Abstract
Mutations in leucine-rich repeat kinase 2 (LRRK2) are the most common cause of familial Parkinson’s disease (PD). In this work, we demonstrate that LRRK2 can be degraded in lysosomes by chaperone-mediated autophagy (CMA), whereas the most common pathogenic mutant form of LRRK2, G2019S, is poorly degraded by this pathway. In contrast to typical CMA substrates, lysosomal binding of both wild-type and several pathogenic mutant LRRK2 proteins is enhanced in the presence of other CMA substrates, which interferes with the organization of the CMA translocation complex, resulting in defective CMA. Cells respond to such LRRK2-mediated CMA compromise by increasing levels of the CMA lysosomal receptor as seen in neuronal cultures and brains of LRRK2 transgenic mice, iPSC-derived dopaminergic neurons, and brains of mutant LRRK2 PD patients. This novel LRRK2 self-perpetuating inhibitory effect on CMA could underlie toxicity in PD by compromising the degradation of alpha-synuclein, another PD-related protein degraded by this pathway.
Keywords: chaperones, lysosomal membrane proteins, lysosomes, neurodegeneration, Parkinson’s disease, proteotoxicity, induced pluripotent stem cells
Mutations in leucine-rich repeat kinase 2 (LRRK2 or dardarin) are to date the most common cause of familial Parkinson’s disease (PD), a neurodegenerative motor disorder characterized by the presence of alpha synuclein (α-syn) containing Lewy Body inclusions, particularly in dopaminergic neurons of the substantia nigra that die over the course of the disease1. Most pathogenic PD mutations are localized to the LRRK2 kinase and GTPase domains, and an increase in the kinase activity of LRRK2, such as that observed in the G2019S mutant variant, has been proposed to be responsible for its neurotoxic effect2. The specific mechanisms by which mutant LRRK2 causes neuronal toxicity in PD, however, remain elusive.
Dysregulation of cellular proteostasis due to alterations in the ubiquitin-proteasome and the autophagic-lysosomal systems, has been described in many neurodegenerative disorders including PD3. Alterations in two of the most common forms of autophagy – macroautophagy and chaperone-mediated autophagy (CMA) – have been described in PD4–8. Several studies have reported possible interactions of LRRK2 with macroautophagy, a process that involves sequestration of portions of cytosol in double membrane vesicles or autophagic vacuoles that then fuse with lysosomes9. LRRK2 can localize to the endosomal/lysosomal compartments10, 11 and expression of LRRK2 mutants in different experimental models increases the presence of autophagic compartments11–14. The interplay of LRRK2 with macroautophagy may however be complex, because while LRRK2 knock-out mice exhibited reduced levels of macroautophagy, at least in kidney15, partial knock-down of LRRK2 in cultured kidney cells led to an increase in autophagic markers12.
In contrast to the multiple studies reporting effects of LRRK2 on macroautophagy, effects of LRRK2 on CMA are unknown. CMA involves the direct transport of cytosolic soluble proteins across the lysosomal membrane in a selective fashion16, 17. CMA substrates contain a pentapeptide motif18 that is recognized by the heat shock cognate protein of 70 kDa (hsc70). Hsc70 targets substrate proteins to the lysosomal membrane19, where they interact with the lysosome-associated membrane protein type 2A (LAMP-2A)20. Once the substrate binds, LAMP-2A multimerizes to form the translocation complex21. LAMP-2A dynamics are also regulated by mobilization in and out of discrete lipid microdomains where degradation of LAMP-2A takes place22, 23.
Previous studies have established a connection between CMA and PD, as wild-type α-syn is a substrate for CMA8, whereas pathogenic forms of this protein bind abnormally to the lysosomal receptor for CMA, thereby preventing their own degradation and the degradation of other CMA substrates7, 8, 24. Another PD associated protein, ubiquitin C-terminal hydrolase L1 (UCH-L1), interacts with CMA components, and this interaction is abnormally enhanced in a UCH-L1 mutant25. Alterations in CMA may in fact be a common feature in many forms of PD, as examination of postmortem brain samples from PD patients show reduced levels of LAMP-2A in the substantia nigra26. We noted that the LRRK2 protein bears eight putative CMA motifs in its amino acid sequence, and decided to determine whether LRRK2 might be a CMA substrate, and how the most common LRRK2 mutation (G2019S) could impact its interaction with CMA.
We have now found that a fraction of cellular LRRK2 can be degraded by CMA, but also that a range of pathogenic mutants, including the common G2019S LRRK2 allele, as well as high concentrations of wild-type LRRK2, inhibit CMA. This inhibition occurs through a novel mechanism, involving a blockage in the formation of the CMA translocation complex at the lysosomal membrane. Cells respond to the LRRK2-mediated blockage of CMA by increasing the amount of the essential component of the translocation complex, LAMP-2A, a phenomenon we also observed in the brains of LRRK2 transgenic mice and in iPSC-derived dopaminergic neurons and brains from LRRK2 mutant PD patients. We demonstrate that one of the proteins affected by the inhibitory effect of LRRK2 on CMA is α-syn. Thus, two of the dominant mutations that cause PD converge mechanistically by inhibiting the normal degradation of cytosolic proteins by CMA.
Results
LRRK2 can be degraded in lysosomes
The contribution of the lysosomal system to the degradation of LRRK2 itself has not been systematically explored. Upon treatment of neuroblastoma cells with lactacystin (a proteasome inhibitor), 3-methyladenine (3MA; to inhibit macroautophagy), or a combination of ammonium chloride and leupeptin (NL; to inhibit total lysosomal proteolysis), we confirmed that a portion of cellular LRRK2 is degraded by the proteasome27, but also found that part of LRRK2 is degraded in lysosomes, as intracellular levels of LRRK2 increased when lysosomal proteolysis was blocked (Fig. 1a). Lysosomal degradation of LRRK2 did not occur through macroautophagy, because LRRK2 levels did not increase upon 3MA treatment (Fig. 1a and Supplementary Fig 1a,b).
