Abstract
The recent discovery of introner-like elements (ILEs) in six fungal species shed new light on the origin of regular spliceosomal introns (RSIs) and the mechanism of intron gains. These novel spliceosomal introns are found in hundreds of copies, are longer than RSIs and harbor stable predicted secondary structures. Yet, they are prone to degeneration in sequence and length to become undistinguishable from RSIs, suggesting that ILEs are predecessors of most RSIs. In most fungi, other near-identical introns were found duplicated in lower numbers in the same gene or in unrelated genes, indicating that intron duplication is a widespread phenomenon. However, ILEs are associated with the majority of intron gains, suggesting that the other types of duplication are of minor importance to the overall gains of introns. Our data support the hypothesis that ILEs’ multiplication corresponds to the main mechanism of intron gain in fungi.
Keywords: ILE, intron duplication, intron gain, intron loss, intron origin, introner, spliceosomal retrohoming
The Proposed Mechanisms for Intron Gain Cannot Explain the High Intron Density in Present Day Eukaryotic Genomes
Eukaryotic genes consist of exons that contain the coding sequence, and of introns that are non-coding and are removed from premature mRNA after transcription. The spliceosome machinery, a large ribonucleoprotein that recognizes specific intronic features, catalyzes two consecutive transesterification reactions that result in splicing of the nuclear introns and ligation of adjacent exons.1 Such a mosaic gene structure is certainly one of the most important features that allowed the appearance of complex organisms during evolution of higher Eukaryotes.2 Indeed, land plants and animals, including humans, have intron-rich genomes (> 3 introns per kb coding sequence) as compared with more simple organisms such as most fungi (< 3 introns per kb coding sequence).3,4 Yet, more than 30 y after their discovery, the origin of spliceosomal introns is still unknown. Analyses of gain and loss of introns in diverse eukaryotic lineages kept the mystery on introns’ origin alive because there was less evidence for gains as compared with losses.4,5 In many Eukaryotes, the estimated rates for intron gain and loss cannot explain the high intron density in many present-day genomes. Indeed, a higher intron loss rate would ultimately result in the disappearance of spliceosomal introns. However, some lineages such as fungi have experienced more balanced rates of intron gains and losses,6,7 suggesting that intron gains can still occur to a large extent in present days. In addition to fungi,6-9 extensive recent intron gains have been reported in the micro-crustacean Daphnia pulex.10
Several mechanisms have been proposed for intron gains and have been recently reviewed in detail.11 The model that has received most support in the scientific community is referred to as intron transposition. It involves reverse splicing of a spliced intron into mRNA of another gene, followed by reverse transcription and homologous recombination at the gene locus. This model is, almost identical to the main mechanism proposed for intron loss by reverse transcription and homologous recombination after intron splicing.11,12 Observations of intron losses occurring more frequently at the 3′ end of the genes support this mechanism.6,12,13 However, according to these models, the difference in rates of intron gain and loss solely depends on the rate of reverse splicing, which is expected to occur at low frequency.14 Thus, the balanced rates of intron gain and loss in certain lineages challenge the intron transposition model. Roy and Irimia proposed two new models to resolve this paradox: spliceosomal retrohoming (reverse splicing of an intron directly into DNA followed by reverse transcription) and template switching during reverse transcription.14 Other mechanisms have also been suggested including: (1) recombination between two paralogs, one containing an intron and the other one intronless (intron transfer); (2) insertion of a transposable element followed by conversion to an intron; (3) intronization of an exon by acquisition of splicing sites; (4) mobilisation and propagation of a self-splicing group II intron from an organelle into the nucleus; (5) insertion during DNA double-strand breaks repair; and finally (6) duplication of a genomic segment that contains cryptic splicing sites.11 However, only the last mechanism has been experimentally proven.15 All the other models, including intron transposition, only rely on indirect evidence and fail to describe how the vast majority of introns were gained.11 It is likely that all proposed mechanisms contribute to intron gains to some extent, but the frequencies at which they occur cannot explain the high number of introns present in numerous Eukaryotes. Therefore, it has been suggested that the mechanism of intron gain in ancestral lineages might differ from those that occur in modern Eukaryotes.5
Intron Duplication is a Widespread Phenomenon in Fungi
