Abstract
Background
Solar radiation should be avoided in melanoma patients. Nevertheless, this is the main means by which the body produces vitamin D. Evidence suggests a protective role against cancer for vitamin D. Since vitamin D performs its function by binding the receptor encoded by the vitamin D-receptor gene (VDR), most studies have focused on polymorphisms (SNPs) within this gene. However, the gene encoding the vitamin D-binding protein (GC) appears in recent studies as a major player in the role of a serum vitamin D level regulator and in Cutaneous Melanoma (CM) predisposition.
Methods
We performed a case-control study of 12 polymorphisms on GC and 9 on VDR among 530 cases and 314 controls from Spanish population.
Results
We found association between SNP rs12512631, located 3′downstream of GC, and risk of CM that seems to fit a dominant model (OR 1.63 95%CI 1.23–2.17 p-value 7×10−4). This association remained Bonferroni’s correction and after adjustment for potential confounders (p-value 3×10−3) and even after increasing the sample size to 1729 individuals (p-value 0.0129). Moreover, we confirmed evidence of an association between CM susceptibility and the linkage disequilibrium block marked by tag-SNP rs222016 (p-value 0.032). This block covers the GC intron 1 region, with probable regulatory functions.
Conclusion
To our knowledge, this is the first vitamin D pathway-related polymorphism study in melanoma risk conducted in the Spanish population. Furthermore, we show an association between polymorphisms in GC and melanoma risk, confirming recent studies in different populations.
Introduction
Cutaneous Melanoma (CM) is caused by the malignant transformation of melanocytes, pigment producer cells, located within epidermal basal cells. These cells produce melanin as a response to UV radiation. Melanoma incidence has been increasing at higher rates than any other malignant tumour in recent decades, and it causes the greatest number of skin-cancer-related deaths worldwide [1]. The main reason for the recent rise in CM incidence is attributed mainly to increased intermittent exposure to UV radiation.
The data about whether sun exposure in Caucasian populations results in health risks or benefits is controversial. Solar radiation is the main source of vitamin D synthesis in humans; however, uncontrolled and intensive sun exposure is dangerous to health and contributes to the development of melanoma [2], [3]. Given that occupational sun exposure has been reported to protect against melanoma, that solar radiation is the main means by which the body produces vitamin D, and that vitamin D is reported to have anti-proliferative effects in melanoma cells, interest has arisen over a possible role for vitamin D in melanoma prevention [4].
The most studied gene in the vitamin D pathway is the vitamin D receptor gene (VDR) located on chromosome 12. The gene has 11 exons and encodes the receptor of the calcitriol form of vitamin D. This receptor is a nuclear transcription and regulating factor that belongs to the steroid hormone superfamily of receptors. Nuclear receptors integrate hormonal, dietary, and other extracellular signals into cell fate decisions via regulation of gene expression and repression of a host of common gene targets. Moreover, many studies have addressed the polymorphisms of the vitamin D receptor in several cancers including ovarian carcinoma [5], breast cancer [6], colorectal cancer [7], non-Hodgkin lymphoma [8], renal cancer [9], oral squamous cell carcinoma [10], esophageal adenocarcinoma [11], non-small cell lung cancer [12], prostate cancer [13], and melanoma [14], [15].
Most recently, genome-wide association studies (GWAS) reported that a SNP in the GC gene is associated with serum levels. This gene encodes for the group-specific component or VDBP (vitamin D-binding protein), with the additional probable involvement of genes related to the production of the active form of vitamin D [16]. VDBP belongs to the albumin family, together with human serum albumin and alpha-fetoprotein. Located on chromosome 4q11–q13, the GC gene is 42.5 kb long and contains 13 exons. At least six non-synonymous SNPs are described, two of them with common frequency (rs7041 and rs4588). The VDBP greatly facilitates vitamin D actions by carrying vitamin D metabolites to various sites of action, while polymorphic VDBP proteins differ in their affinity for 1,25(OH)2 D metabolite [17]. VDBP has been related to multiple sclerosis [18], its association with various lung diseases including asthma, chronic obstructive pulmonary disease and tuberculosis has also been studied [19]. Recent studies, including GWAS, have shown that allelic variation in the GC gene is associated with both VDBP and serum 25(OH)D concentrations, as well as a higher affinity of the VDBP to vitamin D metabolites [16], [20].
Despite the large number of studies evaluating the association between VDR variants and CM, the conclusions on its role in the aetiology are still indecisive. The association between GC variants and vitamin D levels in plasma has already been tested and proved [16], [20], but only one study supports the association between polymorphisms on GC and CM [21]. In the present study, we show a comprehensive analysis of VDR and GC association with CM using data from a wide case-control study (530 melanoma cases and 314 controls) in a Spanish population. Data from VDR have been enlarged in sample size and density of SNPs from previous results [22]. We consider that the present study increases our knowledge of the relationship between vitamin D levels and CM predisposition in a Southern European country such as Spain, where sun incidence is higher than in northern European countries.
Materials and Methods
Ethics Statement
All participants gave written informed consent for this study. The study was approved by the Ethics Committee of Gregorio Marañón University General Hospital.
Study Subjects and Data Collection
A total of 530 sporadic CM patients were recruited from 1st September 2004 to the present, at the departments of Dermatology of three different hospitals in Madrid: Gregorio Marañón University General Hospital, La Paz University Hospital and Ramón y Cajal University Hospital. A total of 314 volunteer cancer-free control samples, frequency matched to cases by sex and age in ten-year categories, were recruited from the Madrid College of Lawyers, the National Cancer Research Centre (CNIO) and from the Gregorio Marañón University General Hospital (Table 1). All participants were non-related Caucasians of Spanish origin, with the same ethnic background [23].