Figure 1. LRRK2 is degraded in lysosomes.
(a) SH-SY5Y cells untreated or incubated with 5µM lactacystin (Lact), 10mM 3-methyladenine (3MA), or 20mM ammonium chloride and 100µM leupeptin (NL) for 9.5 hours. Top: Immunoblots for the indicated proteins. Bottom: Quantification of changes in total levels of LRRK2 (n=6). (b) Brain slices from mouse cortex, midbrain, or cerebellum were incubated without additions or with NL or 3MA for 2 hours at 37°C. Top: Representative immunoblots for the indicated proteins. Bottom: Quantification of changes in levels of LRRK2 relative to those in untreated samples. (n= 3–4). All values are mean+s.e.m. (*p<0.05). (c,d) Homogenate (Hom) and lysosomal fractions (Lys) isolated from SH-SY5Y cells (c) or the indicated mouse brain regions (d) immunoblotted for the indicated proteins. (e) HEK293 cells expressing GFP tagged LRRK2 and transduced with lentivirus control (Ctr) or carrying shRNA against LAMP-2A (L2A(–)) were incubated or not with NL for 12 hours and subjected to immunoblot for the indicated proteins. (f) Immunoblots for LRRK2 and LAMP-2A of homogenates and lysosome-enriched fractions isolated from fibroblasts control (Ctr) or stably interfered for LAMP-2A (L2A(–)) maintained in the presence or absence of serum. Actin and LAMP-1 (L-1) are shown as loading controls of homogenate and lysosomes, respectively. Full-length blots/gels are presented in Supplementary Figure 12.
Lysosomal degradation of LRRK2 also occurs in vivo, as we found a marked increase in LRRK2 levels in slices of mouse cortex, midbrain, and cerebellum after incubation with NL (Fig. 1b). In further support of these findings, we could detect LRRK2 in lysosomes isolated from neuronal cells in culture (Fig. 1c) and from mouse and rat brain regions (Fig. 1d and data not shown). The lack of inhibition of LRRK2 degradation by 3MA in the brain slices (Fig. 1) and the absence of LRRK2 in autophagosomes isolated from the brains of these animals (Supplementary Fig. 1c) suggest that macroautophagy does not contribute considerably to LRRK2 degradation in brain, at least under normal conditions.
Instead, we found that cells knocked-down for LAMP-2A, the CMA lysosomal receptor, exhibited increased intracellular levels of LRRK2 due, at least in part, to reduced LRRK2 degradation by lysosomes, as shown by the lack of effect of NL treatment on LRRK2 levels in these cells (Fig. 1e). Furthermore, the amount of LRRK2 bound to lysosomes isolated from LAMP-2A knock-down cells was strongly reduced in comparison to control cells (Fig. 1f; notice that persistent LAMP-2A knock-down reduces LRRK2 initial accumulation, likely through activation of other proteolytic systems). We conclude that a fraction of intracellular LRRK2 is normally degraded in lysosomes probably via CMA.
Selective degradation of wild-type LRRK2 by CMA
The presence in the amino acid sequence of LRRK2 of 8 putative CMA targeting motifs28 (Fig. 2a) further increased our confidence that CMA may contribute to degradation of this protein. However, the most convincing test to confirm a protein as a CMA substrate is to directly demonstrate its uptake into isolated lysosomes29. We first used lysosomes isolated from rat liver to avoid possible interference from the endogenous LRRK2 detected in the brain lysosomes (Fig. 1d). Incubation of purified LRRK2 with intact liver lysosomes untreated or pre-treated with protease inhibitors revealed that LRRK2 binds to the lysosomal membrane and is selectively taken up by lysosomes (uptake is calculated as the increase in lysosome-associated LRRK2 when lysosomes are pre-treated with protease inhibitors) (Fig. 2b). As noted for other CMA substrate proteins, increasing concentrations of LRRK2 compete with the lysosomal uptake and degradation of a pool of cytosolic proteins substrates for CMA (Fig. 2c) and the binding to the lysosomal membrane of the well-established CMA substrate glyceraldehyde-3-phosphate dehydrogenase (GAPDH)30 (Fig. 2d).
Figure 2. LRRK2 behaves as an atypical CMA substrate.
(a) Putative CMA targeting motifs in LRRK2. (b) Starved rat liver lysosomes untreated or pre-treated with protease inhibitors (PI) incubated with LRRK2. Inset: representative immunoblot. LAMP-2A is shown as lysosomal loading control. Inp: input, 1/5 of LRRK2 added. (n= 7). Uptake: difference between LRRK2 associated with lysosomes with and without PI. (c) Effect of increasing concentrations of LRRK2 on the degradation of a radiolabelled pool of cytosolic proteins by intact lysosomes. Values are percentage of the degradation without LRRK2 (n= 5–6). (d) Association of GAPDH to rat liver lysosomes in the presence of increasing concentrations of LRRK2. Values are percentage of GAPDH associated with lysosomes in the absence of LRRK2 (n = 4–6 in duplicate). Inset: representative immunoblot. Inp: 1 µg of GAPDH. (e, f) Association of LRRK2 and RNase A (e) or LRRK2 and GAPDH (f) with PI treated rat liver lysosomes incubated with increasing concentrations of GAPDH (e) or RNase A (f) or ovalbumin (Ov). Left: representative immunoblots. Right: Percentage of protein association when incubated with lysosomes alone (n = 5–6). Inp: RNase A (0.4 µg), LRRK2 (0.04µg) or GAPDH (1 µg). (g,h) Association of LRRK2 to lysosomes incubated with GAPDH (g) or RNase A (h) at 4°C. (n= 3). (i,j) Immunoblots of brain lysosomes incubated with LRRK2 and increasing concentrations of GAPDH (i) or RNase A (j). All values are mean+s.e.m. (differences with lysosomes incubated with the protein alone were significant for *p<0.05). Full-length blots/gels are in Supplementary Figure 12.
