Abstract
Burkitt lymphoma (BL) can often be cured by intensive chemotherapy, but the toxicity of such therapy precludes its use in the elderly and in patients with endemic BL in developing countries, necessitating new strategies. We used high throughput RNA sequencing and RNA interference screening to discover essential regulatory pathways that cooperate with MYC, the defining oncogene of this cancer. In 38% of sporadic BL (sBL) cases, oncogenic CCND3 mutations produced highly stable cyclin D3 isoforms that drive cell cycle progression. In 70% of sBL cases, mutations affecting the transcription factor TCF3 (E2A) or its negative regulator ID3 fostered TCF3 dependency. TCF3 activated the pro-survival PI(3) kinase pathway in BL, in part by augmenting constitutive B cell receptor signaling. These findings suggest opportunities to improve therapy for patients with BL.
Introduction
Burkitt lymphoma (BL) is an aggressive form of non-Hodgkin lymphoma derived from germinal center B cells1. Translocation of the oncogene MYC is the hallmark of this lymphoma subtype2–4, although similar translocations can occur in other aggressive lymphomas such as diffuse large B cell lymphoma (DLBCL). By gene expression profiling, BL is distinct from the germinal center B cell-like (GCB) subtype of DLBCL despite a common cell of origin for these cancers, suggesting that BL may have a unique pathogenesis5,6. BL is subdivided into a sporadic form (sBL) that is diagnosed in developed countries, an EBV-associated endemic form (eBL) in Africa, New Guinea, and parts of South America, and an HIV-associated form (hivBL). Although these BL subtypes share a common gene expression signature, sBL can be distinguished from eBL and hivBL by gene expression profiling, potentially reflecting distinct pathogenetic mechanisms7.
Intensive chemotherapeutic regimens, delivered both systemically and intrathecally, can cure many patients with sBL, albeit with considerable morbidity and some mortality, especially in elderly patients1. Similar regimens are not typically administered to patients with eBL since therapy-associated immune suppression and other toxicities are not easily managed in this setting8. Hence, less toxic yet effective therapies are desired for both sBL and eBL.
In the present study, we sought to define new pathogenetic mechanisms in BL that might suggest alternative treatment strategies. We used structural genomics to discover recurrent single nucleotide variants (SNVs), several of which were preferentially associated with BL. These data were cross-referenced with functional genomics data from RNA interference screens of BL cell lines to discover pathways that BL cells rely on for proliferation and survival.
Results
We developed an analytic pipeline to retrieve SNVs using massively parallel RNA resequencing (RNA-seq) data that relied on the combined results from three different sequence alignment tools (see Methods). As an initial check of this algorithm, we compared these putative SNVs with those called based on complete genome sequencing of 2 DLBCL tumors and observed an 89% true-positive rate (data not shown). We performed RNA-seq on 28 sBL patient biopsy samples and 13 BL cell lines. For comparison, we used our analytic pipeline to reanalyze previously published RNA-seq data from biopsy samples of 52 GCB DLBCL cases and 28 activated B cell-like (ABC) DLBCL cases9. SNVs present in 7 samples derived from normal B cell subpopulations were eliminated as false positives as were known single nucleotide polymorphisms, leaving a remaining set of putative SNVs (pSNVs) (Supplemental Table 1). By Sanger resequencing of genomic DNA, 95% of pSNVs (495/518) were confirmed as true SNVs (Supplemental Table 2). On average, each BL sample harbored 1641 pSNVs, with the majority being base pair transitions. The enzyme AID is expressed in BL and has the potential to mutate the genome, with a bias towards “hotspot” DNA (DGYW/WRCH) motifs within the first 2 kb of a transcriptional start site. BL pSNVs coincided with AID hotspots in 2.3% of instances, a lower frequency than observed in GCB or ABC DLBCL, but higher than the frequency of AID hotspots at SNPs (Supplemental Figure 1A). Thus, while some BL mutations may be introduced by AID, most are likely due to different mutational mechanisms.
Among BL samples, we searched for genes with recurrent non-synonymous pSNVs in their coding regions, and evaluated whether the prevalence of pSNVs in these genes differed between BL and DLBCL (Table 1). Several genes were mutated recurrently in both BL and DLBCL (TP53, GNA13, MKI67, CCND3), although mutations affecting the p53 DNA binding domain were significantly more common in BL, in keeping with previous work10 (Supplemental Figure 1B). Several genes that are frequently mutated in GCB DLBCL9,11,12 (EZH2, SGK1, BCL2) or ABC DLBCL13,14 (CD79B, MYD88) were rarely if ever mutated in BL. Conversely, many pSNVs were significantly more frequent in BL, indicating a divergent pathogenesis for this lymphoma subtype. At the top of this list was MYC, a gene known to be subject to AID-dependent somatic hypermutation, frequently generating Myc variants that have increased oncogenic potency15 (Supplemental Fig. 1C). A striking finding was that the genes encoding the transcription factor TCF3 (E2A) and its negative regulator ID3 were among the most frequently mutated genes in BL (see below). The helicase DDX3X was mutated in over 30% of BL samples, including many inactivating mutations, but only one GCB DLBCL had a pSNV in this gene. SMARCA4, encoding a component of the SWI/SNF chromatin remodeling complex was frequently mutated (Table 1), as were other components of this complex (ARID1A, DPF1, ACTL6A) (Supplemental Table 2). Overall, this mutational survey suggests that BL is pathogenetically distinct from other aggressive lymphoma subtypes.
Table 1.
| Gene symbol
|
Samples with pSNV (%)
|
p-value
|
||
|---|---|---|---|---|
| BL (n=41)
|
GCB DLBCL (n=52)
|
ABC DLBCL (n=28)
|
||
| MYC | 70.7 | 0 | 0 | 1.9E-20* |
| ID3 | 58.5 | 0 | 0 | 4.2E-16* |
| TP53 | 34.1 | 11.5 | 3.6 | 1.7E-04* |
| DDX3X | 31.7 | 1.9 | 0 | 3.6E-07* |
| TCF3 | 29.3 | 3.8 | 0 | 3.1E-05* |
| SMARCA4 | 26.8 | 1.9 | 3.6 | 1.0E-04* |
| NCOR2 | 19.5 | 7.7 | 0 | 7.2E-03* |
| PDCD 11 | 17.1 | 5.8 | 3.6 | 1.8E-02* |