Table 1. Sample distribution.
| Phase | Institution of origin for samples | Cases N (%) | Controls N (%) |
| Set I | Gregorio Marañon Hospital | 194 (36.61) | 94 (29.94) |
| Ramón y Cajal Hospital | 172 (32.45) | 0 (0.00) | |
| La Paz Hospital | 120 (22.64) | 10 (3.18) | |
| Madrid College of Lawyers/CNIO | 0 (0.00) | 237 (75.48) | |
| Total | 530 | 314 | |
| Set II | Valencian Institute of Oncology (IVO) | 344 (66.80) | 158 (42.70) |
| Dr. Negrín Hospital | 211 (40.97) | 172 (46.49) | |
| Total | 515 | 370 | |
| Total | 1045 | 684 |
CNIO, National Cancer Research Centre, Madrid.
All hospital participants in Set I are from Madrid. The IVO is in Valencia and Dr Negrín Hospital is located in Las Palmas de Gran Canaria, on Canary Islands, as are the populations sampled for each hospital.
The percentage is calculated from the total of cases or controls, respectively, for each phase.
A standardized questionnaire was used to collect information on pigmentation characteristics such as eye, hair and skin colour, number of naevi, presence of lentigines, sun exposure habits, and personal and family history of melanoma, cancer or any other skin disease. Each individual questionnaire has been guided by an expert clinician or a trained nurse. For cases only, tumour characteristics were added and medical data were obtained via medical exploration, those patients with acral or multiple melanoma were excluded from the study, as well as control individuals with suspected personal or family history of melanoma. We selected the following variables: age at diagnosis, sex, eye colour categorized as blue, grey and light green (light eye colour), hazel, light brown, brown and black (dark eye colour), hair was grouped as light colour (very light blonde and red-haired) and dark colour (light brown, medium brown and black) the skin colour stratification was made by the clinicians implicated in the project as very light and never tans, light, medium and dark, however to compare between cases and controls we only show two main categories, fair skin color (very light and never tans and light) and dark skin color (medium and dark), number of naevi (less than 25, between 25–50, between 50 and 100 and more than 100), presence of solar lentigines (yes, no and only on shoulders) and childhood sunburn events (categorization of these variables as well as the distribution of the Spanish population sampled are shown in Table 2).
Table 2. Classification of the Spanish samples studied by age, sex and phenotype.
| Characteristic | Controls (N = 314) n (%) | Cases (N = 530) n (%) | P-Fisher | OR (95% CI) | P-value* |
| Age at diagnosis (years) | 0.223 | 1.23 (0.90–1.69) | 0.201 | ||
| <Mean | 126 (40.13) | 233 (43.96) | |||
| ≥Mean | 97 (30.89) | 226 (42.64) | |||
| Unknown | 91 (28.98) | 71 (13.40) | |||
| Mean (SD) | 52.94 (16.02) | 52.32 (15.96) | |||
| Sex | 0.667 | 0.96 (0.72–1.27) | 0.751 | ||
| Men | 143 (45.54) | 237 (44.72) | |||
| Women | 166 (52.87) | 288 (54.34) | |||
| Unknown | 5 (1.59) | 5 (0.94) | |||
| Eye colour | 2.67×10 −07 | 2.44 (1.77–3.36) | 4.96×10 −08 | ||
| Dark eye colour | 235 (74.84) | 307 (57.92) | |||
| Light eye colour | 68 (21.66) | 217 (40.94) | |||
| Unknown | 11 (3.50) | 6 (1.13) | |||
| Hair colour | 1.13×10 −08 | 3.37 (2.15–5.28) | 8.43×10 −08 | ||
| Brown/Black | 284 (90.45) | 398 (75.09) | |||
| Blond/Red | 25 (7.96) | 122 (23.02) | |||
| Unknown | 5 (1.59) | 10 (1.89) | |||
| Skin colour | 0.076 | 1.16 (0.83–1.49) | 0.452 | ||
| Dark skin colour | 132 (42.04) | 213 (40.19) | |||
| Fair skin colour | 171 (54.46) | 308 (58.11) | |||
| Unknown | 11 (3.50) | 9 (1.70) | |||
| Lentigines | 1.54×10 −14 | 3.39 (2.50–4.59) | 3.86×10 −15 | ||
| No | 157 (50.00) | 142(26.79) | |||
| Yes | 124 (39.49) | 380(71.70) | |||
| Unknown | 33 (10.51) | 8(1.51) | |||
| Number of Naevi | 0.100 | 1.53 (0.98–2.40) | 0.059 | ||
| <50 | 245 (78.03) | 441(83.21) | |||
| ≥50 | 30 (9.55) | 83(15.66) | |||
| Unknown | 39 (12.42) | 6(1.13) | |||
| Childhood sunburn | 1.21×10 −14 | 6.30 (4.52–8.77) | 1.11×10 −27 | ||
| No | 206 (65.61) | 157 (29.62) | |||
| Yes | 70 (22.29) | 336 (63.40) | |||
| Unknown | 38 (12.10) | 37 (6.98) |
Fisher's exact test. P-value, excluding unknown values.
Bold denotes statistically significant results.
SD, standard deviation.
We used an additional pool of samples to enlarge the sample size. Samples were obtained from 334 CM patients and 158 control individuals from the Valencian Institute of Oncology (IVO) and 171 cases and 212 controls from Dr Negrín Hospital, Las Palmas de Gran Canaria. A total set of 515 cases and 370 control individuals (Table 1).
Genomic DNA from cases and controls was isolated from peripheral blood lymphocytes and diluted to a final solution of 50 ng/µl using the traditional saline method or the DNAzol procedure (Invitrogen, Eugene, OR, USA). DNA concentration was quantified in samples using Quant-iT PicoGreen dsDNA Reagent (Invitrogen, Eugene, OR, USA). Further concentration measures were obtained using a Nanodrop 2000 spectrophotometer. Genomic DNA was amplified using the GenomiPhi DNA Amplification Kit (GE Healthcare Bio-Sciences AB, Uppsala, Sweden).
