Abstract
Malaria is still a devastating disease caused by the mosquito-transmitted parasite Plasmodium, particularly Plasmodium falciparum. During the last few years the situation has worsened in many ways, mainly due to malarial parasites becoming increasingly resistant to several anti-malarial drugs. Thus there is an urgent need to find alternate ways to control malaria and therefore it is necessary to identify new drug targets and new classes of anti-malarial drugs. A malaria vaccine would be the ultimate weapon to fight this deadly disease but unfortunately despite encouraging advances a vaccine is not likely soon. DNA helicases from the PcrA/UvrD/Rep (PUR) subfamily are important for the survival of the various organisms, mainly pathogenic bacteria. Members from this subfamily can be targeted and inhibited by a variety of synthetic compounds. Using bioinformatics analysis we have shown that UvrD from this subfamily is the only member present in the P. falciparum genome, while PcrA and Rep are absent in the genome. UvrD from the parasite shows no homology to any protein or enzyme from human and thus can be considered as a strong potential drug target. In the present study we report an in silico analysis of this important enzyme from a variety of Plasmodium species. The results suggest that among all the species of Plasmodium, P. falciparum contains the largest UvrD and this enzyme is variable at the sequence and structural level.
Keywords: DNA repair, Plasmodium species, helicase, malaria parasite, unwinding
Introduction
Malaria caused by Plasmodium, particularly Plasmodium falciparum, is the most serious and widespread parasitic disease of humans.1,2 Each year approximately three hundred million people become infected with malaria and two to three million die as a result in the tropical and subtropical areas of the world. Among the four species of Plasmodium, P. falciparum causes the most fatal form of malaria.1,2 With time, the parasite has developed insecticide and drug resistance.3 The malaria parasite P. falciparum has 14 chromosomes, more than 7000 genes and a four-stage life cycle as it passes from humans to mosquitoes and back again.4 It is very efficient at evading the human immune response. Although the mechanisms by which malaria parasites develops resistance to drugs are unclear, in other organisms, defects in DNA mismatch repair have been linked to increased mutation rates and drug resistance.5 Though the P. falciparum genome sequence data has provided us important insight in the biology of the parasite, the challenge lies ahead in understanding the function of a large number of malaria genes with unknown function. A malaria vaccine would be the ultimate weapon to fight this deadly disease but unfortunately despite encouraging advances a vaccine is not likely to be available in near future. The search for novel effective, safe and affordable anti-malarial drugs for malaria caused by P. falciparum is one of the most important tasks to pursue.6
DNA lesions occur frequently in living cells as a result of spontaneous events or external insults. One mechanism central to genomic stability and the control of mutagenesis is DNA repair. The primary function of DNA repair is to remove potentially deleterious lesions through either damage reversal or damage excision.7 The damage excision processes are characterized by the mechanism of excision employed nucleotide excision repair (NER), base excision repair (BER) and mismatch repair (MMR).7,8 The various repair pathway components have been identified in the genome of the major malaria-causing parasite P. falciparum. Chloroquine, one of the most widely used antimalarials, has been shown to inhibit DNA excision repair processes and increase mutagenesis in the malaria parasite. It is well established that the underlying cause of drug resistance in malaria is the development of specific genetic mutations. There are several sequences identified in PlasmoDB, that are homologous to genes involved in repair pathways from other organisms, indicating that this pathway is likely present in the parasite.4