| GNA13 | 17.1 | 13.5 | 0 | 1.5E-01* |
| MKI67 | 17.1 | 13.5 | 7.1 | 4.8E-01* |
| EXOSC6 | 14.6 | 0 | 0 | 5.5E-04* |
| WDR90 | 14.6 | 1.9 | 0 | 9.8E-03* |
| CCND3 | 14.6 | 1.9 | 10.7 | 8.6E-02* |
| FLYWCH1 | 12.2 | 0 | 0 | 2.0E-03* |
| FOXO1 | 12.2 | 1.9 | 0 | 9.3E-03* |
| ELP2 | 12.2 | 1.9 | 0 | 9.3E-03* |
| MYO18A | 12.2 | 0 | 0 | 9.3E-03* |
| GTSE1 | 12.2 | 0 | 0 | 9.3E-03* |
| YY1AP1 | 12.2 | 0 | 3.6 | 9.3E-03* |
| PCBP1 | 12.2 | 1.9 | 0 | 9.3E-03* |
| HERC1 | 9.8 | 0 | 0 | 1.1E-02* |
| REV3L | 9.8 | 0 | 0 | 7.2E-03* |
| ZNF85 | 9.8 | 0 | 0 | 7.2E-03* |
| ERAP1 | 9.8 | 0 | 0 | 7.2E-03* |
| WHAMM | 9.8 | 0 | 0 | 7.2E-03* |
| C4orf14 | 9.8 | 0 | 0 | 7.8E-03* |
| KANK2 | 9.8 | 0 | 0 | 7.2E-03* |
| DHCR7 | 9.8 | 0 | 0 | 7.2E-03* |
| T OP2A | 9.8 | 0 | 0 | 7.2E-03* |
| C16orf4 8 | 9.8 | 0 | 0 | 7.2E-03* |
| MYD88 | 4.9 | 0 | 28.6 | 1.2E-02 ¶ |
| CD79B | 0 | 1.9 | 17.9 | 8.7E-03 ¶ |
| SGK1 | 0 | 23.1 | 0 | 4.3E-03 † |
| EZH2 | 0 | 36.5 | 3.6 | 1.6E-03* |
| BCL2 | 0 | 38.5 | 7.1 | 1.6E-02* |
p-value for B L vs. all DLBCL
p-value for B L vs. GCB DLBCL
p-value for B L vs. ABC DLBCL
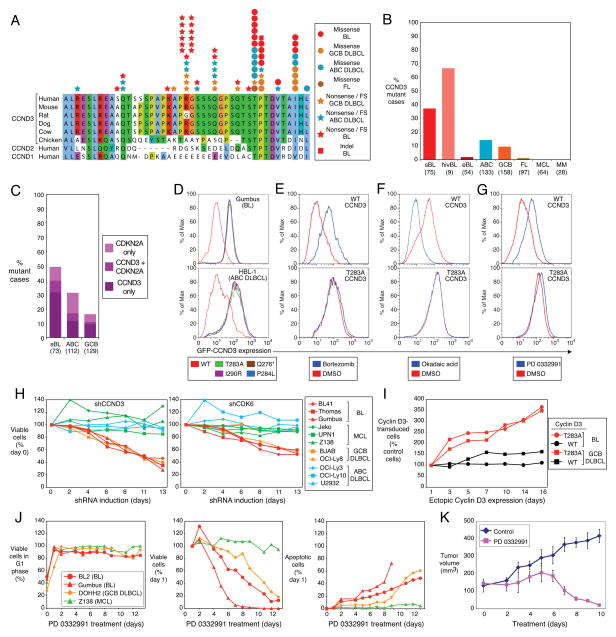
We were intrigued by the mutations in CCND3 since this gene encodes a D-type cyclin that regulates the G1-S cell cycle transition in germinal center B cells, the cell of origin of BL16,17. Sanger resequencing of genomic DNA confirmed that CCND3 incurred multiple nonsense and frame shift mutations that removed up to 41 amino acids from the cyclin D3 carboxy-terminus (Figure 1A, B, Supplemental Table 3). In addition, recurrent missense mutations affected a threonine residue at position 283 (T283), known to be involved in cyclin D3 phosphorylation18, as well as nearby proline (P284) and isoleucine (I290) residues. T283, P284, and I290 are conserved among cyclin D3 homologues in different species and the same or related amino acids occur at corresponding positions in cyclin D1 and cyclin D2 (Figure 1A). At each of these positions, multiple amino acid substitutions occurred, and most mutations were heterozygous. The incidence of CCND3 mutations among sBL samples was 38%, with a potentially higher frequency in hivBL samples. CCND3 mutations were significantly less prevalent among eBL samples (1.8%), indicating a distinct genetic pathogenesis for this BL subtype. CCND3 mutations were also recurrent in ABC and GCB DLBCL, as previously reported9, albeit at a lower frequency than in BL (Figure 1B). In 3 BL and 2 DLBCL cases for which germline DNA was available, the CCND3 mutations (R271fs, T283P, T283A, V287D) were confirmed to be of somatic origin.
Figure 1. OncogenicCCND3 mutations in Burkitt lymphoma.
a, CCND3 mutations in BL, GCB DLBCL, ABC DLBCL and follicular lymphoma (FL) are shown with respect to amino acid positions 250–292 of protein accession NP_001751. b, Frequencies of CCND3 mutations in different lymphoma subtypes. c, Occurrence of CDKN2A and CCND3 mutations in BL, ABC DLBCL and GCB DLBCL Number of samples analyzed are indicated in parentheses. d, CCND3 mutations increase protein stability. FACS analysis of lymphoma cell lines transduced with mutant or wild type GFP-CCND3 fusion proteins. Cells expressing the different cyclin D3 isoforms had equivalent expression of Lyt2-reporter encoded by the same mRNA as the GFP-CCND3 proteins. e, Mutant cyclin D3 proteins are not degraded by the proteasome. GFP-CCND3-transduced HBL1 cells were cultured overnight in the presence of the proteasomal inhibitor bortezomib (20 nM) and analyzed by FACS. f, Mutant cyclin D3 proteins are not destabilized in response to phosphatase inhibition. BL41 cells expressing GFP-CCND3 proteins were treated for 30 min with the pan-phosphatase inhibitor okadaic acid (750 nM) and analyzed by FACS. g, Stability of mutant cyclin D3 proteins is not regulated in the cell cycle. BL41 cells expressing GFP-CCND3 isoforms were treated overnight with the CDK4/6 inhibitor PD 0332991 (1.5 μM), causing arrest at the G1 phase of the cell cycle. FACS analysis indicated that wild type, but not mutant cyclin D3 fusion proteins were stabilized in G1 phase. h, CCND3 and CDK6 shRNAs have selective toxicity for BL and GCB DLBCL cell lines. Shown is the fraction of GFP+, shRNA-expressing cells relative to the GFP−, shRNA-negative fraction at the indicated times, normalized to the day 0 values. i, Mutant CCND3 confers a proliferation advantage in lymphoma cells. Endogenous CCND3 was knocked down by induction of a CCND3 shRNA in Gumbus (BL) and BJAB (GCB DLBCL) cells, while different isoforms of GFP-CCND3 were ectopically expressed. The relative number of GFP-CCND3 expressing cells is plotted versus time after shRNA and GFP-CCND3 induction, normalized to day 0. j, Cell cycle block in G1 phase is lethal to BL and GCB DLBCL cell lines. BL, GCB DLBCL and mantle cell lymphoma (MCL) cell lines were treated with PD 0332991 (1 μM) over indicated time course. G1 phase arrest of viable cells was confirmed using Propidium Iodide (PI) staining and analyzed by FACS (left panel); viable cells were counted using the trypan blue method (middle panel); apoptotic cells were quantified by cleaved PARP/active caspase 3 FACS-staining (right panel). Survival of cells and the fraction of apoptotic cells after PD 0332991 treatment is shown normalized to day 1. Time courses of G1 phase arrest and apoptotic cells were discontinued after day 9 for Gumbus cell line due to the lack of viable cells. All analyses were performed in triplicate. k, Therapeutic potential of the CDK4/6 inhibitor PD 0332991 revealed using a BL xenograft model. Immunodeficient mice bearing established subcutaneous BL xenografts were treated with PD 0332991 (150 mg/kg/day p.o.) for the indicated times. Tumor volumes were estimated by quantitative imaging of luciferase luminescence. Error bars are s.e.m. (n=3).
FS: frameshift
The G1-S phase transition is additionally regulated by the cyclin-dependent kinase inhibitor p16, encoded by the CDKN2A gene, prompting us to search for mutations and homozygous deletions in this gene in BL and DLBCL samples. BL harbored CDKN2A aberrations in 17% of samples, with 8% having both CCND3 and CDKN2A mutations, suggesting that these genetic abnormalities might cooperatively deregulate the cell cycle in some aggressive lymphomas (Figure 1C; Supplemental Tables 3, 4).