We next assessed the effect of the CMA substrate proteins GAPDH (Fig. 2e) and ribonuclease A (RNase A; Fig. 2f)30 and the negative control that lacks any lysosomal targeting motif, on the CMA of LRRK2. To our surprise, and in clear contrast with the competition observed between conventional CMA substrates (GAPDH and RNase A shown here), CMA substrates did not compete with LRRK2’s lysosomal association, but markedly increased the amount of LRRK2 bound to lysosomes in a dose-dependent manner (Fig. 2e,f). In contrast, the non-CMA substrate proteins ovalbumin (Fig. 2e,f) and cyclophilin A (Supplementary Fig. 2a), did not affect the association of LRRK2 with lysosomes. The increased LRRK2 binding was not due to a proportional increase in uptake, as differences between samples treated or not with protease inhibitors were similar (Supplementary Fig. 2b), and increased binding in the presence of GAPDH (Fig. 2g) or RNase A (Fig. 2b) was still observed at low temperatures, which does not allow for translocation.
We confirmed that this unusual binding of LRRK2 in a CMA substrate-dependent manner also occurs in lysosomes isolated from rat brain and that this behavior was, again unique for LRRK2, as other CMA substrates produced the expected competitive response in brain lysosomes (Fig. 2i,j). Immunoblot revealed that the CMA-substrate and LRRK2 were both bound to the lysosomes and that enhanced binding of LRRK2 to brain lysosomes was not observed in the presence of non-CMA substrates such as ovalbumin (Fig. 2i).
In summary, although LRRK2 displays characteristics common to CMA substrates when presented alone to lysosomes, CMA substrates enhanced, rather than inhibited, binding of LRRK2 to lysosomes. These findings reveal the existence of a novel interplay between LRRK2 and CMA substrate proteins at the lysosomal membrane that enhances LRRK2 binding to this membrane but not its translocation into lysosomes.
Compromised degradation of mutant LRRK2
The most common pathogenic LRRK2 mutation is G2019S in the kinase domain that results in a toxic gain of kinase function2. We used LRRK2-tet-on cell lines31 in which expression of wild-type (WT) or G2019S (G/S-LRRK2) can be activated by doxycycline (Supplementary Fig. 3a), induced their expression by a 12–24h pulse of doxycycline, and followed the decrease in levels of the recombinant proteins 48h after the pulse (to confirm that synthesis was no longer occurring). As with endogenous LRRK2 in neuroblastoma cells (Fig. 1a), levels of the WT recombinant protein (Fig. 3a) and of the endogenous LRRK2 in these cells (Supplementary Fig. 3b) were increased by proteasome inhibitors (MG132) and lysosome inhibitors (NL), indicating degradation of LRRK2 by both pathways. Levels of G/S-LRRK2 were also higher after addition of proteasome and lysosome inhibitors, but the effect of blocking lysosomal degradation was comparatively smaller (Fig. 3a). Treatment with 3MA had a similar effect on G/S-LRRK2 as treatment with inhibitors of lysosomal proteolysis, suggesting that the residual lysosomal degradation observed for the mutant was likely taking place mainly by macroautophagy. Higher sensitivity to 3MA was also detected in slices from brains of G/S-LRRK2 transgenic mice when compared with control or WT-LRRK2 transgenic mice (Supplementary Fig. 1d). WT-LRRK2 displayed a half-life of approximately 38h while the half-life of G/S-LRRK2 was nearly doubled (60h) (Fig 3b). Knock-down for LAMP-2A reduced the degradation of WT-LRRK2 by 50% but only had a modest effect on the degradation of G/S-LRRK2 (Fig. 3b). Together, these findings suggest that the degradation of G2019S LRRK2 by CMA is compromised and that the apparent switch of the mutant protein toward degradation by the proteasome and a small fraction by macroautophagy, is not sufficient to maintain the rates of degradation normally observed for the WT protein.
Figure 3. Differences in the degradation of wild-type and G2019S mutant LRRK2.
(a) Immunoblot for GFP of Tet-on-HEK293 cells expressing GFP-tagged wild-type (WT) or G1209S mutant LRRK2 (G/S) pulsed 24h with doxycycline and 48h later (no longer de novo synthesis), incubated without additions (–) or with 20 mM ammonium chloride and 100 µM leupeptin (NL), 5 µM MG132 (MG), or 10mM 3-methyladenine (3MA). Bottom: Percentage of inhibition (n =3). (b) Cells control (Ctr) or RNAi for LAMP-2A (L2A(–)) pulsed as in a, sequentially collected and immunoblotted for GFP. Left: Representative immunoblot. Actin is shown as a loading control. Quantification is shown at the bottom corrected by levels of knock-down (n = 3–4). Right: Immunoblot for LAMP-2A. Bottom shows the percentage of LRRK2 degraded per hour in the same samples. (c) Starved rat livers lysosomes untreated or pre-treated with protease inhibitors (PI) incubated with WT or G/S LRRK2. Inset: Representative immunoblot. (n = 9). (d) Association of GAPDH (5 µg) to starved rat liver lysosomes in the presence of increasing concentrations of WT or G/S LRRK2. Inset: Representative immunoblot. (n=4–6). (e, f) Binding of WT and G/S LRRK2 to lysosomes in the presence of GAPDH (e) or RNase A (f). Top: Representative immunoblots. Bottom: Lysosome-bound LRRK2 expressed as fold-levels when added alone. Trends of mean values (n=2). (g) Rat brain lysosomes incubated with WT or G/S LRRK2 as in c. Inputs: 1/10 of protein added. (h,i) Rat brain lysosomes incubated as in d and e, respectively. Left: Representative immunoblots. Right: Quantifications (n=3–4). All values are expressed as mean+s.e.m. (differences with untreated samples (*) and between WT and G/S LRRK2 (§) were significant for p<0.05). Full-length blots/gels are in Supplementary Figure 12.