MMR is highly conserved in prokaryotes and higher organisms. The most well characterized MMR pathway is of Escherichia coli. Yeast, humans and other eukaryotes have multiple homologs of bacterial MutS (MSH) and MutL (MLH), but no MutH.8 DNA repair processes are involved in maintenance of genomic integrity in response to DNA damaging agents such as irradiation, chemicals and oxygen radicals, as well as errors in DNA metabolism such as misincorporation during DNA replication. The P. falciparum genome encodes at least some components of the major DNA repair processes that have been found in other eukaryotes.4 The core proteins of eukaryotic repair machinery are present in P. falciparum genome but some highly conserved proteins with more accessory roles could not be found (for example, XPA/Rad4, XPC).4 These observations suggest that there are fundamental differences in the components of the repair machinery of P. falciparum and its human host. The presence of UvrD, MutL and MutS homologs including possible orthologs of MSH2, MSH6, MLH1 and PMS1 suggests that P. falciparum can perform post-replication MMR.4,9
P. falciparum genome has an unusual complement of putative MMR proteins based on homology to functionally characterized MMR protein sequences and motifs. It is most likely that defective DNA MMR may play a role in generating genetic diversity and drug resistance in malaria parasites. The unusual complement of genes in the Plasmodium MMR system may reveal the parasites’ particular needs for genetic diversity and replication fidelity. It has also been reported that the parasite genome contains a homolog of UvrD helicase which is involved in MMR.9 In a recent study we have expressed and extensively characterized the UvrD homolog from P. falciparum.10 We have reported that the N-terminal and first half of the C-terminal of PfUvrD contain the characteristic helicase and ATPase activities and the direction of unwinding is 3′ to 5′.10 We have also shown that PfUvrD colocalizes with MLH (MutL homolog) and modulates its activity through physical interaction.10,11 Furthermore the alignment with UvrD from E. coli showed the presence of all the conserved motifs in the UvrD of P. falciparum which suggests that PfUvrD is more homologous to prokaryotic system.10
UvrD from E. coli is a 73-kDa protein and was characterized as DNA helicase II.12 UvrD and Rep belong to the SF1 family, which shares 40% amino-acid identity and these are remarkably similar to the PcrA helicase of Gram-positive bacteria.13,14 The UvrD helicase is essential for the replication of Gram-negative rolling-circle plasmids.15 It has been suggested that PcrA and Rep or UvrD share a common important function in vivo. The Srs2 helicase of Saccharomyces cerevisiae is the closest found ortholog to UvrD, Rep and PcrA in eukaryotes.16 UvrD deletion mutants in E. coli have been shown to be sensitive to UV irradiation, are more prone to mutation (hypermutable) and recombination (hyper-recombination). Similarly, in vitro experiments also show that MutL can load UvrD onto DNA during methyl-directed MMR.17 Once loaded on the DNA, it is thought that UvrD uses its helicase activity to dissociate a fragment of ssDNA from its complementary strand.18 In E. coli MutL has been characterized as the master regulator of MMR because it interacts with and modulates the activity of several other proteins such as MutS, MutH and UvrD involved in MMR.17 The studies in other systems suggest that UvrD is an essential enzyme.19 It has been reported that helicases are feasible drug targets.20 It was also reported that various DNA helicases from the PcrA/UvrD/Rep (PUR) subfamily are essential for the survival of different pathogenic bacteria and they can be inhibited with small synthetic molecules. Altogether these observations suggest that these enzymes are potential novel drug targets.21 It is interesting to note that only UvrD from PcrA/UvrD/Rep (PUR) subfamily is present in the Plasmodium genome. In the present study we report an in silico comparative analysis of UvrD helicase from a variety of Plasmodium species. Since the human host contains no homolog of UvrD9 these studies will aid in determining the importance of UvrD as potential drug target.
Results and Discussion
UvrD helicase of different Plasmodium species are described in the following sections.