A striking observation in the animal Oikopleura dioica16 and in the alga Micromonas pusilla17 was the presence of introns that are nearly identical at the sequence level. In M. pusilla, these near-identical introns are present in thousands of copies and were named introner elements (IE). Near-identical introns were also reported to occur in the fungus Mycosphaerella graminicola.8 Recently, we reported on the occurrence of near-identical introns in five additional fungal species, where they are present in up to five hundred copies.9 We named these high-copy introns introner-like elements (ILE) to refer to IEs found in M. pusilla. Like regular spliceosomal introns (RSIs), ILEs have typical splicing features including canonical acceptor and donor sites, branch point sequence and polypyrimidine tracts, which suggest that they can be spliced by the spliceosome machinery. However, in addition to being present in many near-identical copies, we also found that ILEs have features completely different from RSIs. They are significantly longer and have lower predicted Gibbs free energy (ΔG) values that were ascribed to stable predicted secondary structures. A robust gain analysis showed that up to 90% of gained introns are ILEs. Because our data showed that ILEs quickly degenerate in length and sequence to become undistinguishable from RSIs, we hypothesized that non-ILE-associated gains are highly degenerated ILEs. Thus, most RSIs might originate from ILEs in at least six fungal species.9
In this study, the very first step of the pipeline that was developed to identify ILEs involved a simple BlastN search and clustering method, which retrieved three different types of near-identical introns.9 Depending on the number of introns with a near-identical sequence and whether they were duplicated within the same gene or in different genes, these multi-copy introns were classified as same gene duplications (SGD; 82 members), low-copy introns (LCI; 302 members) and high-copy introns (1226 members) that were subsequently named ILEs. This search revealed that intron duplication is a widespread phenomenon in fungi because it was found in all species included in the study except Aspergillus nidulans (Table 1). However, the contribution of each category to the observed duplication events varies. Nine species contain only LCIs, while both SGDs and LCIs are found in five other species. In the latter, SGDs occur less frequently and contribute to 25–54% of the observed duplications (Table 1). The remaining six fungal species have all three types of duplicated introns, but they also have a very high number of ILEs (24 to 377), which contribute between 60% and 92% to all duplication events (Table 1). Noteworthy, Rhytidhysteron rufulum, Fusarium graminearum and Sclerotinia sclerotiorum contain near-identical introns in high numbers but they correspond to repetitive elements that inserted within RSIs and were not retrieved as ILEs in the subsequent and more stringent steps of ILE identification (Table 1).9
Table 1. Identification of multi-copy introns in 24 fungal species.
| Fungal species | Total | SGD a | LCI a | ILE a |
|---|---|---|---|---|
|
Cladosporium fulvum |
408 |
3 (1) |
28 (7) |
377 (92) |
|
Mycosphaerella graminicola |
344 |
16 (5) |
22 (6) |
306 (89) |
|
Dothistroma septosporum |
322 |
7 (2) |
17 (5) |
298 (93) |
|
Hysterium pulicare |
188 |
16 (9) |
28 (15) |
144 (77) |
|
Mycosphaerella fijiensis |
97 |
14 (14) |
22 (23) |
61 (63) |
|
Stagonospora nodorum |
40 |
0 |
16 (40) |
24 (60) |
|
Fusarium oxysporum |
37 |
0 |
37 (100) |
0 |
|
Coccidioides immitis |
24 |
6 (25) |
18 (75) |
0 |
|
Histoplasma capsulatum |
18 |
0 |
18 (100) |
0 |
|
Rhytidhysteron rufulum |
17 |
5 (29) |
8 (47) |
4b (24) |
|
Leptosphaeria maculans |
13 |
0 |
13 (100) |
0 |
|
Septoria musiva |
13 |
4 (31) |
9 (69) |
0 |
|
Nectria hematococca |
13 |
7 (54) |
6 (46) |
0 |
|
Fusarium graminearum |
12 |
0 |
2 (17) |
10b (83) |
|
Cryptococcus neoformans |
12 |
0 |
12 (100) |
0 |
|
Sclerotinia sclerotiorum |
10 |
0 |
8 (80) |
2b (20) |
|
Cochliobolus heterostrophus |
8 |
0 |
8 (100) |
0 |
|
Botrytis cinerea |
8 |
2 (25) |
6 (75) |
0 |
|
Neurospora crassa |
6 |
0 |
6 (100) |
0 |
|
Trichoderma atroviridae |
6 |
0 |
6 (100) |
0 |
|
Verticillium aalbo-atrum |
6 |
0 |
6 (100) |
0 |
|
Magnaporthe oryzae |
6 |
2 (33) |
4 (67) |
0 |
|
Verticillium dahliae |
2 |
0 |
2 (100) |
0 |
|
Aspergillus nidulans |
0 |
0 |
0 |
0 |
| Total | 1610 | 82 (5) | 302 (19) | 1226 (76) |
For each intron of a given fungal species, a BlastN analysis was performed using the complete intronome. Then, intron clusters were built by grouping a given intron with its near-identical introns. Introns that were duplicated only within the same gene were classified as same gene duplications (SGD). Near-identical introns found in unrelated genes were classified as low-copy introns (LCI) when a search using hidden Markov models did not increase the number of members by more than 2-fold; they were classified as high-copy introns when this search increased the number of members by more than 2-fold. These high-copy introns were subsequently named introner-like elements (ILE).9 aNumber of introns. Contribution of a duplication type to the total number of duplications is indicated as percentage in brackets; bThese high-copy introns were not retrieved as ILEs by additional more stringent analyses.