We then compared the direct uptake of purified WT and G/S-LRRK2 by isolated liver lysosomes after verifying that G/S-LRRK2 did not compromise lysosomal integrity (Supplementary Fig. 2c) and had comparable susceptibility to degradation by the lysosomal hydrolases (Supplementary Fig. 2d). G/S-LRRK2 bound to the lysosomal membrane with a lower efficiency than WT-LRRK2 and its lysosomal uptake was also significantly lower (Fig. 3c). Despite its lower CMA, G/S-LRRK2 exerted a more pronounced inhibitory effect on uptake of other CMA substrates than that observed for WT-LRRK2 (Fig. 3d). Notably, G/S-LRRK2’ s binding to lysosomes was also increased by CMA substrate proteins and this effect was more prominent than that of the WT protein (Fig. 3e,f). Studies with isolated brain lysosomes revealed a similar reduction in lysosomal binding and uptake of G/S-LRRK2 (Fig. 3g), a significantly stronger inhibitory effect on CMA (Fig. 3h) and a slightly higher association to lysosomal membranes in the presence of CMA substrates (Fig. 3i) when compared to WT-LRRK2. We attribute the small differences between liver and brain lysosomes to the presence of endogenous WT-LRRK2 in the brain lysosomes (Fig. 3g). Independent of these small tissue-specific differences, overall these results indicate that G/S-LRRK2 is less efficiently degraded by CMA but exerts greater inhibition over this pathway.
Inhibitory effect of LRRK2 on CMA
To determine the cellular consequences of higher association of LRRK2 with the lysosomal membrane in the presence of CMA substrates (Fig. 2), the exacerbation of this binding for G/S-LRRK2 (Fig. 3e,f, i), and the greater inhibition by the LRRK2 mutant on lysosomal uptake of CMA substrates (Fig. 3d,h), we analyzed the status of the autophagic system in cells expressing WT and G/S-LRRK2. Long-lived proteins, which are typically degraded by the lysosomal system, were degraded more slowly when either form of LRRK2 was overexpressed, although this inhibitory effect was more pronounced for G/S-LRRK2 (Fig. 4a, left). Serum deprivation, used to stimulate autophagy, did not increase protein degradation in the cells expressing WT or G/S-LRRK2 and the inhibitory effect of G/S-LRRK2 was still more pronounced than that of the WT protein (Fig. 4a, right).
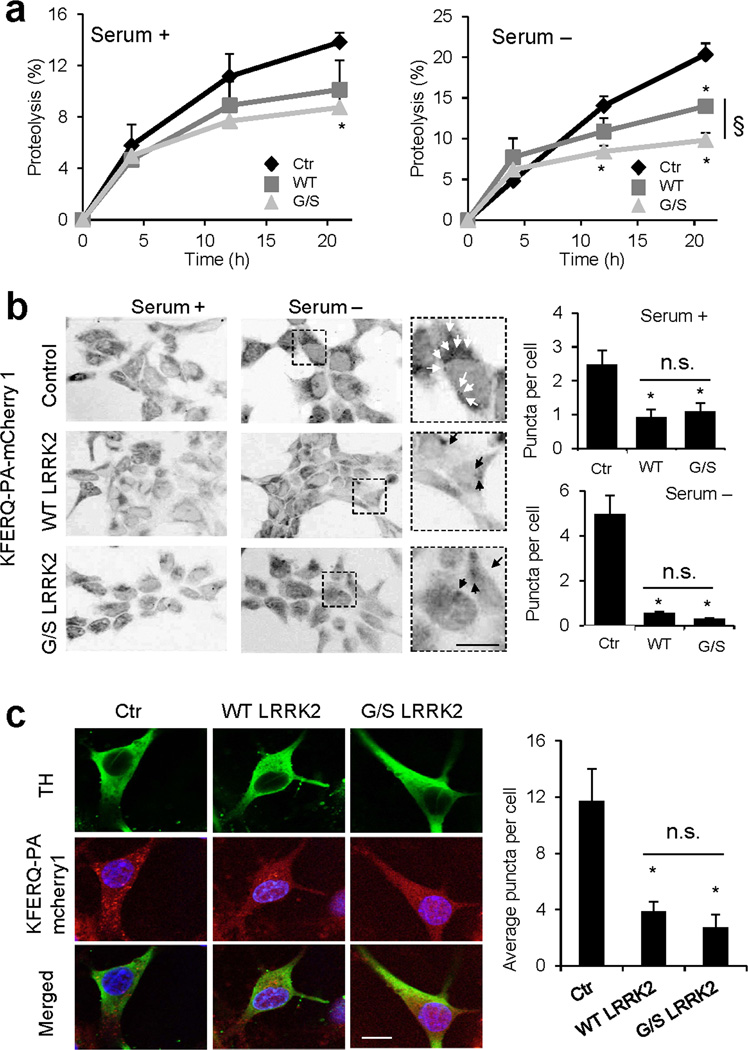
Figure 4. Overexpression of LRRK2 proteins exerts an inhibitory effect on CMA.
(a) HEK293 cells stably transfected with an empty plasmid (Ctr) or plasmids expressing wild-type (WT) or G2019S mutant (G/S) LRRK2 under the control of tetracycline, were treated with doxycycline to activate protein expression and then labeled with 3H-leucine for 48h. After extensive washing, protein degradation at the indicated times was measured as the amount of acid precipitable radioactivity (amino acids and small peptides) released into the media. Values are expressed as percentage of radioactivity in proteins (acid precipitable) at time 0 (n=4). (b) The same cells were transiently transfected with a KFERQ-PA-mCherry1 fluorescent reporter for CMA activity. After photoactivation, cells were maintained for 16h in the presence or absence of serum. Left: Representative micrographs. Insets show higher magnification images. Right: Quantification of the average number of puncta per cell in each condition measured in >150 cells. (c) Ventral midbrain dopaminergic neurons from control (Ctr), WT, or G/S LRRK2 transgenic mice were transduced with the same reporter as in b and co-stained with anti-TH. Left: Representative images. Right: Average number of puncta/cell in >50 TH-positive cells. Scale bars: 10 µm Values are all mean+s.e.m. (differences with control (*) and between WT and G/S LRRK2 (§) were significant for p<0.05).