To explore the function of the CCND3 mutations, we constructed fusion proteins linking green fluorescent protein (GFP) to either wild type or mutant cyclin D3. When expressed in either a BL or ABC DLBCL cell line, all mutant isoforms accumulated to more than 10-fold higher levels than the wild type isoform (Figure 1D). Pulse-chase analysis demonstrated that the mutant CCND3 isoforms had shorter half lives than wild type CCND3 (Supplemental Figure 6G), again suggesting that the CCND3 mutations increase protein stability (Figure 1D). Consistent with this interpretation, the abundance of wild type cyclin D3-GFP was increased by proteasome inhibition, but the mutant isoforms were not further stabilized under this condition (Figure 1E and data not shown). These findings are consistent with previous work linking T283 to cyclin D3 protein stability18, and further suggested that P284 and I290 are also involved in cyclin D3 protein turnover. The degradation of cyclin D3 involves phosphorylation of T283, although the nature of the kinase is unclear. In keeping with this model, treatment of BL cells with the pan-phosphatase inhibitor okadaic acid, which generally increases protein phosphorylation, caused a marked destabilization of wild type cyclin D3-GFP, but not T283A, P284L, Q276* or I290R cyclin D3-GFP (Figure 1F, Supplemental Figure 2E). D-type cyclins are destabilized during S phase of the cell cycle and are relatively stabile in G1 phase19. When BL cells were trapped in G1 phase by treatment with the CDK4/CDK6 inhibitor PD 0332991, wild type cyclin D3-GFP levels increased whereas mutant cyclin D3 isoforms were not further stabilized (Figure 1G and data not shown), suggesting that the CCND3 mutations prevent the physiologic regulation of cyclin D3 protein levels during the cell cycle.
In a parallel effort, an RNA interference screen of lymphoma cell lines revealed that BL lines depend on CDK6, a kinase that partners with D-type cyclins (Supplemental Figure 2A, Supplemental Table 11). Knockdown of CDK6 caused a time-dependent reduction in cell numbers in all BL lines, irrespective of CCND3 mutational status, as well as in some GCB DLBCL lines (Figure 1H; Supplemental Figures 6A, F). Cyclin D3 knockdown was similarly toxic (Figure 1H; Supplemental Figures 6A, C) and was associated with a G1 phase arrest, as expected (Supplemental Figure 2B). In contrast, knockdown of cyclin D1, cyclin D2 or CDK4 had little if any effect on BL lines but was detrimental to cell line models of other lymphoma subtypes (Supplemental Figures 2C, 6B, D, E, F). To test the oncogenic potential of the cyclin D3 mutants, we transduced GFP-tagged wild type or T283A mutant cyclin D3 into two lymphoma cell lines in which the endogenous wild type cyclin D3 was knocked down by RNA interference. In both lines, cells transduced with the cyclin D3 mutant had a marked proliferative advantage over untransduced cells, but overexpression of wild type cyclin D3 had little effect (Figure 1I). Hence, BL lines rely on cyclin D3/CDK6 for cell cycle progression and cyclin D3 mutants augment this effect.
To explore the therapeutic potential of the CDK4/6 inhibitor PD 0332991, we treated BL, GCB DLBCL and mantle cell lymphoma (MCL) cell lines with the drug daily for 2 weeks. After an initial G1 phase arrest, the BL and GCB DLBCL cell lines began to die by day 2, with a steady accumulation of apoptotic cells over the course of treatment (Figure 1J). By contrast, the MCL cell line arrested in G1 phase but did not undergo apoptosis. We next treated a BL xenograft model with PD 0332991 after the establishment of tumors, and observed a profound reduction in tumor volume after 6 days, resulting in the virtual disappearance of tumor cells by day 10 (Figure 1K, Supplemental Figure 2D).
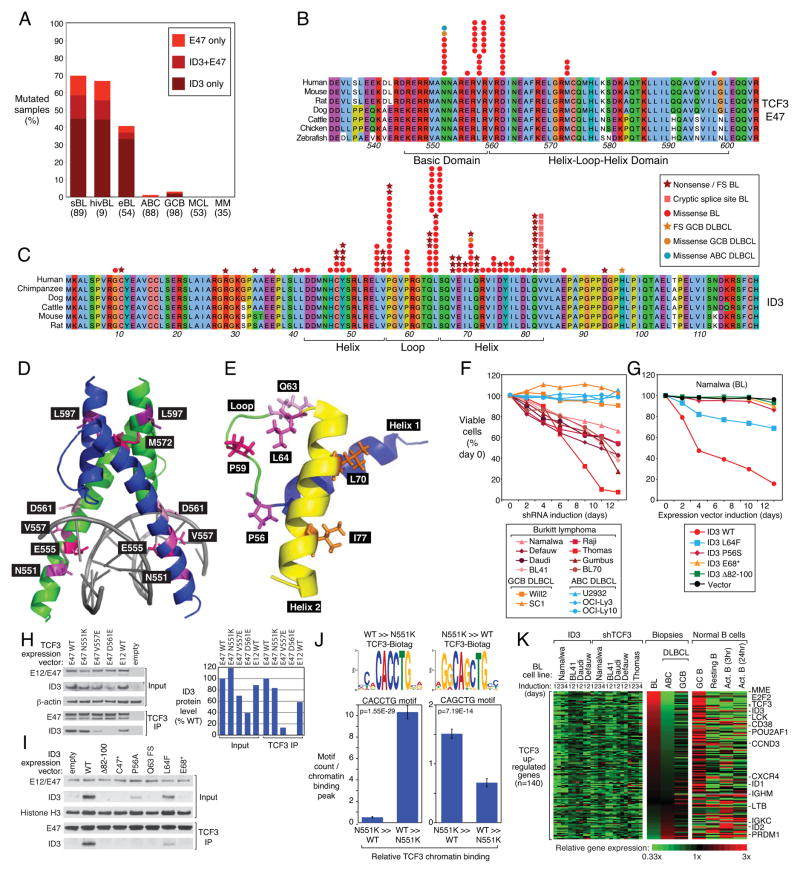
The highly recurrent mutations in TCF3 and ID3 suggested that the transcriptional regulators encoded by these genes play a central role in BL pathogenesis, as they do in normal B cell development (reviewed in ref. 20). Sanger resequencing of genomic DNA confirmed ID3 and/or TCF3 mutations in sBL, hivBL, and eBL, in 70%, 67%, and 40% of samples, respectively, but these mutations were rare or absent in other lymphoma subtypes and in multiple myeloma (Figure 2A). ID3 mutations were more common than TCF3 mutations, occurring in 58% of sBL samples, with 13% harboring mutations in both genes. The majority of cases with ID3 mutations had 2 or more separate mutations and in all tested cases (n=26), the mutations were on separate alleles. By contrast, the majority (63%) of the TCF3 mutations were present on only one allele (Supplemental Figure 3A; Supplemental Table 5). Resequencing of available germ line DNA confirmed the somatic origin of 11 ID3 mutants (C10stop, Y49stop, L54V, V55 frame shift, P56T, L64F, L70P, Q71stop, V82-Q100 deletion, D93 frame shift, A102D) and 3 TCF3 mutants (V557E, D561E, M572K).
