Abstract
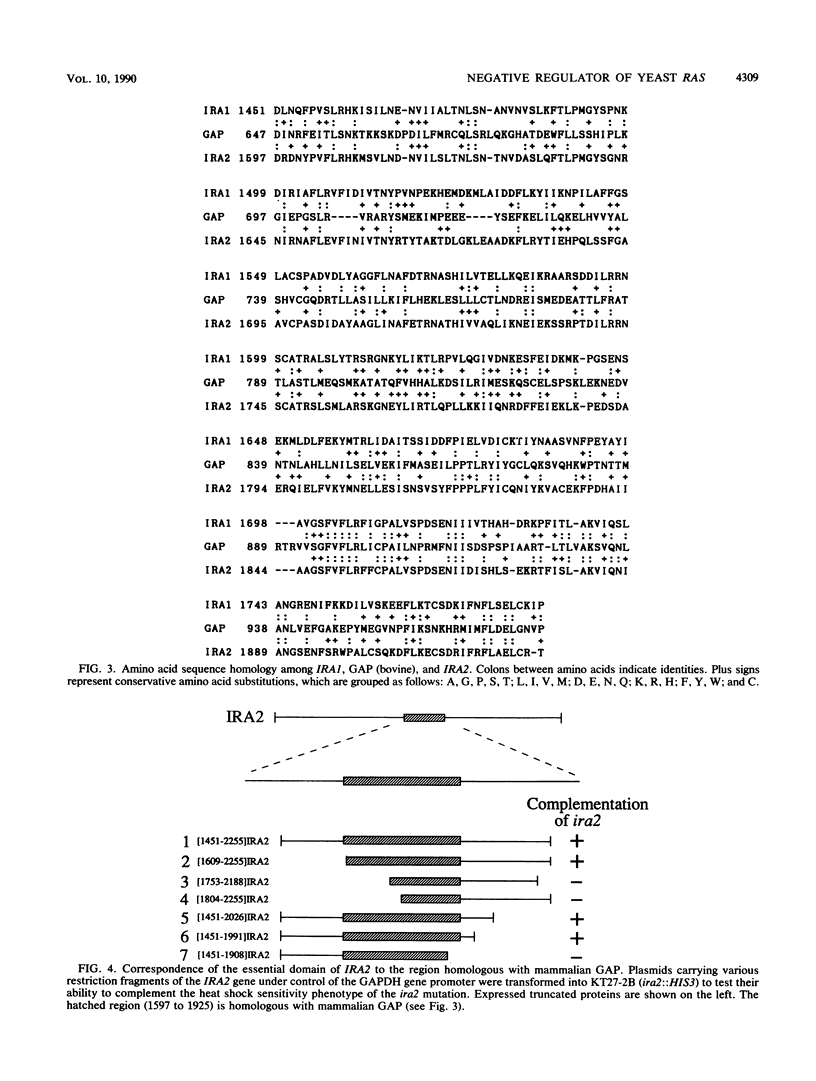
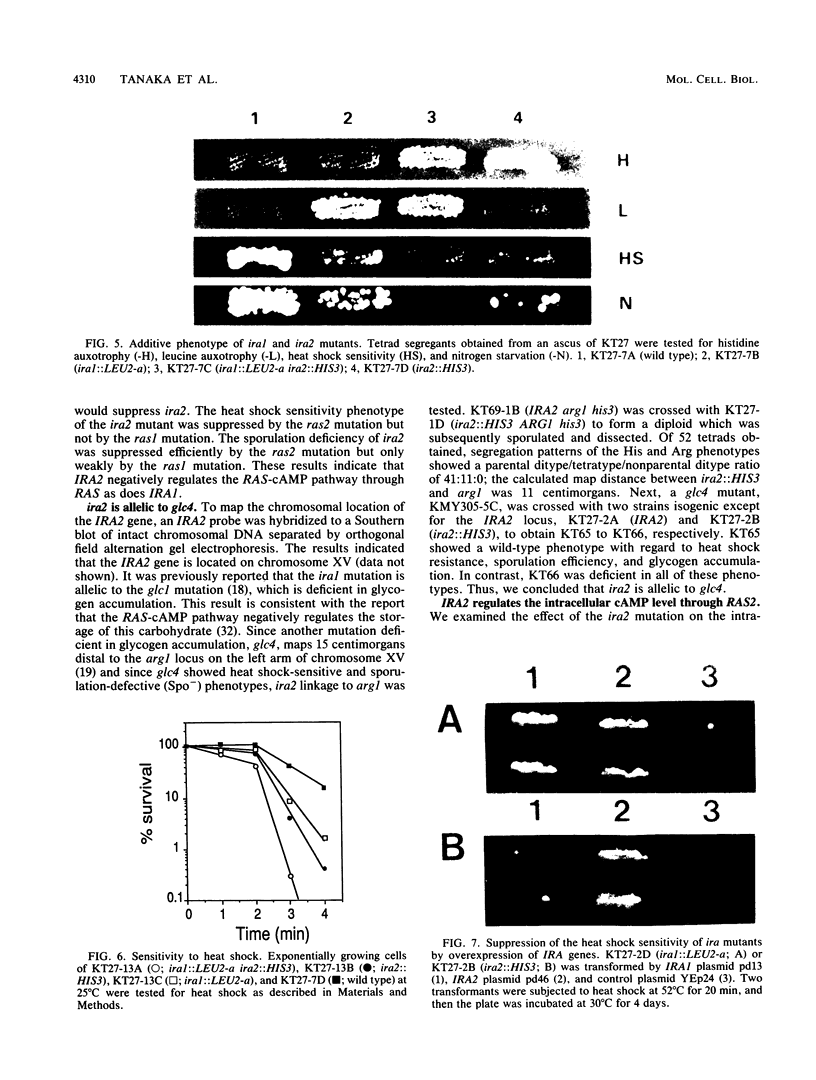
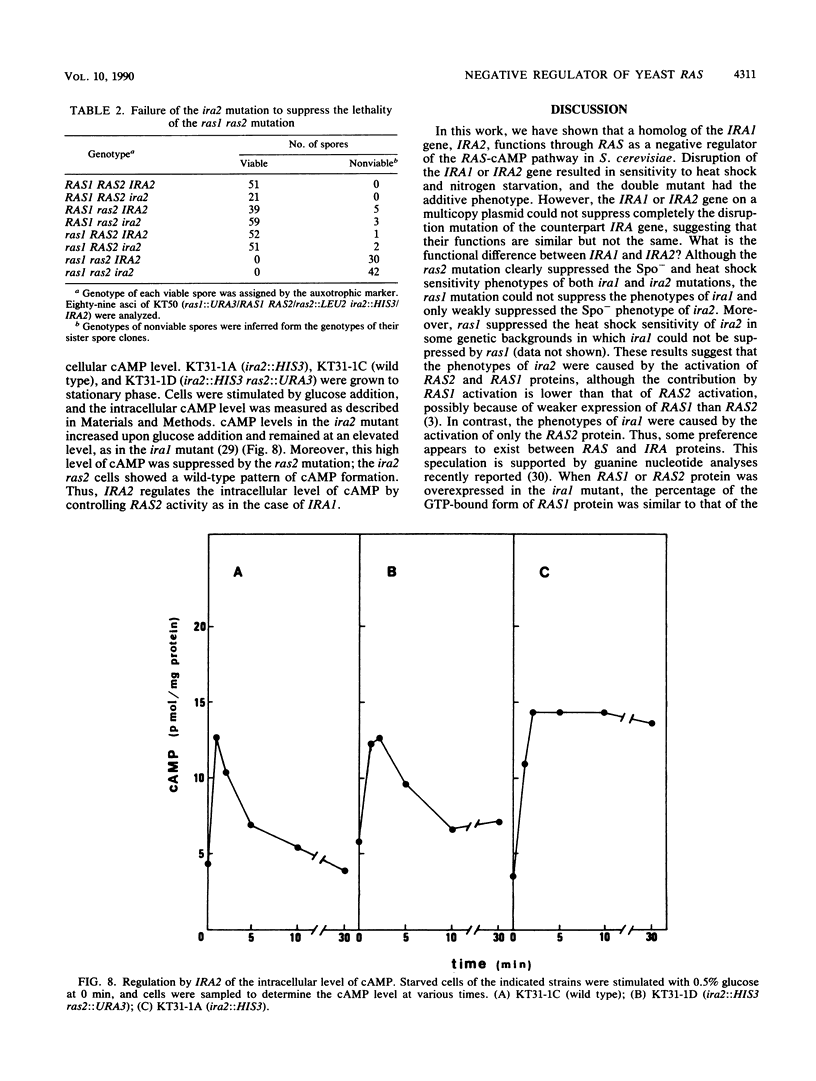
The IRA1 gene is a negative regulator of the RAS-cyclic AMP pathway in Saccharomyces cerevisiae. To identify other genes involved in this pathway, we screened yeast genomic DNA libraries for genes that can suppress the heat shock sensitivity of the ira1 mutation on a multicopy vector. We identified IRA2, encoding a protein of 3,079 amino acids, that is 45% identical to the IRA1 protein. The region homologous between the IRA1 protein and ras GTPase-activating protein is also conserved in IRA2. IRA2 maps 11 centimorgans distal to the arg1 locus on the left arm of chromosome XV and was found to be allelic to glc4. Disruption of the IRA2 gene resulted in (i) increased sensitivity to heat shock and nitrogen starvation, (ii) sporulation defects, and (iii) suppression of the lethality of the cdc25 mutant. Analysis of disruption mutants of IRA1 and IRA2 indicated that IRA1 and IRA2 proteins additively regulate the RAS-cyclic AMP pathway in a negative fashion. Expression of the IRA2 domain homologous with GAP is sufficient for complementation of the heat shock sensitivity of ira2, suggesting that IRA down regulates RAS activity by stimulating the GTPase activity of RAS proteins.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ballester R., Michaeli T., Ferguson K., Xu H. P., McCormick F., Wigler M. Genetic analysis of mammalian GAP expressed in yeast. Cell. 1989 Nov 17;59(4):681–686. doi: 10.1016/0092-8674(89)90014-7. [DOI] [PubMed] [Google Scholar]
- Barbacid M. ras genes. Annu Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- Breviario D., Hinnebusch A. G., Dhar R. Multiple regulatory mechanisms control the expression of the RAS1 and RAS2 genes of Saccharomyces cerevisiae. EMBO J. 1988 Jun;7(6):1805–1813. doi: 10.1002/j.1460-2075.1988.tb03012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broek D., Samiy N., Fasano O., Fujiyama A., Tamanoi F., Northup J., Wigler M. Differential activation of yeast adenylate cyclase by wild-type and mutant RAS proteins. Cell. 1985 Jul;41(3):763–769. doi: 10.1016/s0092-8674(85)80057-x. [DOI] [PubMed] [Google Scholar]