To separately analyze the impact of these proteins on the different autophagic pathways, we first monitored levels and degradation of LC3, a well-established marker for macroautophagy. The increase in cellular LC3 observed when lysosomal proteolysis is inhibited is indicative of the flux of substrates through macroautophagy32. In cells expressing WT-LRRK2, LC3 flux trended slightly higher than in control cells (Supplementary Fig. 4a), which along with the reduced number of LC3-positive puncta observed by immunofluorescence (autophagosomes) (Supplementary Fig. 4b), supported a discrete stimulation of macroautophagy by LRRK2. LC3 flux in cells expressing G/S-LRRK2 was close to that in control cells, suggesting that the decrease in protein degradation observed in these cells was not due to inhibition of macroautophagy.
We then analyzed CMA in the three groups of cells by transfecting them with a photoactivatable (PA) fluorescent reporter (KFERQ-PA-mCherry1)33 that provides visualization of CMA as a change in the distribution of the fluorescent protein from the cytosol (diffuse fluorescent pattern) to lysosomes (punctate fluorescent pattern). Cells expressing WT and G/S-LRRK2 both revealed a marked decrease in the number of fluorescent puncta per cell in basal conditions (Fig. 4b), and failed to induce CMA upon removal of serum from the culture media (Fig. 4b). We conclude that the decrease in degradation of long-lived proteins observed in cells expressing WT-LRRK2 and G/S-LRRK2 was due, for the most part, to their inhibitory effect on CMA.
We found a similar inhibitory effect on CMA for G/S-LRRK2 in neurons. Analysis of CMA activity with the same fluorescent reporter in primary neuronal cultures revealed a significant reduction in CMA activity in neurons from G/S-LRRK2 transgenic mice (Fig. 4c). Notably, although a compromise of LRRK2 lysosomal degradation was not evident in brains of WT-LRRK2 transgenic mice (Supplementary Fig. 1d), we found that CMA activity was significantly reduced in these neurons (Fig. 4c).
Together, these results indicate that both G/S-LRRK2 and also high levels of WT-LRRK2 inhibit CMA in neuronal and non-neuronal cells.
Effect of LRRK2 on CMA components
Recent studies in Drosophila have described that expression of G2019S LRRK2 results in perinuclear clustering of endosomal/lysosomal compartments34. While CMA does not occur in endosomes, we analyzed whether LRRK2-induced changes also affected CMA active lysosomes. Labeling of late endosomes and lysosomes with lysoTracker or LAMP-1 (Supplementary Fig. 5a,b, respectively), confirmed the described abnormal expansions of these vesicular structures not only in cells expressing G/S-LRRK2, but also in cells expressing the WT-LRRK2 protein. While we did not note expansion or clustering of lysosomes active for CMA (defined as those positive for both hsc70 and LAMP-2A (Supplementary Fig. 6a)), immunofluorescence and immunoblot (Supplementary Fig. 6b) revealed that levels of LAMP-2A were about 2-folds higher in cells expressing either WT or G/S-LRRK2. This contrasted with the marked decrease in CMA activity observed in these cells (Fig. 4b) and likely reflects a compensatory response of these cells similar to that described in other conditions with compromised CMA, such as aging or other models of PD35–37. We did not find significant changes in the levels of LAMP-2A in lysosomes isolated from cells knocked-down for LRRK2 or by immunofluorescent analysis of these cells (Supplementary Fig. 6c), or in lysosomes isolated from LRRK2 null mice (Supplementary Fig. 6d), suggesting that the changes in CMA observed in the G/S-LRRK2 are more likely a result of a gain of toxic function rather than a loss of physiological function.
The lack of marked changes in the number or distribution of CMA active lysosomes led us to hypothesize that the inhibitory effect of LRRK2 on CMA did not occur at the level of formation and maturation of the CMA lysosomes, but rather was a result of the direct interaction of LRRK2 with these lysosomes. To dissect the inhibitory effect of G/S-LRRK2 and that of high levels of WT-LRRK2 on CMA, we used the in vitro system with isolated lysosomes. GTP did not affect binding of WT or G/S-LRRK2 to the lysosomal membrane (Fig. 5a) or their increase in the presence of the CMA substrate GAPDH (Fig. 5a), indicating that the GTPase activity of LRRK2 does not modulate its binding to the lysosomal membrane.
Figure 5. Interplay of LRRK2 with CMA components.