Plasmodium berghei
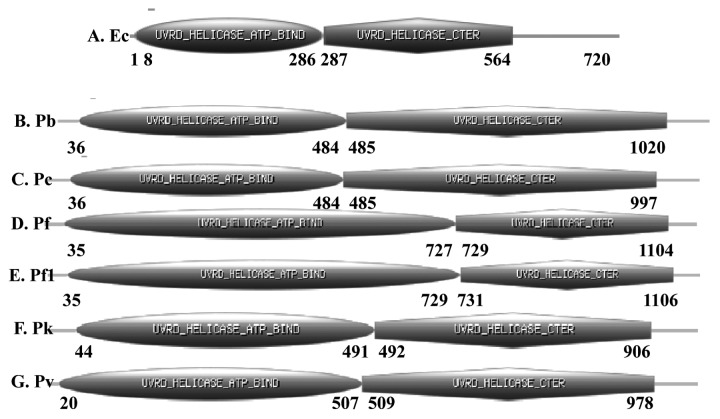
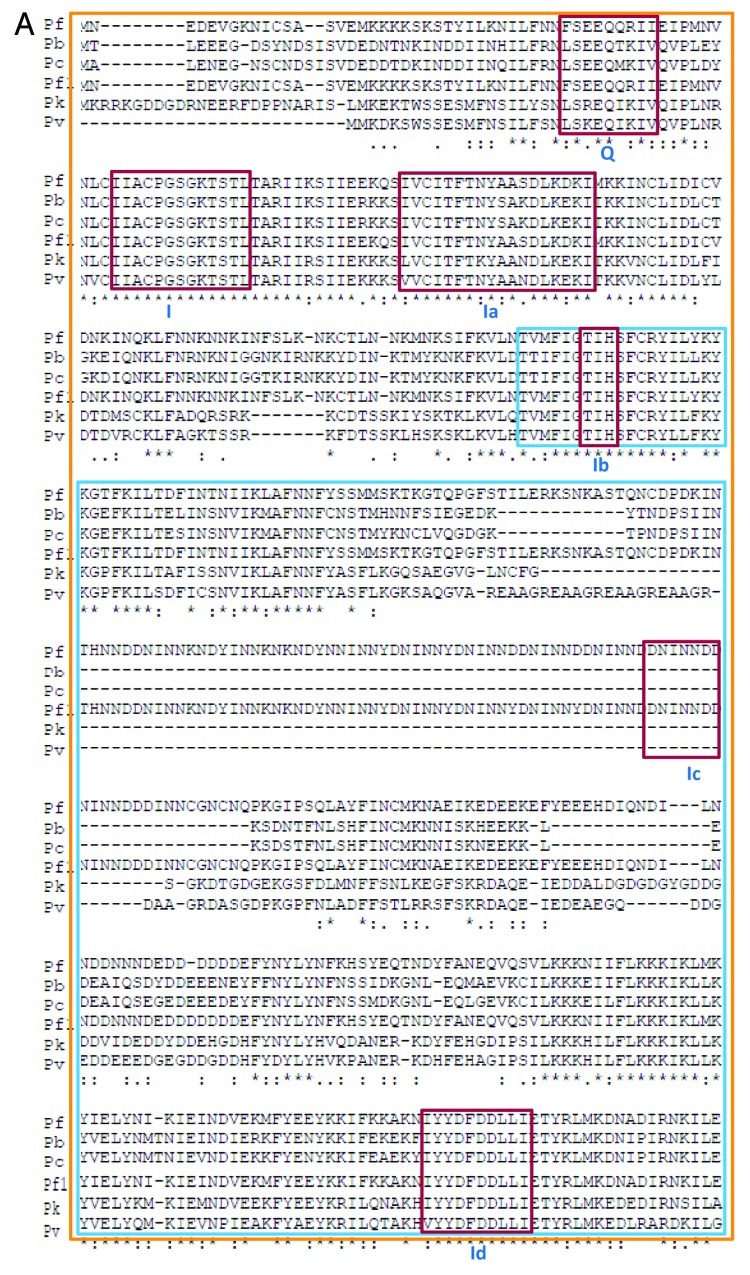
P. berghei UvrD is a 1090 amino acid protein and its molecular weight is ~128 kDa. It is larger than the E. coli UvrD, which is only 720 amino acids (Fig. 1A). The PlasmoDB number for PbUvrD is PBANKA_111380 and it contains the characteristic ATP binding and helicase C-terminal domains from amino acids 36–484 and 485–1020 respectively (Fig. 1B). PbUvrD also contains all the conserved motifs of UvrD helicases except motif Ic (Figs. 2A, 2B and 2C).