Figure 2. Transcriptional activity of TCF3 is essential for Burkitt lymphoma viability.
a, Fre quencies of TCF3 and ID3 mutations in different lymphoma subtypes. b, TCF3 missense mutations in the E47 isoform in BL, ABC DLBCL and GCB DLBCL. Amino acid positions 532–604 of E47 are shown according to protein accession NP_001129611. c, ID3 mutations in BL. Amino acid positions of full length ID3 protein are shown according to protein accession NP_002158. d, Location of TCF3 mutations in the three-dimensional structure of the dimeric E47 basic-helix-loop-helix domain from reference 44. e, Location of ID3 mutations in the three-dimensional structure the ID3 helix-loop-helix domain (pdb accession 2LFH). f, Selective toxicity of TCF3 shRNA for BL cell lines. A vector co-expressing a TCF3 shRNA and GFP was transduced into the indicated cell lines. shRNA expression was induced for the indicated times and the fraction of live GFP+, shRNA+ cells was normalized to the value on day 0. Data represent at least four independent experiments. g, Toxicity of wild type but not mutant ID3 isoforms in ID3-mutant Namalwa cell line. A vector co-expressing ID3 and GFP was transduced into the indicated cell lines. ID3 expression was induced for the indicated times and the fraction of live GFP+, ID3+ cells was normalized to the value on day 0. h, TCF3 E47 mutants with reduced ability to bind ID3. Wild type or mutant TCF3 isoforms were coexpressed with wild type ID3 in 293T cells, and the indicated proteins were assessed either by Western blot analysis in total cellular extracts or after immunoprecipatation (IP) with anti-TCF3 antibodies. ID3 protein levels based on densitometric quantitation of the Western blot results were normalized to E47 protein levels and graphed in the right panel. i, BL-derived mutant ID3 proteins accumulate to lower levels than wild type ID3 and have decreased ability to bind to TCF3. Mutant or wild type ID3 isoforms were expressed in the ID3-deficient Namalwa cell line and analyzed by Western blot for total ID3 protein or ID3 protein immunoprecipitated with TCF3. See text for details. j. DNA sequence motifs enriched under TCF3 genomic binding peaks. Shown at the top are the motifs that were most enriched under peaks that were bound > 4-fold more or less by N551K TCF3 compared to wild type TCF3. The mean number (+/−SEM) of the indicated motifs per peak is plotted below. k, A TCF3 gene expression signature is highly expressed in BL and in normal germinal center cells. Changes of gene expression were profiled in ID3-mutant BL cell lines following TCF3 knockdown or wild type ID3 overexpression. Shown are genes that were downregulated by at least 0.33 log2 in >70% of samples following TCF3 knockdown or following ID3 overexpression. Previously published gene expression profiling datasets from BL, GCB DLBCL, and ABC DLBCL were used5. Expression in normal B cell subpopulations was based on RNA-seq. Genes were ranked according to the difference in expression between BL and ABC DLBCL. FS: frameshift, Δ : Deletion
The TCF3 locus encodes two alternatively spliced isoforms – E47 and E12 – that differ in their basic-helix-loop-helix (B-HLH) DNA binding domains and have distinct functions during normal B cell development21,22. All TCF3 mutations in BL affected the E47 B-HLH domain (Figure 2B), with no mutations affecting E12, suggesting a non-redundant role for E47 in BL pathogenesis. The E47 isoform was more highly expressed than the E12 isoform in BL cases with E47 mutations (Supplemental Figure 3B), suggesting that the TCF3 mutations are gain-of-function. Four evolutionarily conserved amino acids were the target of most of the E47 missense mutations, which occurred in both the basic DNA recognition region (N551K, V557E/G) and in the HLH region (D561E/V/N, M572K) (Figure 2D; Supplemental Table 5). The V557 is adjacent to D561 on a helical surface facing away from DNA, suggesting that mutations affecting these positions could disrupt intra- or intermolecular interactions. The B-HLH domain may also be distorted by lysine substitutions affecting M572 and L597, which lie next to each other in the 3-dimensional structure.
A variety of nonsense and frameshift mutations inactivate ID3 in BL tumors, in the majority of the cases on both alleles, suggesting a tumor suppressor mechanism (Figure 2C; Supplemental Table 5). Highly recurrent missense mutations target the ID3 loop region, which is critical for the ability of ID proteins to block E47 DNA binding23. Two loop region amino acids that border the helices are affected most often: leucine 64 (L64), which makes van-der-Waals interactions with residues in the second HLH helix, and proline 56, which kinks the loop region (Figure 2E). Additional loop region mutants affect proline 59, which also kinks the loop, as well as glutamine 63, which makes hydrogen bonds with arginine 52 of the first helix. Hence, each of the loop region mutants has the potential to distort the tertiary structure of the HLH domain and impair its ability to inhibit E47. In addition, numerous ID3 missense mutations alter residues in the second helix of the HLH domain and, to a lesser extent, the first helix, potentially compromising the ability of these mutants to inhibit E47. Finally, multiple mutations disrupt a splice donor site in ID3, resulting in the utilization of a cryptic splice donor and the deletion of 19 amino acids (Figure 2C; Supplemental Figure 3C).
The RNA interference screen revealed that TCF3 is an essential gene in 4 BL cell lines (Supplemental Figure 3D, Supplemental Table 11), supporting the notion that the TCF3 and ID3 mutations in BL promote E47 action. Subsequent shRNA experiments revealed that knockdown of TCF3 caused a time-dependent toxicity in all BL lines tested, including lines with and without ID3 mutations, but had no effect on cell line models of ABC and GCB DLBCL (Figure 2F; Supplemental Figures 3F, 7A, 7B). Wild type TCF3 was able to rescue BL cells from shTCF3 toxicity (Supplemental Figure 3E). The TCF3 mutants were roughly equivalent to wild type TCF3 in their ability to reverse TCF3 shRNA-mediated toxicity, although the N551K mutant was somewhat less efficient in some cell lines. These results underscore that the TCF3 mutants are not loss-of-function.
Introduction of wild type ID3 into BL cell lines with ID3 mutations was similarly lethal (Figure 2G; Supplemental Figures 3G, 7C). By contrast, BL-derived ID3 mutants had less (L64F) or no toxicity (P56S, E68*, Δ82–100) for the same BL cell lines, arguing that the ID3 mutations are complete or partial loss-of-function alleles. To evaluate the ID3 mutants biochemically, we inducibly expressed wild type or mutant ID3 isoforms in an ID3-deficient BL cell line and analyzed both total ID3 protein levels and the ability of the ID3 isoforms to interact with TCF3. Although the mRNA levels for each ID3 isoform were equivalent, no detectable protein was observed for the three nonsense and frameshift mutants (C47*, E68*, Q63fs), and the splice site mutant (Δ82–100) was barely detectable as a shorter ID3 isoform (Figure 2I; Supplemental Figure 7E). The two common missense mutants were detectable, but were expressed at lower levels than wild type ID3. Most of the ID3 mutant proteins could not be detected by co-immunoprecipitation with TCF3 with the exception of L64F, which associated with TCF3 much less robustly than wild type ID3 (Figure 2I). We conclude that the recurrent ID3 mutations in BL serve to destabilize the protein and prevent its inhibitory interaction with TCF3 E47.
To investigate the influence of mutations in TCF3 on its ability to interact with ID3, we expressed wild type ID3 in HEK293T cells together with wild type or mutant forms of TCF3 E47 or E12. In the absence of TCF3, ID3 protein did not accumulate appreciably, but was increased by proteasomal inhibition (Supplemental Figure 7D). Titration of wild type E47 or E12 caused ID3 protein levels to rise (Supplemental Figure 3H). ID3 protein efficiently co-immunoprecipitated with wild type E47 and E12 (Figure 2H). The N551K E47 mutant behaved similarly to wild type E47 in this assay, but the V557E and D561E mutants did not stabilize ID3 as well as wild type E47 and did not co-immunoprecipitate with ID3 efficiently. These results suggest that the most common TCF3 mutants, V557E and D561E, are selected in BL tumors to evade negative regulation by ID3.