- Broek D., Toda T., Michaeli T., Levin L., Birchmeier C., Zoller M., Powers S., Wigler M. The S. cerevisiae CDC25 gene product regulates the RAS/adenylate cyclase pathway. Cell. 1987 Mar 13;48(5):789–799. doi: 10.1016/0092-8674(87)90076-6. [DOI] [PubMed] [Google Scholar]
- Carlson M., Botstein D. Two differentially regulated mRNAs with different 5' ends encode secreted with intracellular forms of yeast invertase. Cell. 1982 Jan;28(1):145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Daniel J., Becker J. M., Enari E., Levitzki A. The activation of adenylate cyclase by guanyl nucleotides in Saccharomyces cerevisiae is controlled by the CDC25 start gene product. Mol Cell Biol. 1987 Oct;7(10):3857–3861. doi: 10.1128/mcb.7.10.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFeo-Jones D., Scolnick E. M., Koller R., Dhar R. ras-Related gene sequences identified and isolated from Saccharomyces cerevisiae. Nature. 1983 Dec 15;306(5944):707–709. doi: 10.1038/306707a0. [DOI] [PubMed] [Google Scholar]
- Field J., Nikawa J., Broek D., MacDonald B., Rodgers L., Wilson I. A., Lerner R. A., Wigler M. Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol Cell Biol. 1988 May;8(5):2159–2165. doi: 10.1128/mcb.8.5.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs J. B., Marshall M. S. The ras oncogene--an important regulatory element in lower eucaryotic organisms. Microbiol Rev. 1989 Jun;53(2):171–185. doi: 10.1128/mr.53.2.171-185.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs E. G., Beavo J. A. Phosphorylation-dephosphorylation of enzymes. Annu Rev Biochem. 1979;48:923–959. doi: 10.1146/annurev.bi.48.070179.004423. [DOI] [PubMed] [Google Scholar]
