Abstract
Terminal proteins (TPs) of bacteriophages prime DNA replication and become covalently linked to the genome ends. Unexpectedly, we have found functional eukaryotic nuclear localization signals (NLSs) within the TP sequences of bacteriophages from diverse families and hosts. Given the role of bacteriophages as vehicles for horizontal gene transfer (HGT), we postulated that viral genomes that have covalently linked NLS-containing terminal proteins might behave as vectors for HGT between bacteria and the eukaryotic nucleus. To validate this hypothesis, we profited from the in vitro Φ29 amplification system that allows the amplification of heterologous DNAs producing linear molecules of DNA with TP covalently attached to both 5' ends. Interestingly, these in vitro-generated TP-DNA molecules showed enhanced gene delivery in mammalian cells, supporting a possible role in HGT by transferring genes between prokaryotes and eukaryotes. Moreover, these TP-DNA molecules are a useful tool to amplify and subsequently deliver genes efficiently into the eukaryotic nucleus. Here, we suggest various possible applications and further developments of the technique with biotechnological and therapeutic purposes.
Keywords: NLS, Gene delivery, bacteriophage, terminal protein, horizontal gene transfer
As more complete genome sequences become available, the role of horizontal gene transfer (HGT) in shaping them becomes clearer.1,2 However, its importance in the evolution of eukaryotic genomes is often overshadowed by the greater prevalence and the more advanced understanding of gene transfer mechanisms in prokaryotes.3 One of the more interesting theories on prokaryotes-to-eukaryotes gene transfer is the so-called “you are what you eat” hypothesis, by which prokaryotic genes from symbiotic or food bacteria could have replaced ancient eukaryotic genes over evolutionary time.4,5 In agreement with this theory, the occurrence of HGT in eukaryotes is higher in unicellular organisms that do not require specific germ lines to be reproduced and to spread the newly acquired genes.6 However, once inside the eukaryotic cell, the nuclear envelope constitutes a barrier that prevents access to the cell genome. Only small molecules can overcome this blockade and most nuclear proteins are gifted with a specific amino acid portion, called “nuclear localization signal” (NLS), that label them as nuclear and as such they are detected by an specific nuclear transport pathway. This is a fine-tuned process that requires one or more carrier proteins that mediate the transport through the nuclear pore complex in an energy dependent manner.7 In the same way, DNA molecules over 300 bp cannot penetrate efficiently through the nuclear envelope, which constitutes an obvious obstacle for foreign DNA uptake, either from natural origin or from human-developed technologies, such as cell transfection.8
We have found a widespread occurrence of predicted NLSs in phage terminal proteins (TPs) from different families of bacteriophages and with diverse hosts and show that the TPs of five different bacteriophages, Φ29, Nf, PRD1, Bam35 and Cp-1, localize to the mammalian cell nucleus.9 Moreover, after the latter work was accepted we noticed that the TP of the recently discovered phage YS61,10 which has been proposed to constitute a new independent subfamily within the Podoviridae virus family, also contains a putative NLS in its sequence (residues 126–155, according to NLStradamus11 prediction), which further confirms the widespread presence of NLSs in TPs.
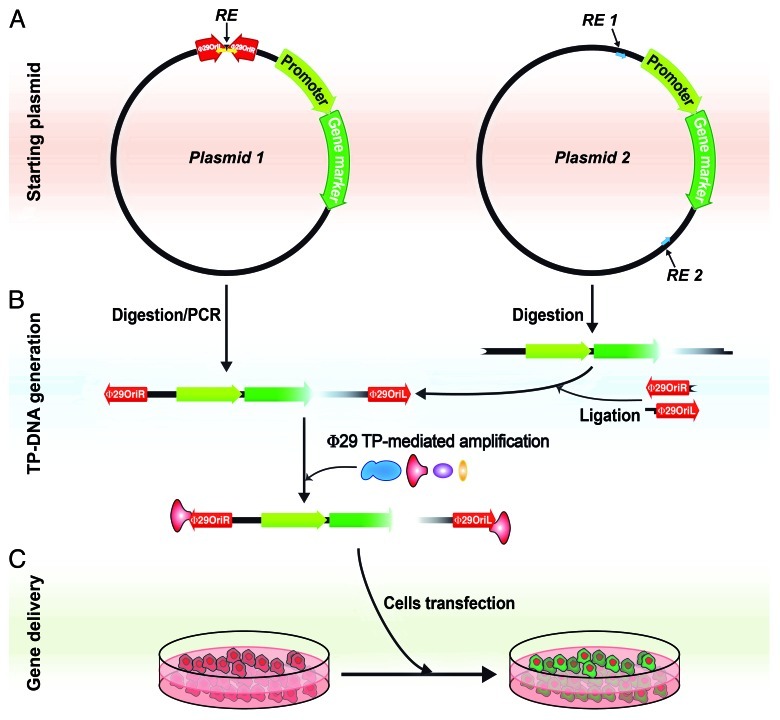
The use of TP-mediated protein-primed DNA replication has been reported or suggested in diverse replicons from all domains of life, including prokaryotic and eukaryotic viruses, linear plasmids and transposons. In this mechanism, the OH group of a specific serine, threonine or tyrosine of the TP is used by the replicative DNA polymerase to start DNA synthesis from both ends of the linear double-stranded genome and thus the TP remains covalently linked to such 5′ ends, giving rise to TP-DNA genome molecules.12 Moreover, since bacteriophages, and viruses in general, are known active vehicles for HGT, we hypothesized that viral genomes that have covalently linked NLS-containing terminal proteins may behave as vectors for HGT between bacteria and the eukaryotic nucleus, thus providing the next step to the “you are what you eat” theory. In order to demonstrate that the linked TP can mediate the nuclear translocation