To gauge the ability of the TCF3 mutants to interact with chromatin, we engineered two BL lines with wild type TCF3 to express a bacterial biotinylation enzyme and then expressed wild type or mutant TCF3 (V557E, D561E, N551K) isoforms that were tagged with an amino acid sequence (“Biotag”) that can be biotinylated by this enzyme. Streptavidin was used to precipitate chromatin bound to each TCF3-Biotag isoform for ChIP-seq analysis. In parallel, we performed ChIP-seq for endogenous TCF3 using anti-TCF3 antibodies in the same BL lines (Supplemental Table 13), and were able to confirm all binding events tested by conventional ChIP analysis (Supplemental Figure 3J). Of 2,117 genes that had endogenous TCF3 binding peaks within 2 kb of the transcriptional start site (TSS) in both BL lines, 93% had overlapping wild type Biotag-TCF3 peaks, indicating that the TCF3-Biotag isoforms interacted with relevant chromatin targets. Both the endogenous and Biotag TCF3 ChIP-seq peaks were enriched for E-box motifs: 5′-CAG(G/C)TG-3′, as expected. Chromatin binding by wild type and mutant TCF3 isoforms overlapped substantially: wild type TCF3-Biotag binding regions had at least 7 overlapping tags in the ChIP-seq data for the V557E, D561E, N551K isoforms in 98%, 98% and 92% of instances, respectively (Supplemental Table 12). Chromatin binding by wild type and mutant TCF3-Biotag to representative genes is shown in Supplemental Figure 8.
Given the somewhat lower overlap between N551K and wild type TCF3 chromatin binding, we identified genomic regions that were 4-fold higher (n=212) or lower (n=139) in their association with wild type and N551K TCF3 in both BL cell lines (p<1E-10) (See Methods; Supplemental Table 14). In these binding regions, the V557E and D561E isoforms behaved similarly to wild type TCF3, again suggesting that these mutations do not alter their chromatin binding specificity (Supplemental Table 14). The peaks bound more effectively by wild type than N551K TCF3 contained multiple copies of the motif 5′-NNCACCTG-3′ whereas the peaks bound preferentially by N551K were enriched for the sequence 5′-GGCAGCTG-3′ (Figure 2J). While both motifs conform to the E-box consensus, these results suggest that N551K TCF3 is an altered specificity mutant that has somewhat different genomic targets than wild type TCF3. Representative genes with differential binding of wild type and N551K TCF3 are shown in Supplemental Figure 8B.
To gain insight into the biological processes controlled by TCF3 in BL, we knocked down TCF3 expression or overexpressed wild type ID3 in ID3-mutant BL lines and profiled changes in gene expression. We identified 139 “TCF3-upregulated” genes that were decreased in expression by both manipulations and 166 “TCF3-downregulated” genes that were increased in expression (Figure 2K; Supplemental Figure 4A). We confirmed the TCF3-dependent regulation of several of these genes by RT-PCR (Supplemental Figure 3I). Many of these TCF3 regulated genes are apparently direct targets of TCF3 based on ChIP-seq analysis of endogenous TCF3 binding. We defined a “whole gene window” as the genomic region encompassing the body of the gene as well as 10 kilobases of its upstream promoter region. Based on this window, 20% (n=4,355) of protein-coding genes had TCF3 peaks in both the BL41 and Namalwa BL lines (Supplemental Table 13). TCF3 binding peaks were enriched among TCF3-upregulated genes (58%; Chi-square p=1.81E-29) and among TCF3–downregulated genes (32%; Chi-square p=1.03E-4). We will refer to the genes that are both bound and regulated by TCF3 as “TCF3 direct targets”.
Expression of TCF3-regulated genes is a prominent characteristic of BL tumors. Using available gene expression profiling datasets from BL, GCB DLBCL, and ABC DLBCL5,24, we observed that TCF3 itself was 3.9-fold and 5.6-fold more highly expressed in BL than in GCB DLBCL and ABC DLBCL, respectively (p<1E-16). Most TCF3-upregulated genes were also more highly expressed in BL than in DLBCL whereas TCF3-downregulated genes were generally expressed at lower levels in BL biopsies, demonstrating that a TCF3-regulatory network contributes substantially to the BL phenotype (p≤0.001; Figure 2K; Supplemental Figures 4A, B). In normal B cell subsets, the TCF3-upregulated genes were more highly expressed in germinal center B cells than in resting or activated blood B cells whereas the TCF3-downregulated genes were at lower levels in germinal center B cells (Figure 2K; Supplemental Figure 4A). Hence, the TCF3 gene expression program in BL appears to be “inherited” from its normal cellular counterpart, in keeping with the requirement for TCF3 in normal germinal center B cell responses25. However, by gene set enrichment analysis, BL tumors with ID3 and/or TCF3 mutations had higher expression of the TCF3-upregulated signature than BL tumors with wild type ID3 and TCF3, and the opposite was true for the TCF3-downregulated signature (p=0.0001; Supplemental Figure 4C). Hence, the transcriptional influence of TCF3 on the BL phenotype appears to be accentuated by ID3/TCF3 mutations.
Biological insights from this analysis included the observation that the negative regulators of TCF3 – ID1, ID2, and ID3 – were each TCF3 direct targets that were upregulated by TCF3, thereby creating an autoregulatory negative feedback loop (Figure 2K; Supplemental Figure 8A). TCF3 also positively regulated several genes that play crucial roles in normal germinal center B cell formation and/or maintenance, including POU2AF1, CXCR4, LTB and CCND3. Interestingly, though, TCF3 also positively regulated PRDM1 (encoding Blimp-1) and XBP1, suggesting that TCF3 may bias germinal center B cells towards plasmacytic differentiation. TCF3 upregulated CCND3 and E2F2, which both promote cell cycle progression, while downregulating RB1, which encodes a tumor suppressor that blocks the G1-S phase transition. It has long been known that TCF3 regulates the antigen-binding components of the BCR, the immunoglobulin heavy (IgH) and light chains22,26. As expected, TCF3 could be readily detected at the immunoglobulin heavy chain (IgH) and κ light chain (Igκ) 3′ enhancers in BL cells and TCF3 increased their IgH and Igκ mRNA expression (Figure 2K; Supplemental Figures 3I, J, 8A). ChIP-seq analysis in BL revealed that TCF3 also binds to genes encoding other BCR components (CD79A, CD79B), BCR coreceptors (CD19, CD21, CD22), and BCR-associated kinases (LYN, FYN, BLK, LCK, SYK), many of which are also TCF3 targets in mouse precursor B cells27 (Supplemental Table 13).
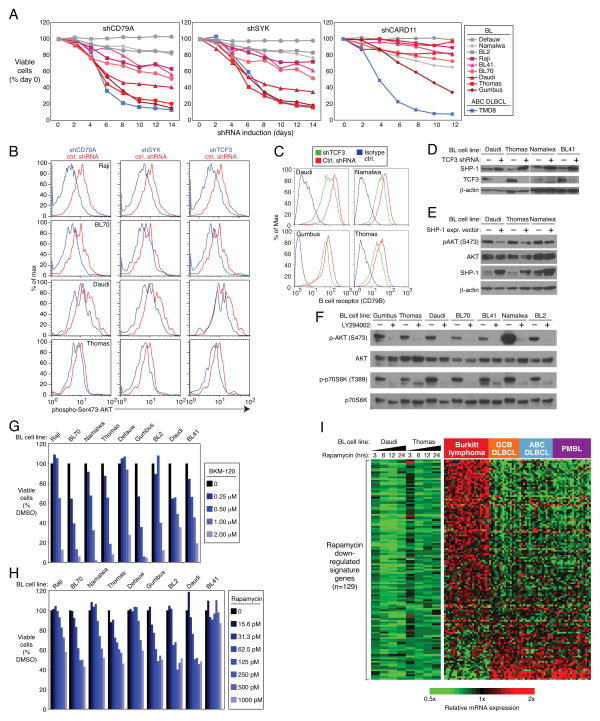
In this regard, it was particularly notable that knockdown of the BCR subunit CD79A was toxic for several BL lines in our RNA interference screen, as well as for ABC DLBCL lines13, but did not affect cell line models of GCB DLBCL and MCL (Supplemental Figure 5A). In a panel of 9 BL cell lines, 6 were clearly BCR-dependent, based on a time-dependent decrease in their viability following knockdown of either CD79A or the BCR-associated tyrosine kinase SYK (Figure 3A). Knockdown of CARD11, an adapter that links the BCR to the NF-κB pathway in ABC DLBCL28, was lethal for one BL cell line but did not affect the other BCR-dependent BL cell lines (Figure 3A; Supplemental Figure 5A). Moreover, BL lines were insensitive to the selective IκB kinase inhibitor MLN120B, whereas ABC DLBCL lines were sensitive, as expected29, confirming that BL lines do not depend strongly on the classical NF-κB pathway for survival (Supplemental Figure 5C). Given this, we speculated that the BCR-dependency of some BL lines might be more akin to “tonic” BCR signaling, as defined in normal mature mouse B cells30,31, a phenomenon that relies on the pro-survival action of the PI(3) kinase pathway more than the NF-κB pathway32. Indeed, knockdown of the BCR component CD79A in BCR-dependent BL cell lines decreased phosphorylation of AKT, a modification that depends on PI(3) kinase signaling (Figure 3B). SYK, a tyrosine kinase that is activated by BCR signaling, was also required to maintain AKT phosphorylation in BL lines (Figure 3B).