(a) Immunoblot (duplicates) of rat liver lysosomes incubated with wild-type (WT) or G2019S mutant (G/S) LRRK2 alone or with 1mM GTP and/or GAPDH. Bottom: Quantification of lysosome-bound LRRK2 expressed as times the binding without additions. (n=6). (b) Effect of kinase inhibitor on WT, G/S, or the kinase-dead mutant D1994A (KiD) LRRK2 incubated with rat liver lysosomes in presence or not of GAPDH (5 µg). Right: Quantification expressed as fold-times the binding without additions. (n=3–5). (c,d) HEK293 cells transiently transfected with the indicated myc-or GFP tagged LRRK2 constructs (see scheme). Full (full length); M (KFERQ motifs (M1–M8)) (c) Binding to GST-hsc70. Lanes 1–6 show 1/10 inputs. NT: non transfected cells. Immunoblot for hsc70 is shown as loading control. Top right: Quantification of LRRK2 bound to hsc70. Where indicated, samples were co-incubated with RNase A to compete the KFERQ-mediated binding of hsc70 (n=3–4). (d) Co-immunoprecipiation (IP) of hsc70 with anti-myc or GFP in the same cells. Inputs (Inp: ¼ of starting volume). (e) Binding or LRRK2 to rat liver lysosomes treated with the indicated concentrations of trypsin. LAMP-1 is shown to confirm the integrity of the lysosomes. (f) Co-immunoprecipiation (IP) of LAMP-2A with anti-LRRK2 in lysosomes incubated with WT or G/S LRRK2. Inp: ¼ of starting sample). IgG: immunoglobulin. Values: LAMP-2A recovered in the IP (IP L2A) corrected for the amount of LRRK2 pulled down and expressed as folds WT. (g, h) LAMP-2A immunoblot of rat liver lysosomes incubated alone (none) or with WT or G/S LRRK2 (g) or lysosomes from control (Ctr) and WT and G/S LRRK2 transgenic mice and subjected to blue native electrophoresis (BNE). Arrow: 700KDa multimeric complex. Duplicates are shown. All values are mean+s.e.m. (differences compared to none or untreated (*) or to full length protein (§) were significant for p<0.05). Full-length blots/gels are in Supplementary Figure 12.
In contrast, SU6656, a selective src family kinase inhibitor, previously used to block the kinase activity of LRRK238, discretely increased binding of WT-LRRK2 to lysosomes both when presented alone or in the presence of other CMA substrates (Fig. 5b). The fact that treatment with the inhibitor under the same conditions did not affect the lysosomal binding of a kinase-dead form of LRRK2 indicates that the changes in WT-LRRK2 binding were indeed a consequence of inhibition of its kinase activity (Fig. 5b). Inhibition of G/S-LRRK2 kinase activity did not affect its lysosomal binding if presented alone but markedly enhanced the GAPDH-induced binding of G/S-LRRK2 to the lysosomal membrane (Fig. 5b). We confirmed that treatment with the kinase inhibitor did not impact lysosomal stability (Supplementary Fig. 2e) and that the WT-LRRK2 did not mimic the MT-LRRK2 behavior even at very high concentration of the inhibitor (Supplementary Fig. 2f). Overall, these results support the finding that binding of G/S-LRRK2 to lysosomal membranes increases in the presence of other CMA substrates, and more so if its kinase activity becomes compromised.
To gain further insight into the regions of LRRK2 important for its CMA targeting and degradation, we investigated the relevance of the different CMA-targeting motifs, the pentapeptide recognized by hsc70, in its sequence (Fig. 5c). Studies in other multi-motif CMA substrates have demonstrated that a single motif is sufficient for lysosomal targeting and that, although the presence of multiple motifs does not increase efficiency of targeting, hsc70 often has a specific order of preference in the binding17. However, once the most favored motif(s) are eliminated, it is not unusual that hsc70 binds to one of the other motifs39. For that reason, we studied binding of this chaperone to previously characterized myc or GFP-tagged truncated forms of LRRK2 that bear different combinations of CMA targeting motifs40, 41. In vitro binding studies to GST-hsc70 revealed that binding to the chaperone of a fragment containing the ROC, COR and Kinase regions (RCK) and that bears the 5 central CMA motifs (M3–M7) but is missing the two N-terminal (M1 and M2) and the most distal c-terminal motifs (M8) was comparable to binding of the full-length LRRK2, suggesting against involvement of these three motifs in chaperone binding (Fig. 5c). In contrast, binding to hsc70 of a truncated form of LRRK2 bearing only M4 to M6 (COR) was markedly reduced, suggesting that either M3 or M7 were important for chaperone binding of LRRK2 (Fig. 5c). We found that M7, the motif in the kinase region of LRRK2, could not be selectively recognized by the chaperone, at least when this region was presented alone (note that the binding to hsc70 of this product could not be competed with a protein bearing a CMA-targeting motif suggesting non-specific binding (Fig. 5c)). However, binding to hsc70 was preserved in a truncated form of LRRK2 (ROC) bearing M3 and M4. Since we did not observe binding to hsc70 of the fragment containing M4 to M6, we concluded that M3, the motif at the ROC region of LRRK2, is a strong candidate for hsc70 binding (Fig. 5c). Co-immunoprecipitation of hsc70 with LRRK2 in cells expressing the different tagged LRRK2 truncated proteins confirmed that binding of hsc70 to the LRRK2 fragment bearing M3 and M4 also occurs in intact cells, but that absence of M3 markedly reduced the amount of hsc70 recovered in the pull-down assay (Fig. 5d). Although future detailed analysis of the contribution of different motifs in LRKK2 binding to hsc70 under different conditions is needed, these results support the requirement of M3 for chaperone binding.
We next analyzed the nature of the binding of LRRK2 to CMA lysosomes and demonstrated that trypsinization of lysosomal membranes gradually decreased binding of LRRK2, supporting protein-to-protein binding of LRRK2 (Fig. 5e; levels of LAMP-1 are shown to demonstrate that trypsin did not reach the lysosomal lumen). The reduced amount of LRRK2 detected in lysosomes from cells knocked-down for LAMP-2A (Fig. 1f) suggested that lysosomal binding of LRRK2 likely occurred through LAMP-2A, and pull-down experiments of lysosome-bound LRRK2 confirmed its binding to LAMP-2A (Fig. 5f). Interestingly, comparison of the amount of LAMP-2A recovered in pull-downs for WT and G/S-LRRK2 revealed that although the mutant protein bound less efficiently to the lysosomal membranes than WT-LRRK2 (Fig. 3c,g), once bound to this receptor, its binding to LAMP-2A was more stable (Fig. 5f).