As was done in our previous study on ILEs, the length and stability of the two other types of near-identical introns were measured. The median length of SGDs and LCIs are in the same range as observed for non-duplicated introns (NDI), but ILEs are about twice as long (Fig. 1A). The ΔG free energy of SGDs and LCIs is not different from that of NDIs, while ILEs have a significantly lower ΔG (Fig. 1B). These results suggest that different mechanisms might be involved in the duplication of each intron type. SGDs are found in only 11 fungal species and are limited in number (maximum of 16 members in a given species). Fifty percent of these duplication events represent segmental duplication within the same gene because exon sequences on each side of these introns are also duplicated. The other 50% might represent intron transpositions within the same transcript or intron transfers between paralogs. Comparable low numbers were also reported in Caenorhabditis elegans in which only three gained introns are SGDs.18 In C. neoformans, a single gene with several putative SGDs was also shown to be most likely the result of a duplication of exonic repeats.19 The two other types of multi-copy introns are found in different unrelated genes, suggesting that they may represent the same type of introns, but differ in multiplication frequency. They have different characteristics (length and ΔG), which suggests that different duplication mechanisms are involved. However, these differences are also consistent with ILE degeneration and LCIs might represent degenerated ILEs. This hypothesis might explain why we could not identify more introns that would have originated from them. Alternatively, LCIs could originate from a low frequency transposition mechanism. Altogether, our results suggest that ILEs are prevailing duplication events in fungi, explaining on average 76% of intron duplications.
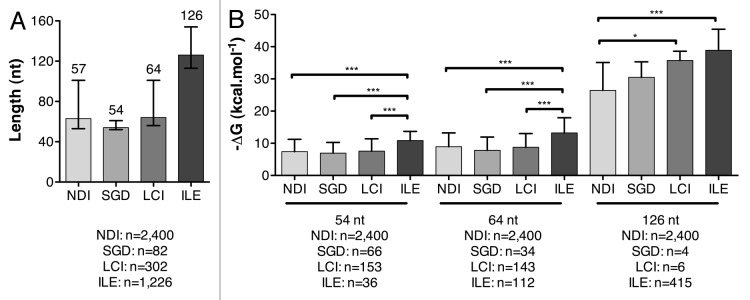
Figure 1. Length and stability of the different types of duplicated introns. The length and predicted Gibbs free energy (∆G) were measured for non-duplicated intron (NDI), same gene duplications (SGD), low-copy introns (LCI) and introner-like elements (ILE) from 24 fungal species included in this study.9 (A) Median length and interquartile range are plotted for each type of intron. The median length is indicated above the bars. (B) Mean and SD of ∆G values of introns with a length corresponding to the median of each type of intron. A non-parametric Kruskall-Wallis test was performed (p < 0.0001), followed by a Dunn’s pairwise comparison test at α = 0.05 significance level. Only significant differences are indicated.
Introner-Like Elements Reconcile the Intron Gain Mechanism in Ancestral and Modern Genomes
Based on the observed degeneration, we speculated that ILEs are at the origin of most RSIs in at least six fungal species, which implies that they should be associated with intron gains. Indeed, ILEs can contribute up to 90% of recent intron gains.9 An intron gain and loss analysis (IGL) in fungal species that contain ILEs showed that gains occur on average 10-fold more frequently than losses (Table 2). Remarkably, this is also true in Septoria musiva, a species that carries highly degenerated ILEs only, which initially could not be identified as such.9 In the IGL analysis shown here, up to 50% of the gains are explained by ILEs, while almost none are explained by SGDs or LCIs (Table 2). The non-explained gains certainly correspond to more ancient gained introns that cannot be recognized as ILEs because of the high level of degeneration.9
Table 2. Single intron gain and loss analysis in fungal species containing ILEs.