Figure 3. PI(3) kinase activity in Burkitt lymphoma.
a, CD79A and SYK knockdown is toxic for a subset of BL cell lines. Vectors co-expressing either CD79A or SYK shRNAs together with GFP were transduced into the indicated cell lines. shRNA expression was induced for the indicated times and the fraction of live GFP+, shRNA+ cells was normalized to the value on day 0. BCR-dependent BL lines are depicted using red colors. The BCR-dependent ABC DLBCL line TMD813 is also shown. Data represent at least three independent experiments. b, Knockdown of CD79A, SYK or TCF3 reduces PI(3) kinase activity. The indicated BL cell lines were transduced with a control shRNA or shRNAs targeting CD79A, SYK, or TCF3. shRNA expression was induced for 2 days, and cells were analyzed by FACS for phospho-S473-AKT staining as a measure of PI(3) kinase activity. c, TCF3 regulates expression of the B cell receptor (BCR) in BL. An shRNA targeting TCF3 or a control shRNA was induced in the indicated BL cell lines for 1 day. Surface expression of the BCR was quantified by FACS analysis of the BCR subunit CD79B. d, TCF3 suppresses PTPN6 (SHP-1) expression. The indicated BL cells were transduced with an shRNA targeting TCF3 and analyzed by Western blot analysis for the indicated proteins. e, Ectopic expression of SHP-1 suppresses phospho-S473-AKT in BL cell lines. Indicated cell lines were transduced with SHP-1 expression vector (+) or an empty vector control (−), whereupon cells were subjected to Western Blot analysis of indicated proteins. f, BL cell lines have constitutively high levels of PI(3) kinase activity. Western blot analysis of BL cell lines revealed high levels of phospho-S473-AKT and phospho-T309-p70S6K, which were reduced by treatment with the PI(3) kinase inhibitor LY294002, as indicated. g, Inhibition of PI(3) kinase activity is toxic to BL cell lines. Shown are viable cells, as quantified by MTS assay, in cultures of BL lines treated with indicated concentrations of the pan-class I PI(3) kinase inhibitor BKM120. h, Inhibition of mTOR activity is lethal to most BL cell lines. Shown are viable cells, as quantified by an MTS assay, in cells treated with the indicated concentrations of rapamycin. i, Genes down regulated by Rapamycin are highly expressed in BL. Changes of gene expression were profiled in BL cell lines following Rapamycin (100 pM) addition for indicated time points. Shown are genes that were downregulated by at least 0.4 log2 in at least 3 of 4 time points in both cell lines. Previously published gene expression profiling datasets from BL, GCB DLBCL, and ABC DLBCL were used5.
Of note, TCF3 knockdown also decreased phospho-AKT levels in BCR-dependent lines tested, as did ID3 overexpression (Figure 3B, Supplemental Figure 5B). This may be explained in part by the downmodulation of surface BCR expression that occurred upon TCF3 depletion (Figure 3C). TCF3 also binds directly to the promoter of PTPN6, encoding the phosphatase SHP-1 that inhibits B cell receptor (BCR) signaling (Supplemental Figures 3J, 8A). Knockdown of TCF3 increased PTPN6 mRNA and protein levels, indicating that TCF3 represses PTPN6 transcription, in accord with previous work demonstrating downregulation of SHP-1 in normal germinal centers and BL tumors33 (Figure 3D; Supplemental Figures 3I, 4A). Ectopic provision of SHP-1 decreased phospho-AKT in BCR-dependent BL lines, suggesting that TCF3-mediated repression of PTPN6 may augment BCR signaling in BL (Figure 3E).
A screen of a larger number of BL cell lines revealed that all had readily detectable AKT phosphorylation that was inhibited by treatment with a PI(3) kinase inhibitor, LY294002 (Figure 3F). Moreover, the mTOR pathway was engaged by PI(3) kinase signaling in BL lines, as judged by phosphorylation of the mTOR substrate p70 S6 kinase (Figure 3F). Treatment of BL cell lines with BKM120, a PI(3) kinase inhibitor in clinical trials34, or rapamycin, an inhibitor of the mTORC1 complex, was toxic to most BL cell lines tested (Figures 3G, H). Hence, PI(3) kinase signaling promotes cell survival in both BCR-dependent and BCR-independent BL cell lines. Other mechanisms to augment PI(3) kinase signaling in BL include mutation of PTEN: 3 BL samples had 4 separate PTEN missense mutations, 3 of which have been documented in other cancers35 (Supplemental Table 2). MYC deregulation in BL contributes to PI(3) kinase signaling by driving expression or MIR17HG, the precursor RNA for miR-19, an inhibitor of PTEN expression36,37. Indeed, MIR17HG expression was 10-fold higher in BL than DLBCL by digital gene expression, which was confirmed by RT-PCR (Supplemental Figure 5D and data not shown). Finally, we used gene expression signatures of mTOR activity to investigate this pathway in BL tumors. We developed gene expression signatures comprised of genes that were either up- or down-regulated following treatment of BL cell lines with the mTORC1 inhibitor rapamycin (Supplemental Table 10). Among tumor biopsies of BL and GCB DLBCL5, rapamycin down-regulated genes were enriched among genes that were more highly expressed in BL tumors (p=0.026; KS statistic), whereas rapamycin up-regulated genes had the opposite enrichment pattern (p=0.007) (Figure 3I; Supplemental Figure 5E). Rapamycin down-regulated genes were also more highly expressed in BL than in other aggressive lymphomas (ABC DLBCL, PMBL) (Figure 3I), supporting the notion that mTORC1 activity is a consistent feature of BL tumors.
Discussion
By merging functional and structural genomic data, we have uncovered previously unappreciated pathways in BL pathogenesis, several of which are amenable to therapeutic attack. We observed striking differences in the frequency of certain genetic abnormalities between BL and GCB DLBCL, despite the fact that both malignancies presumably arise from germinal center B cells (Table 1). The recurrent mutations in ID3, and TCF3 were shared by sBL, hivBL, and eBL, suggesting a common BL pathogenesis irrespective of epidemiology. That said, the frequency of CCND3 mutations was much lower in eBL than in the sBL and hivBL subtypes, suggesting that distinct transforming events may be required in eBL. Our working hypothesis is that these genetic differences are dictated by the occurrence of the MYC translocation early in the pathogenesis of BL as opposed to its acquisition as a progression event in GCB DLBCL. These genetic contrasts are so stark that they could be used as an aid to the diagnostic distinction between BL and GCB DLBCL, which can be difficult in some instances using current methods5,6.