Figure 1. Schematic diagrams showing the domain organization in A-G, E. coli, P. berghei, P. chabaudi, P. falciparum 3D7 (Pf), P. falciparum IT (Pf1), P. knowlesi and P. vivax UvrD helicases. Domain analysis was done using Scan Prosite at (http://expasy.org). The domain structure was taken from the results and used in the figures. UvrD helicase ATP binding and UvrD C-terminal domains are shown. The numbers show the amino acids spanning these motifs. For detailed characterization of P. falciparum 3D7 UvrD helicase please see reference number 10.
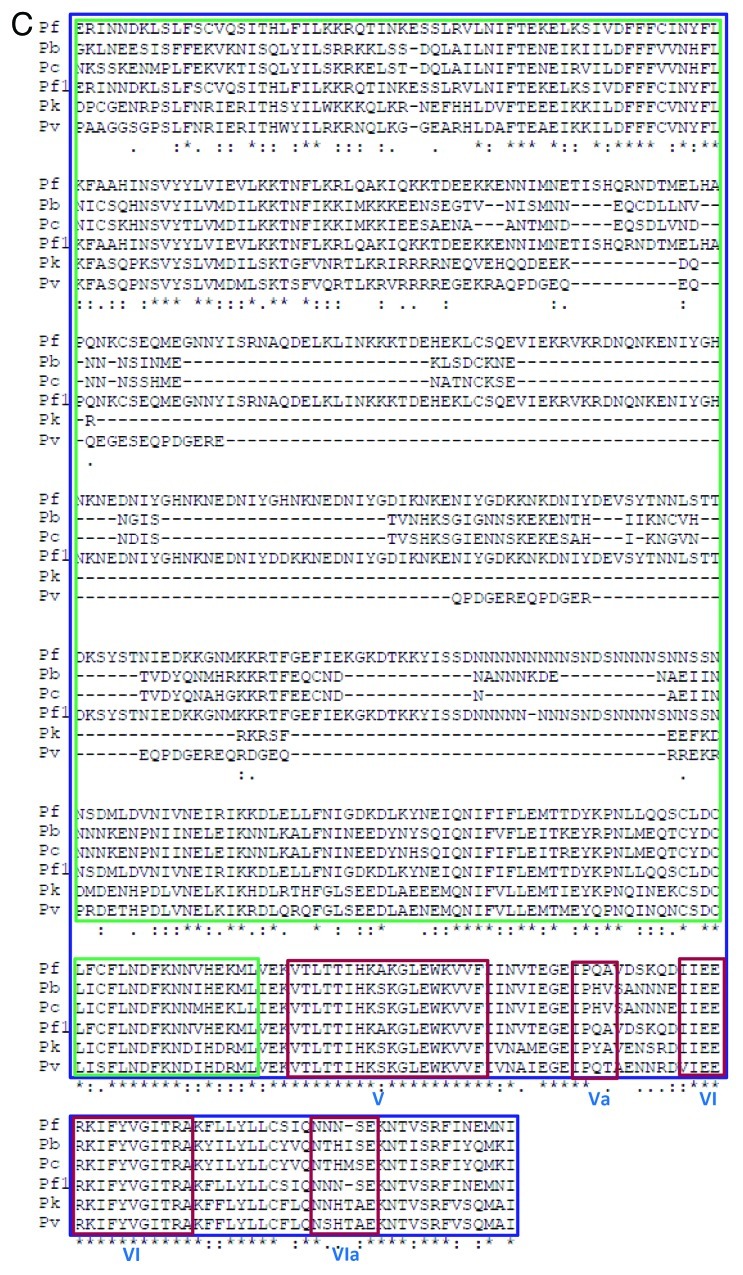
Figure 2A. Comparison of amino acid sequence of P. berghei, P. chabaudi, P. falciparum 3D7 (Pf), P. falciparum IT (Pf1), P. knowlesi and P. vivax UvrD helicases. The alignment was done using clustal omega program (http://www.ebi.ac.uk/Tools/msa/clustalo/). The conserved motifs are boxed in red color and the name of each motif (from Q to VIa) is written in roman numerals. The orange box indicates the domain 1A and the blue box inside it indicates the domain 1B. The purple box denotes the domain 2A and the green box inside it indicates the domain 2B.