SNP Selection
We selected tag-SNPs from vitamin D metabolism-related genes, GC and VDR. These SNPs were chosen using either previous literature information or the HapMap International Project [24] by means of the HaploView v4.2 software forcing Tag-SNPs from the European_CEU subset of data. We selected marker SNPs with a minor allele frequency (MAF) higher than 0.05, with a Hardy-Weinberg equilibrium p-value cutoff of 0.001, we have set an r2 threshold of 0.8. Four linkage disequilibrium (LD) blocks were obtained using HapMap_CEU data for GC and seven blocks for VDR. A total of 21 SNPs were included (12 SNPs belonging to GC and 9 to VDR). Public databases were used to collect additional information about SNPs and genes: NCBI http://www.ncbi.nlm.nih.gov and Ensembl http://www.ensembl.org. Details such as MIM code, location, encoded protein, context sequence, nucleotide changes, MAF for HapMap_CEU and HapMap_TSI (based on a sample population from Northern Europe and Tuscany Italy respectively) and calculated MAF for both cases and controls of all the SNPs studied are provided in Table S1 and in Table 3.
Table 3. Allelic frequencies in cases and controls in spanish population for the GC gene SNPs studied.
| Gene | SNP | MAF HapMap_CEU | MAF HapMap_TSI | MAF Controls | MAF Cases | p HWE | Allelic p-value |
| GC | rs12512631 | 0.35 | 0.32 | 0.31 | 0.37 | 0.027 | 0.0196 |
| rs222049 | 0.08 | 0.10 | 0.09 | 0.08 | 0.341 | 0.324 | |
| rs2282679 | 0.26 | 0.26 | 0.30 | 0.31 | 0.220 | 0.734 | |
| rs705119 | 0.43 | 0.46 | 0.43 | 0.44 | 0.299 | 0.571 | |
| rs4588 | 0.27 | 0.26 | 0.30 | 0.30 | 0.652 | 0.991 | |
| rs7041 | 0.43 | 0.47 | 0.43 | 0.46 | 0.365 | 0.337 | |
| rs188812 | 0.10 | 0.10 | 0.08 | 0.10 | 0.922 | 0.254 | |
| rs222016 | 0.16 | 0.14 | 0.11 | 0.14 | 0.515 | 0.103 | |
| rs1155563 | 0.33 | 0.23 | 0.30 | 0.30 | 0.444 | 0.963 | |
| rs1352844 | 0.14 | 0.13 | 0.10 | 0.11 | 0.976 | 0.912 | |
| rs1352845 | 0.21 | 0.17 | 0.14 | 0.15 | 0.464 | 0.577 | |
| rs3733359 | 0.06 | 0.04 | 0.03 | 0.04 | 0.573 | 0.729 | |
| VDR | rs11574143 | 0.10 | 0.15 | 0.10 | 0.10 | 0.240 | 0.918 |
| rs739837 | 0.43 | 0.40 | 0.49 | 0.49 | 0.243 | 0.779 | |
| rs731236 | 0.43 | 0.41 | 0.39 | 0.38 | 0.884 | 0.639 | |
| rs2228570 | 0.41 | 0.38 | 0.34 | 0.34 | 0.309 | 0.848 | |
| rs4334089 | 0.27 | 0.27 | 0.30 | 0.27 | 0.835 | 0.190 | |
| rs4237855 | 0.42 | 0.35 | 0.48 | 0.47 | 1.09×10 −12 | 0.760 | |
| rs7299460 | 0.31 | 0.34 | 0.34 | 0.31 | 0.666 | 0.374 | |
| rs4760658 | 0.30 | 0.36 | 0.32 | 0.34 | 0.461 | 0.273 | |
| rs4516035 | 0.38 | 0.44 | 0.40 | 0.41 | 0.531 | 0.593 |
Bold marks statistically significant results.
p HWE refers to the p value for Pearson's goodness-of-fit test for deviation from Hardy Weinberg's equilibrium among controls.
MAF, minor allele frequency.
p-value for Pearson's goodness-of-fit chi-square between cases and controls MAF.
HapMap_CEU and HapMap_TSI refers to Caucassian European and Tuscany Italian populations respectively, obtained from HapMap database.
Genotyping Assays
Genotyping was carried out using Kaspar technology (KBiosciences, Hoddesdon, UK). The PCR was performed in a reaction volume of 4 µl containing about 10 ng of genomic DNA, a final concentration of 4× New Kaspar Reaction Mix, and 12 µM of each Kaspar primer.
The PCR assays were performed according to the manufacturer’s instructions. The genotype of each sample was determined by measuring final allele-specific fluorescence in the ABI Prism 7900 HT Detection System, using the SDS 2.3 software for allelic discrimination (Applied Biosystems, Foster City, USA).
As a quality control measure, we included one non-template sample and one sample duplicate per 96-well plate (a total of four per 384-well plate used). Genotypes were provided automatically by the software and were confirmed manually by two different laboratory personnel.