step we took advantage of the described TP-mediated Φ29 minimal amplification system13 that requires only four proteins, DNA polymerase, TP, single-stranded DNA binding protein and double-stranded DNA binding protein. This system generates in vitro DNA molecules containing the TP covalently linked to the 5′ DNA ends that confer resistance to exonuclease degradation. We used a circular plasmid DNA, which contains the yfp gene expression cassette for mammalian cells and the phage Φ29 origins of replication. This plasmid, once linearized, can be amplified in vitro by the phage replication system, thus incorporating the TP covalently linked at both 5′ DNA ends. We found that the attached Φ29 TP increased yfp expression with respect to that of a control linear DNA and, accordingly, this positive effect was reversed by removing the TP with proteinase K treatment.9 These results show that TP-linked DNA enhances gene delivery into the eukaryotic nucleus, supporting a possible role in HGT by transferring genes between prokaryotes and eukaryotes. Furthermore, the in vitro-generated TP-DNA molecules are a useful tool to deliver genes into eukaryotic nucleus. Although the use of TPs as gene delivery vectors had been already suggested,14,15 we have now an amplification system that allows generating the in vitro TP-DNA molecules with increased stability and transfection yield (Fig. 1). This TP-DNA might be generated using either a DNA that already contains the phage origins of replication (Fig. 1A, left side) or any linearized DNA after ligation of linkers containing the required origin sequences13 (Fig. 1A, right side). The possible applications of in vitro-generated TP-DNAs are numerous. On the one hand, we have shown that they can be complexed with standard transfection agents and used for routine cell transfection with an increased yield. On the other hand, the use of fusion proteins with several functional motifs, such as DNA binding domains, NLS, cell penetrating domains (TAT), endosome disruption enhancers (diphteria toxin) or even cell-specific antibodies have been shown to improve gene delivery in cultured cells.16-18 To test whether the Φ29 amplification system is able to use a TP with some fusion domains, we performed in vitro amplification assays with an YFP-TP fusion protein, which has the yellow fluorescent protein (YFP) fused to the N-terminal end of Φ29 TP and found that the YFP-TP protein can be incorporated to the Φ29 genome origins with similar yield than the wild type TP (not shown). Thus, different motifs or proteins could be fused to Φ29 TP in order to obtain TP-DNA molecules with specific features, like the ability to penetrate into an specific cell type and providing expression of the desired gene. It is also interesting to consider further refinements of the system, like the addition of site-specific nuclease domains that may target selected human genes and excise or correct regions of genes implicated in monogenic diseases.19 Altogether, in vitro generation of linear DNA molecules with 5′-linked TPs constitutes a potential novel strategy for the assembly of synthetic therapeutic gene delivery vehicles.
Figure 1. Generation of Φ29 TP-linked DNAs for enhanced gene delivery. (A) Starting plasmid. The transfected DNA should contain a gene marker under a suitable promoter for expression in mammalian cells. In Plasmid 1 (on the left), the Φ29 genome replication origins have been cloned into the plasmid, which allows the generation of linear DNA by cleavage with the appropriate restriction endonuclease (RE) or by PCR amplification with specific primers. Alternatively (on the right), Φ29 replication origins may be linked as specific adapters using incompatible restriction enzyme sites (RE 1 and RE 2). In the latter case, the digested linear DNA fragment may be originated either from an expression plasmid13 or a PCR-generated fragment containing the same restriction sites. Yellow and blue arrows stand for the specific PCR primers in each case. (B) TP-DNA generation. Linear DNAs containing Φ29 replication origins at both ends can be amplified with the addition of four Φ29 DNA replication proteins, DNA polymerase (blue), TP (red), double-stranded DNA binding protein (purple) and single-stranded DNA binding protein (yellow). (C) Gene delivery. The amplification mixture can be directly used for gene delivery experiments, like cell transfection. Evaluation of the gene marker expression should be checked by western blot or suitable activity assays.