Figure 2B. See Figure 2A legend.
Figure 2C. See Figure 2A legend.
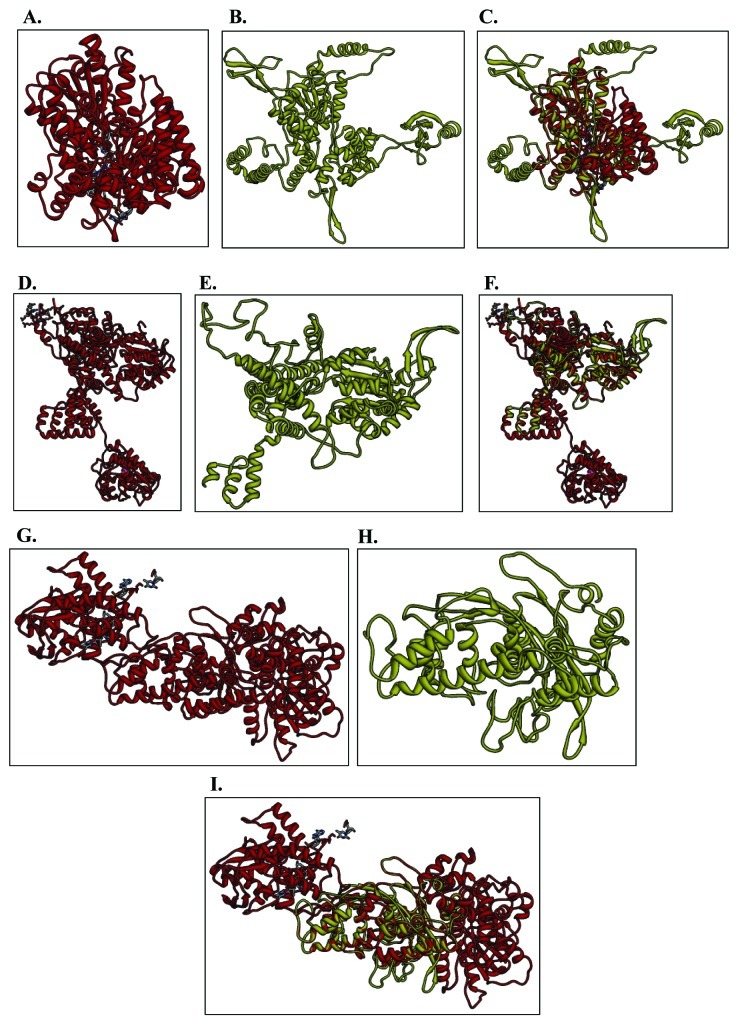
For structural modeling the sequence of full-length PbUvrD was submitted to the Swissmodel homology-modeling server (http://swissmodel.expasy.org/).22 A total of two models were obtained and one model covered only a few amino acids while the second model covered a larger range (amino acid 36–710) of the PbUvrD sequence. Therefore this model was built using E. coli REP helicase as a template.23 PbUvrD primary sequence residues 36 to 710 showed ~15% identity to the E. coli REP helicase.23 The structural modeling of the PbUvrD was therefore done using the known crystal structure of this homolog as the template (PDB number 1uaaA at http://www.rcsb.org/pdb/). The ribbon diagram of the template is shown in Figure 3A and the predicted structure of PbUvrD is shown in Figure 3B. When the modeled structure of PbUvrD and the template were superimposed, it is clear that these structures superimpose partially (Fig. 3C). Molecular graphic images were produced using the UCSF Chimera package.
Figure 3. Structure modeling. The P. berghei, P. chabaudi and P. knowlesi UvrD full-length sequences were submitted to Swissmodel server and the structures were obtained. The molecular graphic images were produced using the UCSF Chimera package from the resource for Biocomputing, Visualization, and Informatics (http://www.cgl.ucsf.edu/chimera) at the University of California, San Francisco (supported by NIH P41 RR-01081). (A) Template for PbUvrD; (B) full-length PbUvrD; (C) superimposed image; (D) Template for PcUvrD; (E) full-length PcUvrD; (F) superimposed image; (G) Template for PkUvrD; (H) full-length PkUvrD; (I) superimposed image.