Statistical Analyses
Analyses were performed using SPSS v17 (SPSS, Chicago, IL, USA). All p-values were two-sided, and those less than 0.05 were considered statistically significant. We analysed the haplotypes using Haploview v4.2 software, we obtained χ2 values by performing a linkage case-control test as described previously [25]. Bonferroni’s correction was used as the method of adjustment for multiple comparisons. For all polymorphisms studied, Fisher's exact test was used both to test for deviations from HWE among controls, and to compare differences in the MAF distributions between cases and controls. We rejected HWE when p-values were lower than 0.0024 according to Bonferroni’s correction for 21 comparisons. In order to assess associations between genotypes, haplotypes and CM risk and between SNPs and each phenotypic characteristic, several analyses were performed. Genotype-related odds ratios (ORs), their corresponding 95% confidence intervals (CIs) and associated p-values were estimated via unconditional logistic regression. This was done for each model: genotypic, dominant and recessive. Same analyses were conducted between SNPs and each phenotypic characteristic. The power of the significant results was obtained using POWER v3.0 software (available at http://dceg.cancer.gov/tools/design/power), with the sample size of this study we are able to obtain a power of 80% as from OR value of 1.49. Odds ratios were then adjusted for known and suspected melanoma risk factors (eye, hair and skin colour, number of naevi, lentigines, and childhood sunburns) in order to assess the potential confounding effects by multivariate logistic regression We use as potential confounders all phenotypic traits that show differences between cases and controls (Table 2), however we added number of naevi and skin color in this study because both are well recognized risk factor for melanoma predisposition [26], [27].
For the second set of samples we obtained genotype-related ORs for genotypic, dominant and recessive models, their corresponding 95% CIs and associated p-values taking into account only SNPs that gave interesting results. These statistics were estimated using unconditional logistic regression.
Functional Analyses
To study the functional implications of SNPs, we used the Pupasuite3.1 software available online at http://pupasuite.bioinfo.cipf.es. We analysed all SNPs on the GC gene with a minor allele frequency higher than 0.05 provided by HapMap (a total of 40 SNPs, provided upon request). We searched for all non-synonymous SNPs, candidate transcription factor (TF) binding sites (through TRANSFAC, JASPAR and ORegAnno), low-flexibility promoter regions with a minimum length of 10 bp, highly conserved regions, splice sites created or disrupted by SNPs, and the presence of miRNA targets. Search criteria were provided at the aforementioned website.
Results
Allelic Distribution of Polymorphisms and Association with CM Risk
After applying Bonferroni’s correction, one SNP on VDR was out of HWE, rs4237855 (p-value 1.09×10−12), and was removed from further analyses. The remaining SNPs complied with HWE. We confirmed that our control population has allele frequencies similar to the HapMap_CEU or HapMap_TSI ones.
Based on unadjusted p-values, we observed evidence of differences between cases and controls in MAF for SNP rs12512631 on the GC gene (p-value 0.0196) which codes for the vitamin D transporter protein. We did not observe differences in the MAF for any other SNP. (Table 3).
Allele frequencies for SNPs (on HapMap_CEU, HapMap_TSI and genotyped cases and controls), p-values for their comparison between 530 CM cases and 314 control individuals, along with p-values for the test of departure from HWE among controls are detailed in Table 3.
Association between Genotypes and Melanoma Risk
The implication of these vitamin D-related genes in melanoma was investigated further by comparing the genotypic distributions of all the SNPs studied. The estimated ORs and associated p-values are shown in Table 4. This was also done for dominant and for recessive models. Relevant results as well as the values adjusted by phenotypic characteristics are shown in Table 5.
Table 4. Genotypic distribution among cases and controls in a Spanish population.
| Controls (N = 314) n (%) | Cases (N = 530) n (%) | ||||||||
| Gene | SNP | Major homozygotes | Heterozygotes | Minor homozygotes | Major homozygotes | Heterozygotes | Minor homozygotes | OR (95% CI) | p-value |
| GC | rs12512631 | 154 (49.04) | 115 (36.62) | 38 (12.10) | 199 (37.55) | 261 (49.25) | 61 (11.51) | 1.29 (1.04–1.60) | 0.0190 |
| rs222049 | 240 (76.43) | 50 (15.92) | 1 (0.32) | 441 (83.21) | 74 13.96) | 2 (0.38) | 0.83 (0.57–1.20) | 0.3150 | |
| rs705119 | 102 (32.48) | 136 (43.31) | 58 (18.47) | 152 (28.68) | 276 (52.08) | 90 (16.98) | 1.06 (0.86–1.31) | 0.5649 | |
| rs4588 | 154 (49.04) | 128 (40.76) | 30 (9.55) | 248 (46.79) | 229 (43.21) | 42 (7.92) | 1.00 (0.81–1.25) | 0.9910 | |
| rs7041 | 101 (32.17) | 140 (44.59) | 60 (19.11) | 145 (27.36) | 271 (51.13) | 100 (18.87) | 1.11 (0.90–1.36) | 0.3333 | |