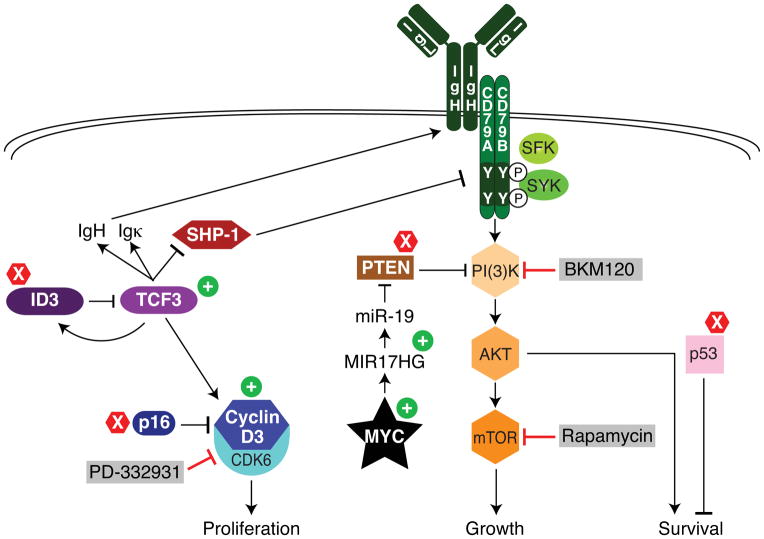
Our findings lead to an integrated model of BL pathogenesis that is summarized in Figure 4. MYC translocations initiate BL, with a majority of these tumors acquiring MYC mutations that may enhance its function, including several novel recurrent mutations discovered here. Clearly, agents targeting MYC, such as those that inhibit its transcription38–40 will be attractive candidates for BL clinical trials. However, our functional and structural genomics analysis revealed several additional oncogenic pathways in BL, suggesting that translocation of MYC is insufficient to cause human BL.
Figure 4. Schematic of recurrent oncogenic pathways in Burkitt lymphoma.
Shown is a model of BL pathogenesis highlighting pathways that regulate proliferation, growth and survival. Gain-of-function mutations are indicated by plus signs and loss-of-mutations by X signs. Grey boxes indicate drugs that can block these deregulated pathways. See text for details.
Aside from MYC translocation, mutations in TCF3 and/or ID3 were the most common genetic events in BL, occurring in more than two thirds of sBL and hivBL cases and in 40% of eBL cases. Most of these mutations block the interaction between TCF3 and ID3, thereby breaking the negative autoregulatory loop created by TCF3 transactivation of ID3. The TCF3 transcriptional program in BL includes many genes that distinguish normal germinal center B cells from other mature B cells subsets, and, in part, this accounts for the distinct gene expression profiles of BL and DLBCL5,6. Interestingly, all BL cell lines were dependent upon TCF3 for survival, including those with and without mutations in TCF3/ID3, which likely reflects the essential role for this transcription factor in normal germinal center responses16,17. Hence, TCF3 appears to act as a “lineage-survival oncogene”41 for BL.
PI(3) kinase signaling emerges from our work as an essential and pervasive pro-survival mechanism in BL. All BL lines had readily apparent PI(3) kinase activity and were killed efficiently by inhibitors of this pathway. Moreover, analysis of gene expression signatures of rapamycin-sensitive mTOR activity indicated that this pathway is likely to be more active in BL than in other aggressive lymphoma subtypes. Multiple mechanisms appear to contribute to PI(3) kinase activity in BL. Although mutations in PTEN may occasionally activate PI(3) kinase signaling in BL, most BL biopsies and cell lines lacked mutations in PTEN or PIK3CA. Transactivation of MIR17HG by MYC in BL, together with amplification of the MIR17HG locus in ~10% of BL tumors42, would lead to miR-19-mediated PTEN inactivation and consequent PI(3) kinase signaling. In two thirds of the BL lines tested, PI(3) kinase signaling was promoted by constitutive BCR signaling that is akin to the “tonic” BCR signaling that sustains normal mouse B cell viability30–32. TCF3 also promoted PI(3) kinase activity, potentially by transactivating the immunoglobulin loci, thereby upregulating surface BCR expression, and by inhibiting expression of PTPN6, encoding the SHP-1 phosphatase that blocks proximal BCR signaling43.
Another key direct target of TCF3 is CCND3, a regulator of the G1-S phase transition that is required for an effective germinal center reaction16,17. Cyclin D3 and its partner CDK6 were required for the proliferation of BL as well as GCB DLBCL cells, even in cell line models with wild type CCND3, probably reflecting the role of cyclin D3 in normal germinal center B cells. Burkitt lymphomas capitalize on this cell cycle regulatory machinery by mutating CCND3 in 38% of sBL cases, resulting in highly stable cyclin D3 isoforms that drive cells to proliferate vigorously. The G1-S phase restriction point is further deregulated by inactivating mutations and deletions targeting p16. Treatment of BL cell lines with a CDK4/6 inhibitor not only caused cell cycle arrest but, unexpectedly, induced apoptosis in vitro and regression of established BL xenografts.
While BL can be cured in developed countries in ~85% of cases occurring in younger patients using high-dose chemotherapy regimens1, these regimens are unsafe in older patients and cannot be deployed in less developed regions due to their immune suppression and due to logistical difficulties that preclude effective delivery8. Hopefully, the new insights into BL pathogenesis described herein will prompt clinical evaluation of drugs targeting the PI(3) kinase pathway, tonic BCR signaling, and cyclin D3/CDK6 in BL. Eventually, the rational combination of such targeted agents may provide more effective and less toxic treatment of BL worldwide.
Methods Summary
RNA-Seq was performed using established Illumina protocols on a HiSeq2000 sequencer. RNA interference screening and cellular toxicity assays were conducted as described14,28. Gene expression profiling was performed using Agilent 4×44K microarrays. Detailed experimental procedures and analytic procedures are presented in Supplemental Methods.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, an NCI SPECS grant (UO1-CA 114778), and by the Foundation for NIH, through a gift from the Richard A. Lauderbaugh Memorial Fund, and by Cancer Research UK. This study was conducted under the auspices of the Lymphoma/Leukemia Molecular Profiling Project (LLMPP). R.S. was supported by the Dr. Mildred Scheel Stiftung für Krebsforschung (Deutsche Krebshilfe). D.J.H is a Kay Kendall Leukaemia Fund Intermediate research fellow. This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, Md. (http://biowulf.nih.gov). We thank Kathleen Meyer for help with the GEO submission and Tom Ellenberger for the E47 crystal structure coordinates. We are grateful to Bao Tran and the Center for Cancer Research Sequencing Facility for expert implementation of next generation RNA sequencing and Kathleen Hartman in NCI sequencing core. We thank the participants in the EMBLEM Study (http://emblem.cancer.gov/) in Uganda. We are grateful to the EMBLEM Study staff for collecting and processing the samples and data and to the Government of Uganda for allowing the study to be done and the samples to be exported for research.