Plasmodium chabaudi
P. chabaudi UvrD is a 1067 amino acid protein and its molecular weight is ~125 kDa. PcUvrD is also larger than the E. coli UvrD, which is only 720 amino acids (Fig. 1A). The PlasmoDB number for PcUvrD is PCHAS_111340 and it contains the characteristic ATP binding and helicase C-terminal domains from amino acids 36–484 and 485–997 respectively (Fig. 1C). PcUvrD also contains all the conserved motifs of UvrD helicases except motif Ic (Fig. 2A−C).
For structural modeling the sequence of full-length PcUvrD was submitted to the Swissmodel homology-modeling server (http://swissmodel.expasy.org/).22 A total of two models were obtained and one model covered only a few amino acids while the second model covered a larger range (amino acid 43–703) of the PcUvrD sequence. Therefore this model was built using E. coli RecBCD helicase chain B as template.24 PcUvrD primary sequence residues 43 to 703 showed ~13% identity to the E. coli RecBCD helicase chain B.24 The structural modeling of the PcUvrD was therefore done using the known crystal structure of this homolog as the template (PDB number 1w36 Chain B at http://www.rcsb.org/pdb/). The ribbon diagram of the template is shown in Figure 3D and the predicted structure of PcUvrD is shown in Figure 3E. When the modeled structure of PcUvrD and the template were superimposed, it is clear that these structures superimpose partially (Fig. 3F). Molecular graphic images were produced using the UCSF Chimera package as described above.
Plasmodium falciparum
P. falciparum 3D7 strain/IT strain UvrD is a 1441/1442 amino acid protein and its molecular weight is ~170 kDa. The PlasmoDB number for PfUvrD 3D7 strain is PF3D7_0514100 and for IT strain is PFIT_0514200 and it is almost twice the size of its E. coli UvrD counterpart, which is only 720 amino acids (Fig. 1A). PfUvrD contains the characteristic ATP binding and helicase C-terminal domains (Fig. 1D and E) and we have recently biochemically characterized the PfUvrD. PfUvrD also contains all the conserved motifs of UvrD helicases including motif Ic (Fig. 2A−C). We have recently reported that the structural model of PfUVrD was built using PcrA DNA helicase from B. stearothermophilus25 and when the modeled structure of PfUvrD and the template were superimposed, these structures superimposed partially.10
Plasmodium knowlesi
P. knowlesi UvrD is a 976 amino acid protein and its molecular weight is ~114 kDa. PkUvrD is also slightly larger than its E. coli counterpart, which is only 720 amino acids (Fig. 1A). The PlasmoDB number for PkUvrD is PKH_101840 and it contains the characteristic ATP binding and helicase C-terminal domains from amino acids 44–491 and 492–906 respectively (Fig. 1F). PkUvrD also contains all the conserved motifs of UvrD helicases except motif Ic (Fig. 2A−C).
The sequence of full-length PkUvrD was also submitted to the Swissmodel homology-modeling server (http://swissmodel.expasy.org/) for structural modeling.22 A total of four models were obtained and three models covered only a few amino acids while the fourth model covered a larger range (amino acid 520–974) of the PkUvrD sequence. Therefore this model was built using E. coli RecBCD helicase chain C as template.24 PkUvrD primary sequence residues 520 to 974 showed ~8% sequence identity to the E. coli RecBCD helicase chain C.24 The structural modeling of the PkUvrD was therefore done using the known crystal structure of this homolog as the template (PDB number 1w36 Chain C at http://www.rcsb.org/pdb/). The ribbon diagram of the template is shown in Figure 3G and the predicted structure of PkUvrD is shown in Figure 3H. When the modeled structure of PkUvrD and the template were superimposed, it is clear that these structures superimpose partially (Fig. 3I). Molecular graphic images were produced using the UCSF Chimera package as described above.