| rs188812 | 261 (83.12) | 44 (14.01) | 2 (0.64) | 425 (80.19) | 97 (18.30) | 1 (0.19) | 1.25 (0.86–1.80) | 0.2422 | |
| rs222016 | 236 (75.16) | 58 (18.47) | 5 (1.59) | 368 (69.43) | 127 (23.96) | 8 (1.51) | 1.30 (0.95–1.77) | 0.1020 | |
| rs1155563 | 150 (47.77) | 121 (38.54) | 30 (9.55) | 252 (47.55) | 217 (40.99) | 45 (8.61) | 0.99 (0.79–1.25) | 0.9633 | |
| rs1352844 | 221 (70.38) | 51 (16.24) | 3 (0.96) | 416 (78.49) | 93 (17.55) | 8 (1.51) | 1.02 (0.73–1.42) | 0.9137 | |
| rs1352845 | 204 (64.97) | 70 (22.29) | 4 (1.27) | 365 (68.87) | 138 (26.04) | 8 (1.51) | 1.09 (0.81–1.48) | 0.5675 | |
| rs3733359 | 254 (80.89) | 18 (5.73) | 0 (0) | 483 (91.13) | 38 (7.17) | 0 (0) | 1.10 (0.62–1.97) | 0.7400 | |
| VDR | rs11574143 | 238 (75.80) | 55 (17.52) | 1 (0.32) | 383 (72.26) | 88 (16.60) | 1 (0.19) | 0.98 (0.68–1.41) | 0.9143 |
| rs739837 | 74 (23.57) | 160 (50.96) | 66 (21.02) | 121 (22.83) | 258 (48.68) | 115 (21.70) | 0.97 (0.79–1.19) | 0.7731 | |
| rs731236 | 109 (34.71) | 141 (44.90) | 44 (14.01) | 186 (35.09) | 248 (46.86) | 64 (12.09) | 0.95 (0.76–1.18) | 0.6325 | |
| rs2228570 | 140 (44.59) | 130 (41.40) | 39 (12.42) | 217 (40.94) | 225 (42.52) | 58 (11.01) | 1.02 (0.82–1.27) | 0.8502 | |
| rs4334089 | 138 (43.95) | 121 (38.54) | 25 (7.96) | 261 (49.25) | 192 (36.23) | 36 (6.79) | 0.86 (0.68–1.08) | 0.1898 | |
| rs4237855 | 102 (32.48) | 76 (24.20) | 91 (28.98) | 165 (31.13) | 129 (24.34) | 140 (26.42) | 0.98 (0.81–1.17) | 0.7977 | |
| rs7299460 | 131 (41.72) | 137 (43.63) | 32 (10.20) | 236 (44.53) | 224 (42.26) | 47 (8.87) | 0.90 (0.73–1.13) | 0.3680 | |
| rs4760658 | 136 (43.31) | 134 (42.68) | 27 (8.60) | 225 (42.45) | 212 (40) | 67 (12.64) | 1.13 (0.91–1.39) | 0.2795 | |
| rs4516035 | 106 (33.76) | 149 (47.45) | 45 (14.33) | 183 (34.53) | 228 (43.02) | 94 (17.74) | 1.06 (0.86–1.29) | 0.5980 | |
Heterozygotes and minor homozygotes individuals count for cases and controls and their percentages are calculated among the total of samples including fails.
OR (CI 95%) means Odds Ratio and its 95% confidence interval in parentheses. OR and p values are calculated via unconditional logistic regression considering differences between cases and controls genotypes.
Bold denotes statistically significant results considering as significant p values lower than 0.05.
Table 5. Genotypic analyses of SNPs with Cutaneous Melanoma risk association.
| Non-adjusted | Adjusted* | Enlarged sample | |||||
| SNP | Statistical model | OR (CI 95%) | p-value | OR (CI 95%) | p-value | OR (CI 95%) | p-value |
| rs12512631 | Genotypic | 1.29 (1.04–1.60) | 0.019 | 1.32 (1.01–1.72) | 0.041 | 1.11 (0.96–1.28) | 0.153 |
| Dominant | 1.63 (1.23–2.17) | 7×10 −4 | 1.71 (1.20–2.45) | 3×10 −3 | 1.28 (1.05–1.56) | 0.012 | |
| Recessive | 0.94 (0.61–1.45) | 0.774 | 0.94 (0.55–1.60) | 0.809 | 0.90 (0.68–1.19) | 0.458 | |
| rs222016 | Genotypic | 1.30 (0.95–1.77) | 0.102 | 1.23 (0.83–1.82) | 0.312 | 1.11 (0.92–1.35) | 0.277 |
| Dominant | 1.37 (0.98–1.93) | 0.068 | 1.30 (0.85–1.98) | 0.222 | 1.22 (0.97–1.52) | 0.083 | |
| Recessive | 0.95 (0.31–2.93) | 0.929 | 0.65 (0.14–3.09) | 0.586 | 0.65 (0.35–1.20) | 0.168 | |
Bold denotes statistically significant results.
OR (CI 95%) means Odds Ratio and its 95% confidence interval.
Adjusted for eye colour, hair colour, skin colour, lentigines, number of naevi and childhood sunburn.
The enlarged sample makes a total of 684 controls and 1045 cases.
One SNP was found to be associated with CM susceptibility risk, rs12512631 on the GC gene (OR 1.29 95%CI: 1.04–1.60; p-value 0.0190) (Table 4). Moreover, the association between rs12512631 and CM risk was highly significant in the dominant model (OR 1.63 95% CI: 1.23–2.17; p-value 7×10−4) (Table 5). This significance remained after Bonferroni’s correction for multiple testing.
A trend to significance was found for rs222016 (OR 1.23 95% CI: 0.83–1.82; p-value 0.102). Furthermore, when performing the statistical analyses considering dominant model, we obtained borderline significance (OR 1.37, 95% CI: 0.98–1.93; p-value 0.068). No association remained statistically significant for any other SNP.
To assess the independence of risk factors associated with CM, we performed a multivariate analysis that took into account phenotypic risk factors such as eye, skin and hair colour, number of naevi, lentigines and childhood sunburn events, along with candidate SNPs. The associated SNP, rs12512631, maintained its significance for both genotypic (p-value 0.041) and dominant models (p-value 0.003), whereas there was no further trend to significance for rs222016 (see adjusted values in Table 5).
We increased the population sample and performed the association analyses for the two significant CM-associated SNPs (rs12512631 and rs222016). We studied a total of 1045 melanoma patients and 684 control individuals, and the statistical significance of rs12512631 remained (OR 1.28 95% CI: 1.05–1.56, p-value 0.013). This association continued to be significant after Bonferroni’s correction for two comparisons (p-value threshold of 0.025) (Table 5).
