Mol Life Sci. 2008;65:3725–3736. doi: 10.1007/s00018-008-8236-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi X, Garry DJ. Muscle stem cells in development, regeneration, and disease. Genes Dev. 2006;20:1692–1708. doi: 10.1101/gad.1419406. [DOI] [PubMed] [Google Scholar]
- 49.Horsley V, Pavlath GK. Forming a multinucleated cell: molecules that regulate myoblast fusion. Cells Tissues Organs. 2004;176:67–78. doi: 10.1159/000075028. [DOI] [PubMed] [Google Scholar]
- 50.Shcherbata HR, Yatsenko AS, Patterson L, Sood VD, Nudel U, Yaffe D, Baker D, Ruohola-Baker H. Dissecting muscle and neuronal disorders in a Drosophila model of muscular dystrophy. EMBO J. 2007;26:481–493. doi: 10.1038/sj.emboj.7601503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bandhuvula P, Honbo N, Wang GY, Jin ZQ, Fyrst H, Zhang M, Borowsky AD, Dillard L, Karliner JS, Saba JD. S1P lyase: a novel therapeutic target for ischemia-reperfusion injury of the heart. Am J Physiol Heart Circ Physiol. 2011;300:H1753–H1761. doi: 10.1152/ajpheart.00946.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao P, Iezzi S, Carver E, Dressman D, Gridley T, Sartorelli V, Hoffman E. Slug is a novel downstream target of MyoD. Temporal profiling in muscle regeneration. J Biol Chem. 2002;277:30091–30101. doi: 10.1074/jbc.M202668200. [DOI] [PubMed] [Google Scholar]
- 53.Plant DR, Colarossi FE, Lynch GS. Notexin causes greater myotoxic damage and slower functional repair in mouse skeletal muscles than bupivacaine. Muscle Nerve. 2006;34:577–585. doi: 10.1002/mus.20616. [DOI] [PubMed] [Google Scholar]
- 54.Olivera A, Eisner C, Kitamura Y, Dillahunt S, Allende L, Tuymetova G, Watford W, Meylan F, Diesner SC, Li L, Schnermann J, Proia RL, Rivera J. Sphingosine kinase 1 and sphingosine-1-phosphate receptor 2 are vital to recovery from anaphy-lactic shock in mice. J Clin Invest. 2010;120:1429–1440. doi: 10.1172/JCI40659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Karliner JS. Sphingosine kinase and sphingosine 1-phosphate in cardioprotection. J Cardiovasc Pharmacol. 2009;53:189–197. doi: 10.1097/FJC.0b013e3181926706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jo SK, Bajwa A, Ye H, Vergis AL, Awad AS, Kharel Y, Lynch KR, Okusa MD. Divergent roles of sphingosine kinases in kidney ischemia-reperfusion injury. Kidney Int. 2009;75:167–175. doi: 10.1038/ki.2008.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pitson SM. Regulation of sphingosine kinase and sphingolipid signaling. Trends Biochem Sci. 2011;36:97–107. doi: 10.1016/j.tibs.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 58.Bulfield G, Siller WG, Wight PA, Moore KJ. X chromosome-linked muscular dystrophy (mdx) in the mouse. Proc Natl Acad Sci U S A. 1984;81:1189–1192. doi: 10.1073/pnas.81.4.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Connolly AM, Keeling RM, Mehta S, Pestronk A, Sanes JR. Three mouse models of muscular dystrophy: the natural history of strength and fatigue in dystrophin-, dystrophin/utrophin-, and laminin a2-deficient mice. Neuromuscul Disord. 2001;11:703–712. doi: 10.1016/s0960-8966(01)00232-2. [DOI] [PubMed] [Google Scholar]
- 60.Cardoso DF, Lopes-Ferreira M, Faquim-Mauro EL, Macedo MS, Farsky SH. Role of crotoxin, a phospholipase A2 isolated from Crotalus durissus terrificus snake venom, on inflammatory and immune reactions. Mediators Inflamm. 2001;10:125–133. doi: 10.1080/09629350124986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Emslie-Smith AM, Harris JB. Lymphopaenia in the inflammatory response caused by notechis II-5. Toxicon. 1989;27:499–500. doi: 10.1016/0041-0101(89)90110-4. [DOI] [PubMed] [Google Scholar]