Plasmodium vivax
P. vivax UvrD is a 1048 amino acid protein and its molecular weight is ~119 kDa. PvUvrD is also larger than its E. coli counterpart, which is only 720 amino acids (Fig. 1A). The PlasmoDB number for PvUvrD is PVX_080535 and it contains the characteristic ATP binding and helicase C-terminal domains from amino acids 20–507 and 509–978 respectively (Fig. 1G). PvUvrD also contains all the conserved motifs of UvrD helicases except motif Ic (Fig. 2A−C).
The sequence of full-length PvUvrD was also submitted to the Swissmodel homology-modeling server (http://swissmodel.expasy.org/) for structural modeling.22 A total of two models were obtained which covered very limited area of less than 100 amino acids of the PvUvrD sequence and were not analyzed further.
Overall this analysis indicates that the UvrD helicase in all the Plasmodium species is not highly conserved. Although in all the species all the motifs except motif Ic are present but in P. falciparum the UvrD is quite large and contains long insertions in between various conserved motifs such as Ib and Id, II and III, IVc and V (Fig. 2A−C). The structural modeling results also suggest that all these UvrD helicases are different at structural level (Fig. 3).
Materials and Methods
All the sequence data used in the analysis were downloaded from Plasmodium genome database available at www.plasmodb.org. Further detailed analysis of the protein sequences was done using Expasy (http://prosite.expasy.org) and the data are presented in figure. The downloaded sequences were aligned using clustal omega program at (http://www.ebi.ac.uk/Tools/msa/clustalo/). All the motifs in conserved domains were manually assigned and the data are presented in figures. The sequences of Plasmodium berghei, Plasmodium chabaudi, Plasmodium knowlesi and Plasmodium vivax were submitted to Swissmodel homology-modeling server (http://swissmodel.expasy.org/) for structural modeling.22 Molecular graphic images were produced using the UCSF Chimera package (http://www.cgl.ucsf.edu/chimera) from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIH P41 RR-01081).26
Acknowledgments
This work in R.T's laboratory is partially supported by the Department of Biotechnology and Department of Science and Technology grants. Infra-structural support from the Department of Biotechnology, Government of India is gratefully acknowledged.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cib/article/23125
References
- 1.Tuteja R. Malaria - the global disease. FEBS J. 2007;274:4669. doi: 10.1111/j.1742-4658.2007.05996.x. [DOI] [PubMed] [Google Scholar]
- 2.Tuteja R. Malaria - an overview. FEBS J. 2007;274:4670–9. doi: 10.1111/j.1742-4658.2007.05997.x. [DOI] [PubMed] [Google Scholar]
- 3.Hyde JE. Drug-resistant malaria - an insight. FEBS J. 2007;274:4688–98. doi: 10.1111/j.1742-4658.2007.05999.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gardner MJ, Shallom SJ, Carlton JM, Salzberg SL, Nene V, Shoaibi A, et al. Sequence of Plasmodium falciparum chromosomes 2, 10, 11 and 14. Nature. 2002;419:531–4. doi: 10.1038/nature01094. [DOI] [PubMed] [Google Scholar]
- 5.Castellini MA, Buguliskis JS, Casta LJ, Butz CE, Clark AB, Kunkel TA, et al. Malaria drug resistance is associated with defective DNA mismatch repair. Mol Biochem Parasitol. 2011;177:143–7. doi: 10.1016/j.molbiopara.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greenwood BM, Fidock DA, Kyle DE, Kappe SHI, Alonso PL, Collins FH, et al. Malaria: progress, perils, and prospects for eradication. J Clin Invest. 2008;118:1266–76. doi: 10.1172/JCI33996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tuteja N, Tuteja R. Unraveling DNA repair in human: molecular mechanisms and consequences of repair defect. Crit Rev Biochem Mol Biol. 2001;36:261–90. doi: 10.1080/20014091074192. [DOI] [PubMed] [Google Scholar]