Associations between Genotypes and Phenotypic Characteristics
We assessed whether SNPs from the GC and VDR genes were associated with various phenotypic characteristics. The SNP rs3733359, located on the 5′UTR of the GC gene, showed a significant association with dark skin colour (OR 0.53, 95% CI: 0.30–0.94, p-value 0.023). We also observed a weak significant association for the 3′UTR SNP rs739837 of the VDR gene with fair skin colour (p-value 0.048). We found strong association between non-synonymous SNP rs2228570, located within exon 4 of the VDR gene, and absence of childhood sunburn (OR 0.065 95% CI 0.49–0.86, p-value 0.003). No more evidence of associations was found.
Haplotype Analysis and Association with Melanoma Risk
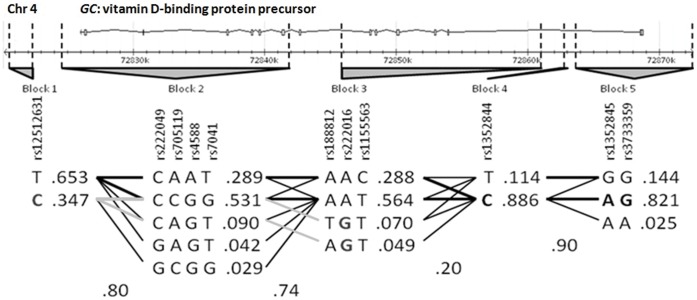
We performed haplotype analyses using the 11 tag-SNPs in GC (the gene on which we found an associated SNP) selected previously from HapMap data. These SNPs were organized into five independent blocks according to the Haploview program for the use of data from a European subset of samples (HapMap_CEU). Block 1 included rs12512631 in a 3′downstream region of GC. Block 2 is represented by one SNP located 3′downstream and three SNPs within the 3′end of the gene, two of them corresponding to exon 9. Blocks 3, 4 and 5 contained six SNPs from various intron regions, including intron 1, and SNP with rs3733359, located on 5′UTR. Block 5 represented the most likely promoter region with consensus sequences for TF binding sites. Data analysis was done according to the block structures detailed in Figure 1. The case-control analysis of the haplotype distribution revealed a statistical association with melanoma susceptibility on two SNPs, rs12512631 (p-value 9×10−3), and rs222016 (p-value 0.031), representing Blocks 1 and 3, respectively (Table S2). These data were coherent with the associations described above.
Figure 1. Haplotype distribution within GC gene according to Tag-SNPs selected by Haploview v4.2.
Genotyped SNPs are indicated by their rs#. LD Blocks are shown in gray. GC gene chromosomic location and ideogram in reverse orientation appears on the top of the figure, where boxes represent exons and horizontal black lines introns. Bold denotes associated risk alleles, and light grey lines indicate connected haplotypes.
Functional Implication
To assess for possible functional implications of polymorphisms, we carried out a prioritization of SNPs based on functional properties using the Pupasuite 3.1 software. Results showed non-synonymous changes of aminoacid on rs4588 (K436T) and rs7041 (E416D). TRANSFAC revealed three SNPs located in consensus sequence affecting the TF binding sites; rs4588 FOXJ2, rs7041 HNF-1 and rs222014 E2F-1 (located within intron 3 and grouped on block 3 with tag-SNP rs222016). None of the other SNPs seemed to have any additional functionality.
Discussion
In this case-control study, we have detected for the first time statistical evidence suggesting that rs12512631 on GC is associated with risk for CM. GC is an essential gene in the vitamin D pathway since it codifies for VDBP, the transporter of all the intermediate and final forms of vitamin D. Variants in this gene may modulate protein expression or activity of this protein and, therefore may affect vitamin D synthesis and distribution. There are few studies of GC polymorphisms, however, to our knowledge all of them are related to the serum vitamin D levels in diseases such as CM, prostate and colorectal cancer. In these, a clear association was found between GC polymorphisms and circulating vitamin D levels, but the association between polymorphisms and the disease itself was not at all clear [21], [28], [29], [30]. Shen et al. reported that one allele of the GC gene may be a risk factor for chronic obstructive pulmonary disease [31], and Abbas et al. observed a lower breast cancer risk associated with the Gc2–2 allele, independent of vitamin D levels [32]. Recently an independent GWAS on vitamin D levels, with further validation on CM patients, has pointed again to the GC gene as a candidate for melanoma susceptibility when vitamin D levels are taken into consideration [16], [21].
Our study suggests that the 3′downstream region of the GC gene, marked through the SNP rs12512631, is associated with CM risk. Furthermore, evidence of association was suggested by the tag-SNP rs222016, which marks a large disequilibrium block within the gene, including intron 1. SNP rs222016 is in LD with SNP rs222014, which could have functional implications through the disruption of a transcription factor element (E2F-1), but we cannot discard other mechanisms such as alternative splicing signalling. Previous studies have shown an association between tag-SNP rs12512631 and susceptibility to prostate and colorectal cancer, which supports our results [28], [29].
We did not observe any evidence of association between CM and VDR variants overall, in accordance with our previous results [22]. Some other controversial results are reported in the literature. A review by Köstner indicates only one variant in VDR associated with CM risk, Fok1 (rs2228570) [33], but a meta-analysis of various studies revealed that the only variant that presents solid evidence is Bsm1 (not considered in our study) and remarked on the need to adjust by phenotype or environmental characteristics [34]. Gapska et al. found an association only when analyzing the haplotypes, but not the variants themselves [14]. More recent studies have not detected an association with CM risk on the VDR variants unless adjusting by vitamin D levels [15], [35], [36]. This last situation might explain why we have not found a significant association; vitamin D levels were not taken into account.
The strength of this study is the homogeneity of the Spanish population sample and the ability to control for established risk factors for CM through a structured questionnaire. We recognize, however, some potential for misclassification of phenotypic characteristics due to the subjective nature of the phenotypic attributes considered. Controls participated on a volunteer basis, which may have introduced some selection bias.