- Marshall M. S., Gibbs J. B., Scolnick E. M., Sigal I. S. Regulatory function of the Saccharomyces cerevisiae RAS C-terminus. Mol Cell Biol. 1987 Jul;7(7):2309–2315. doi: 10.1128/mcb.7.7.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall M. S., Hill W. S., Ng A. S., Vogel U. S., Schaber M. D., Scolnick E. M., Dixon R. A., Sigal I. S., Gibbs J. B. A C-terminal domain of GAP is sufficient to stimulate ras p21 GTPase activity. EMBO J. 1989 Apr;8(4):1105–1110. doi: 10.1002/j.1460-2075.1989.tb03480.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K., Uno I., Ishikawa T. Genetic analysis of the role of cAMP in yeast. Yeast. 1985 Sep;1(1):15–24. doi: 10.1002/yea.320010103. [DOI] [PubMed] [Google Scholar]
- Matsumoto K., Uno I., Kato K., Ishikawa T. Isolation and characterization of a phosphoprotein phosphatase-deficient mutant in yeast. Yeast. 1985 Sep;1(1):25–38. doi: 10.1002/yea.320010104. [DOI] [PubMed] [Google Scholar]
- Mortimer R. K., Schild D. Genetic map of Saccharomyces cerevisiae, edition 9. Microbiol Rev. 1985 Sep;49(3):181–213. doi: 10.1128/mr.49.3.181-213.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakafuku M., Obara T., Kaibuchi K., Miyajima I., Miyajima A., Itoh H., Nakamura S., Arai K., Matsumoto K., Kaziro Y. Isolation of a second yeast Saccharomyces cerevisiae gene (GPA2) coding for guanine nucleotide-binding regulatory protein: studies on its structure and possible functions. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1374–1378. doi: 10.1073/pnas.85.5.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikawa J., Cameron S., Toda T., Ferguson K. M., Wigler M. Rigorous feedback control of cAMP levels in Saccharomyces cerevisiae. Genes Dev. 1987 Nov;1(9):931–937. doi: 10.1101/gad.1.9.931. [DOI] [PubMed] [Google Scholar]
- Powers S., Kataoka T., Fasano O., Goldfarb M., Strathern J., Broach J., Wigler M. Genes in S. cerevisiae encoding proteins with domains homologous to the mammalian ras proteins. Cell. 1984 Mar;36(3):607–612. doi: 10.1016/0092-8674(84)90340-4. [DOI] [PubMed] [Google Scholar]