- 62.Schwab S, Pereira J, Matloubian M, Xu Y, Huang Y, Cyster J. Lymphocyte sequestration through S1P lyase inhibition an disruption of S1P gradients. Science. 2005;309:1735–1739. doi: 10.1126/science.1113640. [DOI] [PubMed] [Google Scholar]
- 63.Spencer MJ, Montecino-Rodriguez E, Dorshkind K, Tidball JG. Helper (CD4(+)) and cytotoxic (CD8(+)) T cells promote the pathology of dystrophin-deficient muscle. Clin Immunol. 2001;98:235–243. doi: 10.1006/clim.2000.4966. [DOI] [PubMed] [Google Scholar]
- 64.Eghtesad S, Jhunjhunwala S, Little SR, Clemens PR. Rapamycin ameliorates dystrophic phenotypeinmdx mouseskeletal muscle. Mol Med. 2011;17:917–924. doi: 10.2119/molmed.2010.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee H, Deng J, Kujawski M, Yang C, Liu Y, Herrmann A, Kortylewski M, Horne D, Somlo G, Forman S, Jove R, Yu H. STAT3-induced S1PR1 expression is crucial for persistent STAT3 activation in tumors. Nat Med. 2010;16:1421–1428. doi: 10.1038/nm.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sekine Y, Suzuki K, Remaley AT. HDL and sphingosine-1-phosphate activate stat3 in prostate cancer DU145 cells via ERK1/2 and S1P receptors, and promote cell migration and invasion. Prostate. 2011;71:690–699. doi: 10.1002/pros.21285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Frias MA, James RW, Gerber-Wicht C, Lang U. Native and reconstituted HDL activate Stat3 in ventricular cardiomyocytes via ERK1/2: role of sphingosine-1-phosphate. Cardiovasc Res. 2009;82:313–323. doi: 10.1093/cvr/cvp024. [DOI] [PubMed] [Google Scholar]
- 68.Calise S, Blescia S, Cencetti F, Bernacchioni C, Donati C, Bruni P. Sphingosine 1-phosphate stimulates proliferation and migration of satellite cells: role of S1P receptors. Biochim Biophys Acta. 2012;1823:439–450. doi: 10.1016/j.bbamcr.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 69.Meacci E, Vasta V, Donati C, Farnararo M, Bruni P. Receptor-mediated ctivation of phospholipase D by sphingosine 1-phosphate in skeletal muscle C2C12 cells. A role for protein kinase C. FEBS Lett. 1999;457:184–188. doi: 10.1016/s0014-5793(99)01033-9. [DOI] [PubMed] [Google Scholar]
- 70.Meacci E, Donati C, Cencetti F, Romiti E, Bruni P. Permissive role of protein kinase C alpha but not protein kinase C delta in sphingosine 1-phosphate-induced Rho A activation in C2C12 myoblasts. FEBS Lett. 2000;482:97–101. doi: 10.1016/s0014-5793(00)02039-1. [DOI] [PubMed] [Google Scholar]
- 71.Meacci E, Cencetti F, Formigli L, Squecco R, Donati C, Tiribilli B, Quercioli F, Zecchi Orlandini S, Francini F, Bruni P. Sphingosine 1-phosphate evokes calcium signals in C2C12 myoblasts via Edg3 and Edg5 receptors. Biochem J. 2002;362:349–357. doi: 10.1042/0264-6021:3620349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Meacci E, Cencetti F, Donati C, Nuti F, Farnararo M, Kohno T, Igarashi Y, Bruni P. Down-regulation of EDG5/S1P2 during myogenic differentiation results in the specific uncoupling of sphingosine 1-phosphate signalling to phospholipase D. Biochim Biophys Acta. 2003;1633:133–142. doi: 10.1016/s1388-1981(03)00106-9. [DOI] [PubMed] [Google Scholar]
- 73.Danieli-Betto D, Peron S, Germinario E, Zanin M, Sorci G, Franzoso S, Sandona D, Betto R. Sphingosine 1-phosphate signaling is involved in skeletal muscle regeneration. Am J Physiol Cell Physiol. 2010;298:C550–C558. doi: 10.1152/ajpcell.00072.2009. [DOI] [PubMed] [Google Scholar]