We show a comprehensive study of genetic variation on the vitamin D pathway genes VDR (vitamin D receptor) and GC (vitamin D transporter), and examine their putative role in CM susceptibility. We observed statistically significant results for CM susceptibility with two variants in the GC gene, but none in the VDR gene. One of these associations remained significant after correction for multiple testing, SNP rs12512631. This association suggests that GC may also play a role in modulating the susceptibility to CM.
We encourage replication of these findings in independent studies since the GC gene may well be a new marker for CM predisposition.
Supporting Information
SNPs on GC and VDR genes considered in this study. GC refers to Vitamin D binding protein gene; VDR refers to Vitamin D receptor gene. Bold in sequence context denotes nucleotide change. Location is described considering as the first Exon 1 of consensus sequence. DWST means downstream, UTR means untranscribed region and UPST means upstream.
(DOCX)
Case-control study based on haplotypes in the GC gene conducted in Spanish population. Bold denotes statistically significant results. Italic Haplotypes on each block mark the risk haplotype. The Marker number indicates the order of the tag-SNP on the gene. LD Block means linkage disequilibrium block. Frequencies are calculated for the association alleles of each haplotype among 530 cases and 314 controls.
(DOCX)
Acknowledgments
We would like to give thank to all the patients and volunteers who gave their consent for the study. We also thank the medical staff of Gregorio Marañón, La Paz and Ramón y Cajal Hospitals for collecting the samples and the Biobank of the Instituto Valenciano de Oncología for providing the material for the genetic analysis. Genotyping with Kaspar was performed at the Unidad Central de Investigación Médica of the Facultad de Medicina of the University of Valencia.
Funding Statement
This study has been supported by a grant from the National Health System Ministry (Institute Carlos III) PI10_0405. MPC is funded by the Generalitat Valenciana Conselleria d’Educació VALi+d program (ACIF/2011/207). MIV is funded by the Spanish Ministry of Science and Education under a FPI grant (BES-2008-009234). GR is funded by the Spanish Ministry of Health (CP08_00069). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jerant AF, Johnson JT, Sheridan CD, Caffrey TJ (2000) Early detection and treatment of skin cancer. Am Fam Physician 62: 357–368, 375–356, 381–352. [PubMed] [Google Scholar]
- 2. Holick MF (2003) Vitamin D deficiency: what a pain it is. Mayo Clin Proc 78: 1457–1459. [DOI] [PubMed] [Google Scholar]
- 3. Holick MF (2003) Evolution and function of vitamin D. Recent Results Cancer Res. 164: 3–28. [DOI] [PubMed] [Google Scholar]
- 4. Field S, Newton-Bishop JA (2011) Melanoma and vitamin D. Mol Oncol. 5: 197–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lurie G, Wilkens LR, Thompson PJ, Carney ME, Palmieri RT, et al. (2011) Vitamin D receptor rs2228570 polymorphism and invasive ovarian carcinoma risk: pooled analysis in five studies within the Ovarian Cancer Association Consortium. Int J Cancer 128: 936–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Alimirah F, Peng X, Murillo G, Mehta RG (2011) Functional significance of vitamin D receptor FokI polymorphism in human breast cancer cells. PLoS One 6: e16024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Touvier M, Chan DS, Lau R, Aune D, Vieira R, et al. (2011) Meta-analyses of vitamin D intake, 25-hydroxyvitamin D status, vitamin D receptor polymorphisms, and colorectal cancer risk. Cancer Epidemiol Biomarkers Prev 20: 1003–1016. [DOI] [PubMed] [Google Scholar]
- 8. Smedby KE, Eloranta S, Duvefelt K, Melbye M, Humphreys K, et al. (2011) Vitamin D receptor genotypes, ultraviolet radiation exposure, and risk of non-Hodgkin lymphoma. Am J Epidemiol 173: 48–54. [DOI] [PubMed] [Google Scholar]
- 9. Karami S, Brennan P, Navratilova M, Mates D, Zaridze D, et al. (2010) Vitamin d pathway genes, diet, and risk of renal cell carcinoma. Int J Endocrinol 2010: 879362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bektas-Kayhan K, Unur M, Yaylim-Eraltan I, Ergen HA, Toptas B, et al. (2010) Association of vitamin D receptor Taq I polymorphism and susceptibility to oral squamous cell carcinoma. In Vivo 24: 755–759. [PubMed] [Google Scholar]
- 11.Chang CK, Mulholland HG, Cantwell MM, Anderson LA, Johnston BT, et al.. (2011) Vitamin D Receptor Gene Variants and Esophageal Adenocarcinoma Risk: A Population-Based Case-Control Study. J Gastrointest Cancer. [DOI] [PubMed] [Google Scholar]
- 12. Srinivasan M, Parwani AV, Hershberger PA, Lenzner DE, Weissfeld JL (2010) Nuclear vitamin D receptor expression is associated with improved survival in non-small cell lung cancer. J Steroid Biochem Mol Biol 123: 30–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kidd LC, Paltoo DN, Wang S, Chen W, Akereyeni F, et al. (2005) Sequence variation within the 5' regulatory regions of the vitamin D binding protein and receptor genes and prostate cancer risk. Prostate 64: 272–282. [DOI] [PubMed] [Google Scholar]
- 14. Gapska P, Scott RJ, Serrano-Fernandez P, Mirecka A, Rassoud I, et al. (2009) Vitamin D receptor variants and the malignant melanoma risk: a population-based study. Cancer Epidemiol 33: 103–107. [DOI] [PubMed] [Google Scholar]