Footnotes
Author contributions
RS, RMY, MC, SJ, MZ, HK, ALS and DJH designed and performed experiments. TAW designed experiments. WX, YY, EB and HZ performed experiments. WX, GW, XL and JP analyzed data. AR, PK, HKM-H, GO, RDG, JMC, LMR, EC, ESJ, JD, EBS, RIF, RMB, RRT, JRC, DDW, WCC, SP, WW, MDO, SJR, SMM, MR and AR supplied BL patient samples or cell lines, and reviewed pathological and clinical data. LMS designed and supervised research and wrote the manuscript.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature
References
- 1.Yustein JT, Dang CV. Biology and treatment of Burkitt’s lymphoma. Curr Opin Hematol. 2007;14:375–381. doi: 10.1097/MOH.0b013e3281bccdee. [DOI] [PubMed] [Google Scholar]
- 2.Dalla-Favera R, et al. Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc Natl Acad Sci U S A. 1982;79:7824–7827. doi: 10.1073/pnas.79.24.7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taub R, et al. Translocation of the c-myc gene into the immunoglobulin heavy chain locus in human Burkitt lymphoma and murine plasmacytoma cells. Proc Natl Acad Sci U S A. 1982;79:7837–7841. doi: 10.1073/pnas.79.24.7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adams JM, Gerondakis S, Webb E, Corcoran LM, Cory S. Cellular myc oncogene is altered by chromosome translocation to an immunoglobulin locus in murine plasmacytomas and is rearranged similarly in human Burkitt lymphomas. Proc Natl Acad Sci U S A. 1983;80:1982–1986. doi: 10.1073/pnas.80.7.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dave SS, et al. Molecular diagnosis of Burkitt’s lymphoma. N Engl J Med. 2006;354:2431–2442. doi: 10.1056/NEJMoa055759. [DOI] [PubMed] [Google Scholar]
- 6.Hummel M, et al. A biologic definition of Burkitt’s lymphoma from transcriptional and genomic profiling. N Engl J Med. 2006;354:2419–2430. doi: 10.1056/NEJMoa055351. [DOI] [PubMed] [Google Scholar]
- 7.Piccaluga PP, et al. Gene expression analysis uncovers similarity and differences among Burkitt lymphoma subtypes. Blood. 2011;117:3596–3608. doi: 10.1182/blood-2010-08-301556. [DOI] [PubMed] [Google Scholar]
- 8.Orem J, Mbidde EK, Weiderpass E. Current investigations and treatment of Burkitt’s lymphoma in Africa. Trop Doct. 2008;38:7–11. doi: 10.1258/td.2007.060093. [DOI] [PubMed] [Google Scholar]
- 9.Morin RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaidano G, et al. p53 mutations in human lymphoid malignancies: association with Burkitt lymphoma and chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 1991;88:5413–5417. doi: 10.1073/pnas.88.12.5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morin RD, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42:181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pasqualucci L, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011;43:830–837. doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis RE, et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 2010;463:88–92. doi: 10.1038/nature08638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ngo VN, et al. Oncogenically active MYD88 mutations in human lymphoma. Nature. 2011;470:115–119. doi: 10.1038/nature09671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hemann MT, et al. Evasion of the p53 tumour surveillance network by tumour-derived MYC mutants. Nature. 2005;436:807–811. doi: 10.1038/nature03845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cato MH, Chintalapati SK, Yau IW, Omori SA, Rickert RC. Cyclin D3 is selectively required for proliferative expansion of germinal center B cells. Mol Cell Biol. 2010 doi: 10.1128/MCB.00650-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peled JU, et al. Requirement for cyclin D3 in germinal center formation and function. Cell Res. 2010;20:631–646. doi: 10.1038/cr.2010.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Casanovas O, Jaumot M, Paules AB, Agell N, Bachs O. P38SAPK2 phosphorylates cyclin D3 at Thr-283 and targets it for proteasomal degradation. Oncogene. 2004;23:7537–7544. doi: 10.1038/sj.onc.1208040. [DOI] [PubMed] [Google Scholar]
- 19.Diehl JA, Cheng M, Roussel MF, Sherr CJ. Glycogen synthase kinase-3beta regulates cyclin D1 proteolysis and subcellular localization. Genes Dev. 1998;12:3499–3511. doi: 10.1101/gad.12.22.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kee BL. E and ID proteins branch out. Nat Rev Immunol. 2009;9:175–184. doi: 10.1038/nri2507. [DOI] [PubMed] [Google Scholar]
- 21.Beck K, Peak MM, Ota T, Nemazee D, Murre C. Distinct roles for E12 and E47 in B cell specification and the sequential rearrangement of immunoglobulin light chain loci. J Exp Med. 2009;206:2271–2284. doi: 10.1084/jem.20090756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murre C, McCaw PS, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- 23.Pesce S, Benezra R. The loop region of the helix-loop-helix protein Id1 is critical for its dominant negative activity. Mol Cell Biol. 1993;13:7874–7880. doi: 10.1128/mcb.13.12.7874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lenz G, et al. Stromal gene signatures in large-B-cell lymphomas. N Engl J Med. 2008;359:2313–2323. doi: 10.1056/NEJMoa0802885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwon K, et al. Instructive role of the transcription factor E2A in early B lymphopoiesis and germinal center B cell development. Immunity. 2008;28:751–762. doi: 10.1016/j.immuni.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 26.Murre C, et al. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cel. 1989;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- 27.Lin YC, et al. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol. 2010;11:635–643. doi: 10.1038/ni.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ngo VN, et al. A loss-of-function RNA interference screen for molecular targets in cancer. Nature. 2006;441:106–110. doi: 10.1038/nature04687. [DOI] [PubMed] [Google Scholar]
- 29.Lam LT, et al. Small molecule inhibitors of IkappaB kinase are selectively toxic for subgroups of diffuse large B-cell lymphoma defined by gene expression profiling. Clin Cancer Res. 2005;11:28–40. [PubMed] [Google Scholar]
- 30.Lam KP, Kuhn R, Rajewsky K. In vivo ablation of surface immunoglobulin on mature B cells by inducible gene targeting results in rapid cell death. Cell. 1997;90:1073–1083. doi: 10.1016/s0092-8674(00)80373-6. [DOI] [PubMed] [Google Scholar]
- 31.Kraus M, Alimzhanov MB, Rajewsky N, Rajewsky K. Survival of resting mature B lymphocytes depends on BCR signaling via the Igalpha/beta heterodimer. Cell. 2004;117:787–800. doi: 10.1016/j.cell.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 32.Srinivasan L, et al. PI3 kinase signals BCR-dependent mature B cell survival. Cell. 2009;139:573–586. doi: 10.1016/j.cell.2009.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Delibrias CC, Floettmann JE, Rowe M, Fearon DT. Downregulated expression of SHP-1 in Burkitt lymphomas and germinal center B lymphocytes. J Exp Med. 1997;186:1575–1583. doi: 10.1084/jem.186.9.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bendell JC, et al. Phase I, Dose-Escalation Study of BKM120, an Oral Pan-Class I PI3K Inhibitor, in Patients With Advanced Solid Tumors. J Clin Oncol. 2011 doi: 10.1200/JCO.2011.36.1360. [DOI] [PubMed] [Google Scholar]
- 35.Forbes SA, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olive V, et al. miR-19 is a key oncogenic component of mir-17–92. Genes Dev. 2009;23:2839–2849. doi: 10.1101/gad.1861409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiao C, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17–92 expression in lymphocytes. Nat Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zuber J, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Delmore JE, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mertz JA, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci U S A. 2011;108:16669–16674. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garraway LA, Sellers WR. Lineage dependency and lineage-survival oncogenes in human cancer. Nat Rev Cancer. 2006;6:593–602. doi: 10.1038/nrc1947. [DOI] [PubMed] [Google Scholar]
- 42.Schiffman JD, et al. Genome wide copy number analysis of paediatric Burkitt lymphoma using formalin-fixed tissues reveals a subset with gain of chromosome 13q and corresponding miRNA over expression. Br J Haematol. 2011;155:477–486. doi: 10.1111/j.1365-2141.2011.08883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cornall RJ, et al. Polygenic autoimmune traits: Lyn, CD22, and SHP-1 are limiting elements of a biochemical pathway regulating BCR signaling and selection. Immunity. 1998;8:497–508. doi: 10.1016/s1074-7613(00)80554-3. [DOI] [PubMed] [Google Scholar]
- 44.Ellenberger T, Fass D, Arnaud M, Harrison SC. Crystal structure of transcription factor E47: E-box recognition by a basic region helix-loop-helix dimer. Genes Dev. 1994;8:970–980. doi: 10.1101/gad.8.8.970. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.