| Fungal species | Orthologs | Introns | ILEs | Ancestral intron a | Single gain b | Single loss b | SGD at gain positions c | LCI at gain positions c | ILE at gain positions c |
|---|---|---|---|---|---|---|---|---|---|
|
Cladosporium fulvum |
3050 |
3483 |
110 |
2209 |
178 |
20 |
0 |
5 (0.028) |
95 (0.534) |
|
Dothistroma septosporum |
3050 |
3516 |
101 |
2209 |
199 |
10 |
0 |
2 (0.010) |
91 (0.457) |
|
Septoria musiva |
2824 |
2084 |
- |
906 |
372 |
60 |
1 (0.003) |
2 (0.005) |
- |
|
Mycosphaerella fijiensis |
2824 |
1951 |
14 |
906 |
236 |
43 |
0 |
1 (0.004) |
14 (0.059) |
| Mycosphaerella graminicola | 2824 | 2240 | 44 | 906 | 388 | 40 | 0 | 1 (0.003) | 43 (0.111) |
Single gains and single losses were determined using only one outgroup clade for each species as described in our previous report.9 Contribution of same gene duplications (SGD), low-copy introns (LCI) and introner-like elements (ILE) to single gains was determined. aIntron position conserved in all analyzed fungal species; b Introns that are present or absent only in the considered species; c Numbers in brackets are numbers of SGDs, LCIs or ILEs at single gain positions divided by the number of single gains.
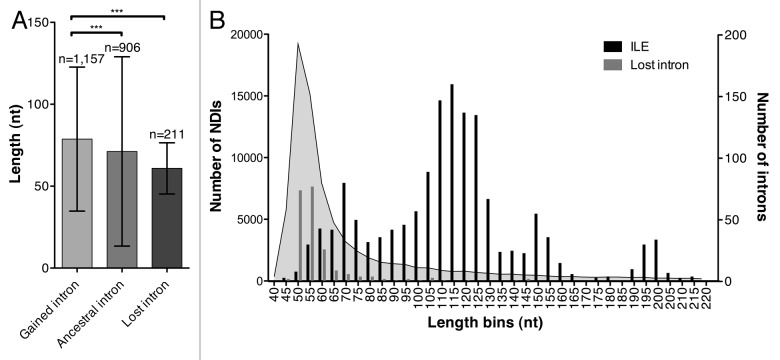
Our analysis also revealed that introns absent in other species are similar in length to ancestral introns that are conserved in all fungal species included in this study, although with a much lower standard deviation (Fig. 2A). Our findings suggest that the majority of new introns originate from ILEs, which subsequently lose their stable secondary structure and shorten toward the optimal intron length, to eventually be lost (Fig. 2B). Accordingly in Aspergillus species, it was found that lost introns are significantly shorter than conserved introns.7 Our proposed model for fungal intron birth, life and death is consistent with the high intron dynamics observed in fungi, but also with lower dynamics in higher Eukaryotes, which is most likely related to the different generation times. Intron-rich genomes usually have longer introns,3 which would hamper their loss.
Figure 2. Birth, life and death of spliceosomal introns in fungi. (A) Gained introns are single gains in Cladosporium fulvum, Dothistroma septosporum, Mycosphaerella graminicola, Mycosphaerella fijiensis or Septoria musiva as determined in Table 2. Ancestral introns are conserved among all fungi included in this study. Lost introns are single losses in one of the five fungal species. Length of lost introns that are still present in the other four species was calculated and corrected for outliers using the formula: (sum-max-min)/(length-2). A non-parametric Kruskall-Wallis test was performed (p < 0.0001), followed by a Dunn’s pairwise comparison test at α = 0.05 significance level. Only significant differences are indicated. (B) Length distribution of non-duplicated introns (NDIs), introner-like elements (ILEs) and lost introns in the five fungal species listed above.