We then analyzed the effect of WT and G/S-LRRK2 on LAMP-2A. We hypothesized that this tight binding to LAMP-2A of G/S-LRRK2 might inhibit CMA by favoring mobilization of LAMP-2A into specific lipid microdomains at the lysosomal membrane where it eventually undergoes degradation23. However, incubation of lysosomes with LRRK2 did not enhance recruitment of LAMP-2A to the lysosomal lipid microdomains, even when other CMA substrate proteins were added to the incubation media (Supplementary Fig. 2g,h and data not shown). We analyzed the effect of LRRK2 on the organization of LAMP-2A into the multimeric complex responsible for translocation of CMA substrates21 which can be visualized as a single 700 kDa band by blue native electrophoresis (BNE) and immunoblotting for LAMP-2A (Fig. 5g). We found that both LRRK2 proteins, at concentrations that inhibit CMA uptake, markedly decreased the amount of LAMP-2A detectable in the multimeric complex (Fig. 5g). Lysosomes from G/S-LRRK2 transgenic mice also revealed a relatively lower abundance of the LAMP-2A translocation complex when compared with lysosomes from control mice (Fig. 5h). In the case of the WT-LRRK2 transgenic mice, there was also a trend of lower levels of multimeric LAMP-2A, although as shown in Fig. 5h, there was more variability from animal to animal than in the case of the mutant protein. Overall, these results support the notion that the WT and G/S-LRRK2 inhibition of LAMP-2A multimerization is responsible for the reduced CMA activity observed in non-neuronal and neuronal cells overexpressing these proteins.
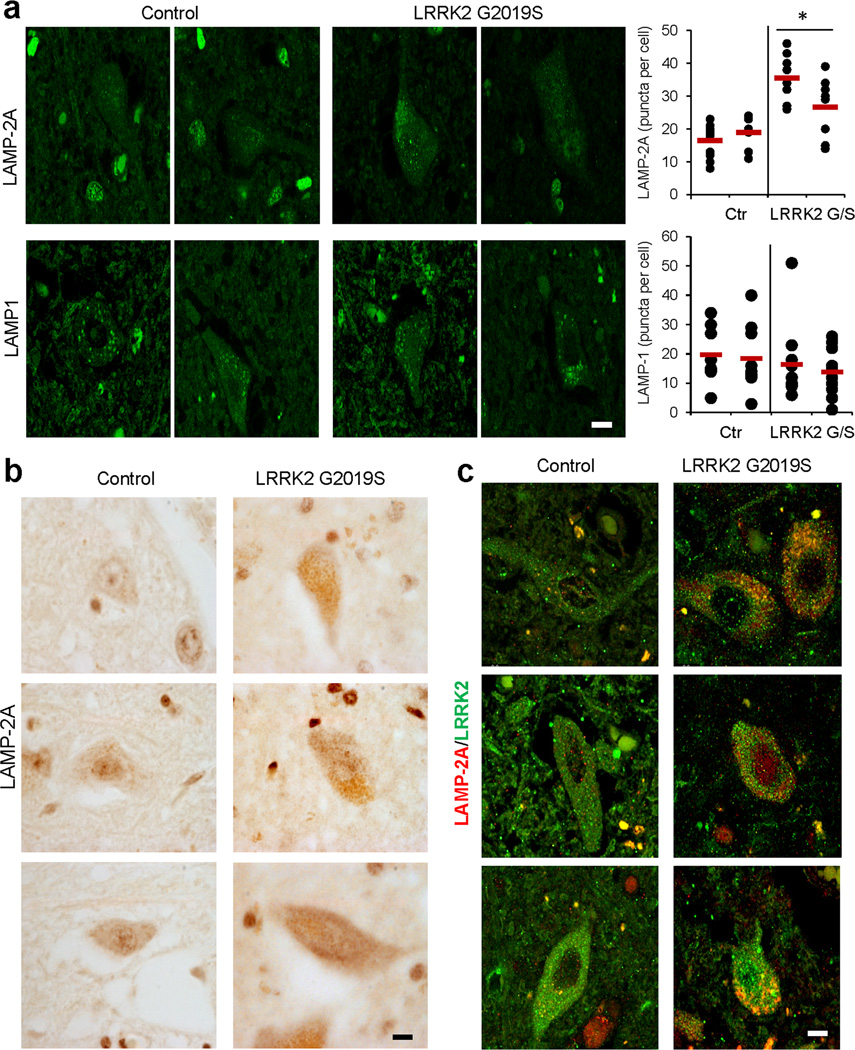
Finally, we confirmed that alterations in LAMP-2A were evident in the dorsal motor nucleus of the vagus (DMV), a region considered to be the first highly targeted area of the central nervous system in PD42, from PD patients with the G2019S LRRK2 mutation. Similar to the upregulation of LAMP-2A observed in cultured cells, when expression of G/S MT-LRRK2 was activated (Supplementary Fig. 6a,b), immunostaining for LAMP-2A revealed a marked increase in the levels of LAMP-2A in the patients’ brains when compared with brains from age-matched control individuals (Fig. 6a,b), also detectable after immunoblot for LAMP-2A of the same brain samples (Supplementary Fig. 7a). The increase in LAMP-2A does not seem a marker of general upregulation of the lysosomal system in the PD brains, because we did not detect significant differences in levels of LAMP-1, a more abundant lysosomal membrane protein, between control and patients’ brains (Fig 6a and Supplementary Fig. 7a). In addition, and in agreement with our findings in vitro, association of LRRK2 with LAMP-2A positive compartments was also markedly increased in the brains of the PD patients (Fig. 6c and Supplementary Fig. 7b). These findings are supportive of selective alterations in CMA in the brains of mutant LRRK2 PD patients.
Figure 6. Altered CMA markers in brains of familial LRRK2 PD patients.