- 8.Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 9.Shankar J, Tuteja R. UvrD helicase of Plasmodium falciparum. Gene. 2008;410:223–33. doi: 10.1016/j.gene.2007.12.015. [DOI] [PubMed] [Google Scholar]
- 10.Ahmad M, Ansari A, Tarique M, Satsangi AT, Tuteja R. Plasmodium falciparum UvrD helicase translocates in 3′ to 5′ direction, colocalizes with MLH and modulates its activity through physical interaction. PLoS One. 2012;7:e49385. doi: 10.1371/journal.pone.0049385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tarique M, Satsangi AT, Ahmad M, Singh S, Tuteja R. Plasmodium falciparum MLH is schizont stage specific endonuclease. Mol Biochem Parasitol. 2012;181:153–61. doi: 10.1016/j.molbiopara.2011.10.012. [DOI] [PubMed] [Google Scholar]
- 12.Kumura K, Sekiguchi M. Identification of the uvrD gene product of Escherichia coli as DNA helicase II and its induction by DNA-damaging agents. J Biol Chem. 1984;259:1560–5. [PubMed] [Google Scholar]
- 13.Petit MA, Dervyn E, Rose M, Entian KD, McGovern S, Ehrlich SD, et al. PcrA is an essential DNA helicase of Bacillus subtilis fulfilling functions both in repair and rolling-circle replication. Mol Microbiol. 1998;29:261–73. doi: 10.1046/j.1365-2958.1998.00927.x. [DOI] [PubMed] [Google Scholar]
- 14.Anand SP, Mitra P, Naqvi A, Khan SA. Bacillus anthracis and Bacillus cereus PcrA helicases can support DNA unwinding and in vitro rolling-circle replication of plasmid pT181 of Staphylococcus aureus. J Bacteriol. 2004;186:2195–9. doi: 10.1128/JB.186.7.2195-2199.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bruand C, Ehrlich SD. UvrD-dependent replication of rolling-circle plasmids in Escherichia coli. Mol Microbiol. 2000;35:204–10. doi: 10.1046/j.1365-2958.2000.01700.x. [DOI] [PubMed] [Google Scholar]
- 16.Aboussekhra A, Chanet R, Zgaga Z, Cassier-Chauvat C, Heude M, Fabre F. RADH, a gene of Saccharomyces cerevisiae encoding a putative DNA helicase involved in DNA repair. Characteristics of radH mutants and sequence of the gene. Nucleic Acids Res. 1989;17:7211–9. doi: 10.1093/nar/17.18.7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matson SW, Robertson AB. The UvrD helicase and its modulation by the mismatch repair protein MutL. Nucleic Acids Res. 2006;34:4089–97. doi: 10.1093/nar/gkl450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mechanic LE, Frankel BA, Matson SW. Escherichia coli MutL loads DNA helicase II onto DNA. J Biol Chem. 2000;275:38337–46. doi: 10.1074/jbc.M006268200. [DOI] [PubMed] [Google Scholar]
- 19.Sinha KM, Stephanou NC, Unciuleac MC, Glickman MS, Shuman S. Domain requirements for DNA unwinding by mycobacterial UvrD2, an essential DNA helicase. Biochemistry. 2008;47:9355–64. doi: 10.1021/bi800725q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tuteja R. Helicases - feasible antimalarial drug target for Plasmodium falciparum. FEBS J. 2007;274:4699–704. doi: 10.1111/j.1742-4658.2007.06000.x. [DOI] [PubMed] [Google Scholar]
- 21.Chène P. PcrA/UvrD/Rep DNA helicases in bacterial genomes. Biochem Syst Ecol. 2008;36:101–9. doi: 10.1016/j.bse.2007.08.001. [DOI] [Google Scholar]
- 22.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 23.Korolev S, Hsieh J, Gauss GH, Lohman TM, Waksman G. Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell. 1997;90:635–47. doi: 10.1016/S0092-8674(00)80525-5. [DOI] [PubMed] [Google Scholar]
- 24.Singleton MR, Dillingham MS, Gaudier M, Kowalczykowski SC, Wigley DB. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature. 2004;432:187–93. doi: 10.1038/nature02988. [DOI] [PubMed] [Google Scholar]
- 25.Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/S0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 26.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]