With the resonance of IEs in M. pusilla, it is very likely that genome invasion by introns could have occurred at least once in an ancestral Eukaryotic lineage to give rise to the present-day intron-rich Eukaryotes. This hypothesis suggests that the mechanisms of intron gains in ancestral and modern genomes are still the same. From the results presented above, multiplication of ILEs in fungi and IEs in M. pusilla is certainly the main mechanism of intron gain in these species. Because of the high frequency of duplication events, ILE and IE multiplication likely involves a mechanism different from those proposed so far. Yet, spliceosomal retrohoming is the model that would comply best with our observations, but additional concepts are required in this model to take into account ILE specific characteristics. The predicted stable secondary structures of ILEs seem to be under selection pressure as suggested by the many compensatory mutations observed in ILEs.9 It is tempting to speculate that ILE secondary structures might significantly contribute to the multiplication mechanism. We are now setting up experiments to find evidence for the mobility of ILEs and deciphering the mechanism of their multiplication.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cib/article/23147
References
- 1.Will CL, Lührmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol. 2011;3:a003707. doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koonin EV. The origin of introns and their role in eukaryogenesis: a compromise solution to the introns-early versus introns-late debate? Biol Direct. 2006;1:22. doi: 10.1186/1745-6150-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rogozin IB, Carmel L, Csuros M, Koonin EV. Origin and evolution of spliceosomal introns. Biol Direct. 2012;7:11. doi: 10.1186/1745-6150-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Csuros M, Rogozin IB, Koonin EV. A detailed history of intron-rich eukaryotic ancestors inferred from a global survey of 100 complete genomes. PLoS Comput Biol. 2011;7:e1002150. doi: 10.1371/journal.pcbi.1002150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roy SW, Gilbert W. Rates of intron loss and gain: implications for early eukaryotic evolution. Proc Natl Acad Sci USA. 2005;102:5773–8. doi: 10.1073/pnas.0500383102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nielsen CB, Friedman B, Birren B, Burge CB, Galagan JE. Patterns of intron gain and loss in fungi. PLoS Biol. 2004;2:e422. doi: 10.1371/journal.pbio.0020422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang LY, Yang YF, Niu DK. Evaluation of models of the mechanisms underlying intron loss and gain in Aspergillus fungi. J Mol Evol. 2010;71:364–73. doi: 10.1007/s00239-010-9391-6. [DOI] [PubMed] [Google Scholar]
- 8.Torriani SF, Stukenbrock EH, Brunner PC, McDonald BA, Croll D. Evidence for extensive recent intron transposition in closely related fungi. Curr Biol. 2011;21:2017–22. doi: 10.1016/j.cub.2011.10.041. [DOI] [PubMed] [Google Scholar]
- 9.van der Burgt A, Severing E, de Wit PJ, Collemare J. Birth of new spliceosomal introns in fungi by multiplication of introner-like elements. Curr Biol. 2012;22:1260–5. doi: 10.1016/j.cub.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 10.Li W, Tucker AE, Sung W, Thomas WK, Lynch M. Extensive, recent intron gains in Daphnia populations. Science. 2009;326:1260–2. doi: 10.1126/science.1179302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yenerall P, Zhou L. Identifying the mechanisms of intron gain: progress and trends. Biol Direct. 2012;7:29. doi: 10.1186/1745-6150-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fink GR. Pseudogenes in yeast? Cell. 1987;49:5–6. doi: 10.1016/0092-8674(87)90746-X. [DOI] [PubMed] [Google Scholar]
- 13.Roy SW, Gilbert W. The pattern of intron loss. Proc Natl Acad Sci USA. 2005;102:713–8. doi: 10.1073/pnas.0408274102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roy SW, Irimia M. Mystery of intron gain: new data and new models. Trends Genet. 2009;25:67–73. doi: 10.1016/j.tig.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 15.Hellsten U, Aspden JL, Rio DC, Rokhsar DS. A segmental genomic duplication generates a functional intron. Nat Commun. 2011;2:454. doi: 10.1038/ncomms1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denoeud F, Henriet S, Mungpakdee S, Aury JM, Da Silva C, Brinkmann H, et al. Plasticity of animal genome architecture unmasked by rapid evolution of a pelagic tunicate. Science. 2010;330:1381–5. doi: 10.1126/science.1194167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Worden AZ, Lee JH, Mock T, Rouzé P, Simmons MP, Aerts AL, et al. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science. 2009;324:268–72. doi: 10.1126/science.1167222. [DOI] [PubMed] [Google Scholar]
- 18.Coghlan A, Wolfe KH. Origins of recently gained introns in Caenorhabditis. Proc Natl Acad Sci USA. 2004;101:11362–7. doi: 10.1073/pnas.0308192101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharpton TJ, Neafsey DE, Galagan JE, Taylor JW. Mechanisms of intron gain and loss in Cryptococcus. Genome Biol. 2008;9:R24. doi: 10.1186/gb-2008-9-1-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]