- Powers S., O'Neill K., Wigler M. Dominant yeast and mammalian RAS mutants that interfere with the CDC25-dependent activation of wild-type RAS in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Feb;9(2):390–395. doi: 10.1128/mcb.9.2.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Ruggieri R., Tanaka K., Nakafuku M., Kaziro Y., Toh-e A., Matsumoto K. MSI1, a negative regulator of the RAS-cAMP pathway in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1989 Nov;86(22):8778–8782. doi: 10.1073/pnas.86.22.8778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Nucleotide sequence and transcriptional mapping of the yeast pet56-his3-ded1 gene region. Nucleic Acids Res. 1985 Dec 9;13(23):8587–8601. doi: 10.1093/nar/13.23.8587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Matsumoto K., Toh-E A. IRA1, an inhibitory regulator of the RAS-cyclic AMP pathway in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Feb;9(2):757–768. doi: 10.1128/mcb.9.2.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Matsumoto K., Toh-e A. Dual regulation of the expression of the polyubiquitin gene by cyclic AMP and heat shock in yeast. EMBO J. 1988 Feb;7(2):495–502. doi: 10.1002/j.1460-2075.1988.tb02837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Nakafuku M., Satoh T., Marshall M. S., Gibbs J. B., Matsumoto K., Kaziro Y., Toh-e A. S. cerevisiae genes IRA1 and IRA2 encode proteins that may be functionally equivalent to mammalian ras GTPase activating protein. Cell. 1990 Mar 9;60(5):803–807. doi: 10.1016/0092-8674(90)90094-u. [DOI] [PubMed] [Google Scholar]
- Tatchell K., Robinson L. C., Breitenbach M. RAS2 of Saccharomyces cerevisiae is required for gluconeogenic growth and proper response to nutrient limitation. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3785–3789. doi: 10.1073/pnas.82.11.3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T., Uno I., Ishikawa T., Powers S., Kataoka T., Broek D., Cameron S., Broach J., Matsumoto K., Wigler M. In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell. 1985 Jan;40(1):27–36. doi: 10.1016/0092-8674(85)90305-8. [DOI] [PubMed] [Google Scholar]
- Trahey M., McCormick F. A cytoplasmic protein stimulates normal N-ras p21 GTPase, but does not affect oncogenic mutants. Science. 1987 Oct 23;238(4826):542–545. doi: 10.1126/science.2821624. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]