- 74.Fan Y, Maley M, Beilharz M, Grounds M. Rapid death of injected myoblasts in myoblast transfer therapy. Muscle Nerve. 1996;19:853–860. doi: 10.1002/(SICI)1097-4598(199607)19:7<853::AID-MUS7>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 75.Beauchamp JR, Pagel CN, Partridge TA. A dual-marker system for quantitative studies of myoblast transplantation in the mouse. Transplantation. 1997;63:1794–1797. doi: 10.1097/00007890-199706270-00015. [DOI] [PubMed] [Google Scholar]
- 76.Zheng B, Cao B, Crisan M, Sun B, Li G, Logar A, Yap S, Pollett JB, Drowley L, Cassino T, Gharaibeh B, Deasy BM, Huard J, Peault B. Prospective identification of myogenic endothelial cells in human skeletal muscle. Nat Biotechnol. 2007;25:1025–1034. doi: 10.1038/nbt1334. [DOI] [PubMed] [Google Scholar]
- 77.Sampaolesi M, Blot S, D'Antona G, Granger N, Tonlorenzi R, Innocenzi A, Mognol P, Thibaud JL, Galvez BG, Barthelemy I, Perani L, Mantero S, Guttinger M, Pansarasa O, Rinaldi C, Cusella De Angelis MG, Torrente Y, Bordignon C, Bottinelli R, Cossu G. Mesoangioblast stem cells ameliorate muscle function in dystrophic dogs. Nature. 2006;444:574–579. doi: 10.1038/nature05282. [DOI] [PubMed] [Google Scholar]
- 78.Sampaolesi M, Torrente Y, Innocenzi A, Tonlorenzi R, D'Antona G, Pellegrino MA, Barresi R, Bresolin N, De Angelis MG, Campbell KP, Bottinelli R, Cossu G. Cell therapy of alpha-sarcoglycan null dystrophic mice through intra-arterial delivery of mesoangioblasts. Science. 2003;301:487–492. doi: 10.1126/science.1082254. [DOI] [PubMed] [Google Scholar]
- 79.Crisan M, Yap S, Casteilla L, Chen CW, Corselli M, Park TS, Andriolo G, Sun B, Zheng B, Zhang L, Norotte C, Teng PN, Traas J, Schugar R, Deasy BM, Badylak S, Buhring HJ, Giacobino JP, Lazzari L, Huard J, Peault B. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell. 2008;3:301–313. doi: 10.1016/j.stem.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 80.Rodriguez AM, Pisani D, Dechesne CA, Turc-Carel C, Kurzenne JY, Wdziekonski B, Villageois A, Bagnis C, Breittmayer JP, Groux H, Ailhaud G, Dani C. Transplantation of a multipotent cell population from human adipose tissue induces dystrophin expression in the immunocompetent mdx mouse. J Exp Med. 2005;201:1397–1405. doi: 10.1084/jem.20042224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dezawa M, Ishikawa H, Itokazu Y, Yoshihara T, Hoshino M, Takeda S, Ide C, Nabeshima Y. Bone marrow stromal cells generate muscle cells and repair muscle degeneration. Science. 2005;309:314–317. doi: 10.1126/science.1110364. [DOI] [PubMed] [Google Scholar]
- 82.De Bari C, Dell'Accio F, Vandenabeele F, Vermeesch JR, Raymackers JM, Luyten FP. Skeletal muscle repair by adult human mesenchymal stem cells from synovial membrane. J Cell Biol. 2003;160:909–918. doi: 10.1083/jcb.200212064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Donati C, Cencetti F, Nincheri P, Bernacchioni C, Brunelli S, Clementi E, Cossu G, Bruni P. Sphingosine 1-phosphate mediates proliferation and survival of mesoangioblasts. Stem Cells. 2007;25:1713–1719. doi: 10.1634/stemcells.2006-0725. [DOI] [PubMed] [Google Scholar]
- 84.Nincheri P, Luciani P, Squecco R, Donati C, Bernacchioni C, Borgognoni L, Luciani G, Benvenuti S, Francini F, Bruni P. Sphingosine 1-phosphate induces differentiation of adipose tissue-derived mesenchymal stem cells towards smooth muscle cells. Cell Mol Life Sci. 2009;66:1741–1754. doi: 10.1007/s00018-009-9181-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.de la Garza-Rodea AS. Doctoral Thesis. Leiden University Medical Center, Leiden University; The Netherlands: 2011. Mesenchymal stem cell in skeletal muscle regeneration; p. 15. [Google Scholar]