- 15.Orlow I, Roy P, Reiner AS, Yoo S, Patel H, et al.. (2011) Vitamin D receptor polymorphisms in patients with cutaneous melanoma. Int J Cancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ahn J, Yu K, Stolzenberg-Solomon R, Simon KC, McCullough ML, et al. (2010) Genome-wide association study of circulating vitamin D levels. Hum Mol Genet 19: 2739–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pani MA, Regulla K, Segni M, Hofmann S, Hufner M, et al. (2002) A polymorphism within the vitamin D-binding protein gene is associated with Graves' disease but not with Hashimoto's thyroiditis. J Clin Endocrinol Metab 87: 2564–2567. [DOI] [PubMed] [Google Scholar]
- 18. Disanto G, Ramagopalan SV, Para AE, Handunnetthi L (2010) The emerging role of vitamin D binding protein in multiple sclerosis. J Neurol 258: 353–358. [DOI] [PubMed] [Google Scholar]
- 19. Chishimba L, Thickett DR, Stockley RA, Wood AM (2010) The vitamin D axis in the lung: a key role for vitamin D-binding protein. Thorax 65: 456–462. [DOI] [PubMed] [Google Scholar]
- 20. Wang TJ, Zhang F, Richards JB, Kestenbaum B, van Meurs JB, et al. (2010) Common genetic determinants of vitamin D insufficiency: a genome-wide association study. Lancet 376: 180–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Davies JR, Chang YM, Snowden H, Chan M, Leake S, et al. (2011) The determinants of serum vitamin D levels in participants in a melanoma case-control study living in a temperate climate. Cancer Causes Control 22: 1471–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Barroso E, Fernandez LP, Milne RL, Pita G, Sendagorta E, et al. (2008) Genetic analysis of the vitamin D receptor gene in two epithelial cancers: melanoma and breast cancer case-control studies. BMC Cancer 8: 385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Laayouni H, Calafell F, Bertranpetit J (2010) A genome-wide survey does not show the genetic distinctiveness of Basques. Hum Genet 127: 455–458. [DOI] [PubMed] [Google Scholar]
- 24. The International HapMap Project. Nature 426: 789–796. [DOI] [PubMed] [Google Scholar]
- 25. Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21: 263–265. [DOI] [PubMed] [Google Scholar]
- 26. Newton Bishop JA, Bishop DT (2005) The genetics of susceptibility to cutaneous melanoma. Drugs Today (Barc) 41: 193–203. [DOI] [PubMed] [Google Scholar]
- 27. Kvaskoff M, Whiteman DC, Zhao ZZ, Montgomery GW, Martin NG, et al. Polymorphisms in nevus-associated genes MTAP, PLA2G6, and IRF4 and the risk of invasive cutaneous melanoma. Twin Res Hum Genet 14: 422–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ahn J, Albanes D, Berndt SI, Peters U, Chatterjee N, et al. (2009) Vitamin D-related genes, serum vitamin D concentrations and prostate cancer risk. Carcinogenesis 30: 769–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Poynter JN, Jacobs ET, Figueiredo JC, Lee WH, Conti DV, et al. (2010) Genetic variation in the vitamin D receptor (VDR) and the vitamin D-binding protein (GC) and risk for colorectal cancer: results from the Colon Cancer Family Registry. Cancer Epidemiol Biomarkers Prev 19: 525–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hibler EA, Hu C, Jurutka PW, Martinez ME, Jacobs ET (2012) Polymorphic Variation in the GC and CASR Genes and Associations with Vitamin D Metabolite Concentration and Metachronous Colorectal Neoplasia. Cancer Epidemiol Biomarkers Prev 21: 368–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shen LH, Zhang XM, Su DJ, Yao SP, Yu BQ, et al. (2010) Association of vitamin D binding protein variants with susceptibility to chronic obstructive pulmonary disease. J Int Med Res 38: 1093–1098. [DOI] [PubMed] [Google Scholar]
- 32. Abbas S, Linseisen J, Slanger T, Kropp S, Mutschelknauss EJ, et al. (2008) The Gc2 allele of the vitamin D binding protein is associated with a decreased postmenopausal breast cancer risk, independent of the vitamin D status. Cancer Epidemiol Biomarkers Prev 17: 1339–1343. [DOI] [PubMed] [Google Scholar]
- 33. Kostner K, Denzer N, Muller CS, Klein R, Tilgen W, et al. (2009) The relevance of vitamin D receptor (VDR) gene polymorphisms for cancer: a review of the literature. Anticancer Res 29: 3511–3536. [PubMed] [Google Scholar]
- 34. Mocellin S, Nitti D (2008) Vitamin D receptor polymorphisms and the risk of cutaneous melanoma: a systematic review and meta-analysis. Cancer 113: 2398–2407. [DOI] [PubMed] [Google Scholar]
- 35. Newton-Bishop JA, Beswick S, Randerson-Moor J, Chang YM, Affleck P, et al. (2009) Serum 25-hydroxyvitamin D3 levels are associated with breslow thickness at presentation and survival from melanoma. J Clin Oncol 27: 5439–5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Randerson-Moor JA, Taylor JC, Elliott F, Chang YM, Beswick S, et al. (2009) Vitamin D receptor gene polymorphisms, serum 25-hydroxyvitamin D levels, and melanoma: UK case-control comparisons and a meta-analysis of published VDR data. Eur J Cancer 45: 3271–3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SNPs on GC and VDR genes considered in this study. GC refers to Vitamin D binding protein gene; VDR refers to Vitamin D receptor gene. Bold in sequence context denotes nucleotide change. Location is described considering as the first Exon 1 of consensus sequence. DWST means downstream, UTR means untranscribed region and UPST means upstream.
(DOCX)
Case-control study based on haplotypes in the GC gene conducted in Spanish population. Bold denotes statistically significant results. Italic Haplotypes on each block mark the risk haplotype. The Marker number indicates the order of the tag-SNP on the gene. LD Block means linkage disequilibrium block. Frequencies are calculated for the association alleles of each haplotype among 530 cases and 314 controls.
(DOCX)