Acknowledgments
This work was supported by Grant BFU2011–23645 and Consolider-Ingenio Grant 2010 24717 from the Spanish Ministry of Economy and Competitiveness (to M.S.) and by an institutional grant from Fundación Ramón Areces to the Centro de Biología Molecular “Severo Ochoa.” I.H. is a holder of the Formación de Profesorado Universitario fellowship from the Spanish Ministry of Education.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cib/article/22829
References
- 1.Abby SS, Tannier E, Gouy M, Daubin V. Lateral gene transfer as a support for the tree of life. Proc Natl Acad Sci U S A. 2012;109:4962–7. doi: 10.1073/pnas.1116871109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Syvanen M. Evolutionary implications of horizontal gene transfer. Annu Rev Genet. 2012;46:341–58. doi: 10.1146/annurev-genet-110711-155529. [DOI] [PubMed] [Google Scholar]
- 3.Keeling PJ, Palmer JD. Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet. 2008;9:605–18. doi: 10.1038/nrg2386. [DOI] [PubMed] [Google Scholar]
- 4.Doolittle WF. You are what you eat: a gene transfer ratchet could account for bacterial genes in eukaryotic nuclear genomes. Trends Genet. 1998;14:307–11. doi: 10.1016/S0168-9525(98)01494-2. [DOI] [PubMed] [Google Scholar]
- 5.Doolittle WF, Boucher Y, Nesbø CL, Douady CJ, Andersson JO, Roger AJ. How big is the iceberg of which organellar genes in nuclear genomes are but the tip? Philos Trans R Soc Lond B Biol Sci. 2003;358:39–57, discussion 57-8. doi: 10.1098/rstb.2002.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andersson JO. Lateral gene transfer in eukaryotes. Cell Mol Life Sci. 2005;62:1182–97. doi: 10.1007/s00018-005-4539-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Görlich D, Mattaj IW. Nucleocytoplasmic transport. Science. 1996;271:1513–8. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 8.Ludtke JJ, Zhang G, Sebestyén MG, Wolff JA. A nuclear localization signal can enhance both the nuclear transport and expression of 1 kb DNA. J Cell Sci. 1999;112:2033–41. doi: 10.1242/jcs.112.12.2033. [DOI] [PubMed] [Google Scholar]
- 9.Redrejo-Rodríguez M, Muñoz-Espín D, Holguera I, Mencía M, Salas M. Functional eukaryotic nuclear localization signals are widespread in terminal proteins of bacteriophages. Proc Natl Acad Sci U S A. 2012;109:18482–7. doi: 10.1073/pnas.1216635109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kleppen HP, Holo H, Jeon SR, Nes IF, Yoon SS. Novel Podoviridae family bacteriophage infecting Weissella cibaria isolated from Kimchi. Appl Environ Microbiol. 2012;78:7299–308. doi: 10.1128/AEM.00031-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nguyen Ba AN, Pogoutse A, Provart N, Moses AM. NLStradamus: a simple Hidden Markov Model for nuclear localization signal prediction. BMC Bioinformatics. 2009;10:202. doi: 10.1186/1471-2105-10-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salas M. Protein-priming of DNA replication. Annu Rev Biochem. 1991;60:39–71. doi: 10.1146/annurev.bi.60.070191.000351. [DOI] [PubMed] [Google Scholar]
- 13.Mencía M, Gella P, Camacho A, de Vega M, Salas M. Terminal protein-primed amplification of heterologous DNA with a minimal replication system based on phage Φ29. Proc Natl Acad Sci U S A. 2011;108:18655–60. doi: 10.1073/pnas.1114397108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tolmachov O, Coutelle C. Covalent attachment of multifunctional chimeric terminal proteins to 5′ DNA ends: A potential new strategy for assembly of synthetic therapeutic gene vectors. Med Hypotheses. 2007;68:328–31. doi: 10.1016/j.mehy.2006.06.055. [DOI] [PubMed] [Google Scholar]
- 15.Tsai HH, Huang CH, Lin AM, Chen CW. Terminal proteins of Streptomyces chromosome can target DNA into eukaryotic nuclei. Nucleic Acids Res. 2008;36:e62. doi: 10.1093/nar/gkm1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uherek C, Fominaya J, Wels W. A modular DNA carrier protein based on the structure of diphtheria toxin mediates target cell-specific gene delivery. J Biol Chem. 1998;273:8835–41. doi: 10.1074/jbc.273.15.8835. [DOI] [PubMed] [Google Scholar]
- 17.Xavier J, Singh S, Dean DA, Rao NM, Gopal V. Designed multi-domain protein as a carrier of nucleic acids into cells. J Control Release. 2009;133:154–60. doi: 10.1016/j.jconrel.2008.09.090. [DOI] [PubMed] [Google Scholar]
- 18.Sobolev AS. Novel modular transporters delivering anticancer drugs and foreign DNA to the nuclei of target cancer cells. J BUON. 2009;14(Suppl 1):S33–42. [PMC free article] [PubMed] [Google Scholar]
- 19.Prieto J, Molina R, Montoya G. Molecular scissors for in situ cellular repair. Crit Rev Biochem Mol Biol. 2012;47:207–21. doi: 10.3109/10409238.2011.652358. [DOI] [PubMed] [Google Scholar]