(a) Immunofluorescence for LAMP-2A and LAMP-1 of sections from the dorsal motor nucleus of the vagus nerve of brains from two different unaffected control individuals and two PD individuals with the LRRK2 G2019S mutation. Left: Images from representative fields. Right: Quantification of the number of fluorescent puncta per cell. Black dots show values in individual cells and red marks show the mean value. (b) Immunohistochemistry for LAMP-2A in the same samples. (c) Immunofluorescence for LAMP-2A (red) and LRRK2 (green) in the same samples. Merged images from both fluorophore channels are shown. Scale bar: 10 µm. (for colocalization quantification see Supplementary Fig. 7b). Values are mean+s.e.m. (*p<0.05).
Interaction of other LRRK2 mutant variants with CMA
We next examined three additional LRRK2 mutant variants, two in the kinase site and one in the GTP-binding region, for possible differences in their degradation by CMA and impact on this autophagic pathway. We selected a mutation in the ROC site of LRRK2 (R1441C LRRK2 or R/C LRRK2) and a mutation in the kinase site of LRRK2 (I2020T or I/T LRRK2) both described in familial PD patients and shown to increase LRRK2 dimerization, and an experimental mutation that ablates LRRK2 kinase activity (D1994A LRRK2 or D/A LRRK2) which displays low dimer stability43.
We found that whereas I/T LRRK2 displayed levels of binding and uptake by isolated lysosomes comparable to those of the WT-LRRK2, binding and in particular uptake of R/C- and D/A-LRRK2 were significantly reduced when compared with WT-LRRK2 (Fig. 7a). Notably, the mutant forms with reduced binding/uptake by lysosomes showed a more pronounced increase in their binding to lysosomes in the presence of CMA substrates (GAPDH and RNase A shown in Fig. 7b). These results support that the inability to translocate to lysosomes may enhance the substrate-induced lysosomal binding of LRRK2 and favor its accumulation in the membrane of these organelles. Pull-down experiments of the LRRK2 variants bound to lysosomes in the in vitro system (Fig. 7c) or co-IP from cells expressing GFP-tagged forms of each of these LRRK2 variants (Fig. 7d) revealed that the enhanced interaction of G/S-LRRK2 with LAMP-2A was also observed for R/C-LRRK2 but not for D/A LRRK2, suggesting that the enhanced substrate-dependent binding of this variant to the lysosomal membrane may involve different mechanisms than the persistent binding to LAMP-2A of G/S and R/C. As expected from the efficient degradation of I/T-LRRK2 by CMA (Fig. 7a) and its lower association to the lysosomal membrane in the presence of substrates (Fig. 7b), we did not find significant differences to WT-LRRK2 in the amount of this protein bound to LAMP-2A (Fig. 7c,d).
Figure 7. Interaction of additional LRRK2 mutant variants with CMA.
(a) LRRK2 immunoblot of rat liver lysosomes incubated with wild-type (WT), G2019S (G/S), D1994A (D/A, kinase-dead), I2020T (I/T), or R1441C (R/C) mutant LRRK2 alone or in the presence of protease inhibitors (PI) Lanes 1–5: ¼ input. Bottom: Quantification of binding and uptake (n=4–5). (b) Immunoblots of lysosomes incubated as in a alone or in the presence of increasing concentrations of RNase A (left) or GAPDH (right). Right: Quantification of LRRK2 bound as folds of binding when incubated with lysosomes alone. (n=3). (c) Co-immunoprecipitation (Co-IP) of LAMP-2A with anti-LRRK2 from lysosomes incubated with LRRK2 proteins as in a, were subjected to immunoprecipitation with LRRK2. Co-immunoprecipitated fractions (Co-IP) were immunobloted for LAMP-2A and LRRK2. Input: Immunoblot for LAMP-2A before the affinity purification. Bottom: Amount of LAMP-2A co-immunoprecipitated expressed as folds the value in WT-LRRK2. (n=3–4). (d) Co-immunoprecipitation of LAMP-2A with anti-GFP in Tet-on-HEK293 cells expressing GFP-LRRK2 proteins and pulsed for 24 hours with doxycycline. Inputs (Inp: ¼ of added) (e) The same cells as in d and control cells transduced with the KFERQ-PA-mCherry1. Left: Representative images, full field and higher magnification panels. Right: Quantification after photactivation and 16 hours in serum deprived media (n > 150 cells). Values are all mean+s.e.m. (Differences with wild type (*) or with the other mutants (§) were significant for p<0.05). Bars: 10 µM. Full-length blots/gels are in Supplementary Figure 12.
To directly analyze the impact of these different LRRK2 mutants on CMA in intact cells, we expressed these variants in cells under the tet-regulated system described before (Supplementary Fig. 3c) and used the fluorescent KFERQ-PA-mCherry1 reporter to quantify CMA activity. In agreement with the lower degradation of R/C and D/A-LRRK2 by CMA (Fig. 7a) and their higher association to lysosomes when in the presence of CMA substrates (Fig. 7b), we found that these two variants significantly reduced CMA activity to levels comparable to those observed for G/S-LRRK2 (Fig. 7e). A reduction in CMA activity, although significantly more discrete, was also observed in cells expressing I/T LRRK2, although in these the predominant feature was a remarkable increase in the size of the lysosomal compartments related to CMA (Fig. 7e). Although the significance of these changes in CMA lysosomes requires further investigation, these findings highlight that different LRRK2 mutations may inhibit CMA through multiple different mechanisms.
Consequences of the unusual interplay of LRRK2 with CMA
Because enhanced binding to LAMP-2A at the lysosomal membrane was shared by two of the LRRK2 mutants, including the most common mutation in familial PD patients (G/S-LRRK2), we set out to analyze the consequences of this abnormal binding. We speculated that by blocking the formation of the receptor complex (Fig. 5g), G/S-LRRK2 and high levels of WT-LRRK2 may lead to higher concentration of substrate proteins bound to lysosomes at a given time because the inability of LAMP-2A to multimerize does not affect substrate binding, but rather, prevents their translocation inside the lysosomal lumen21. This high concentration may be particularly detrimental for pathogenic proteins that tend to organize into abnormal multimeric complexes and oligomers.
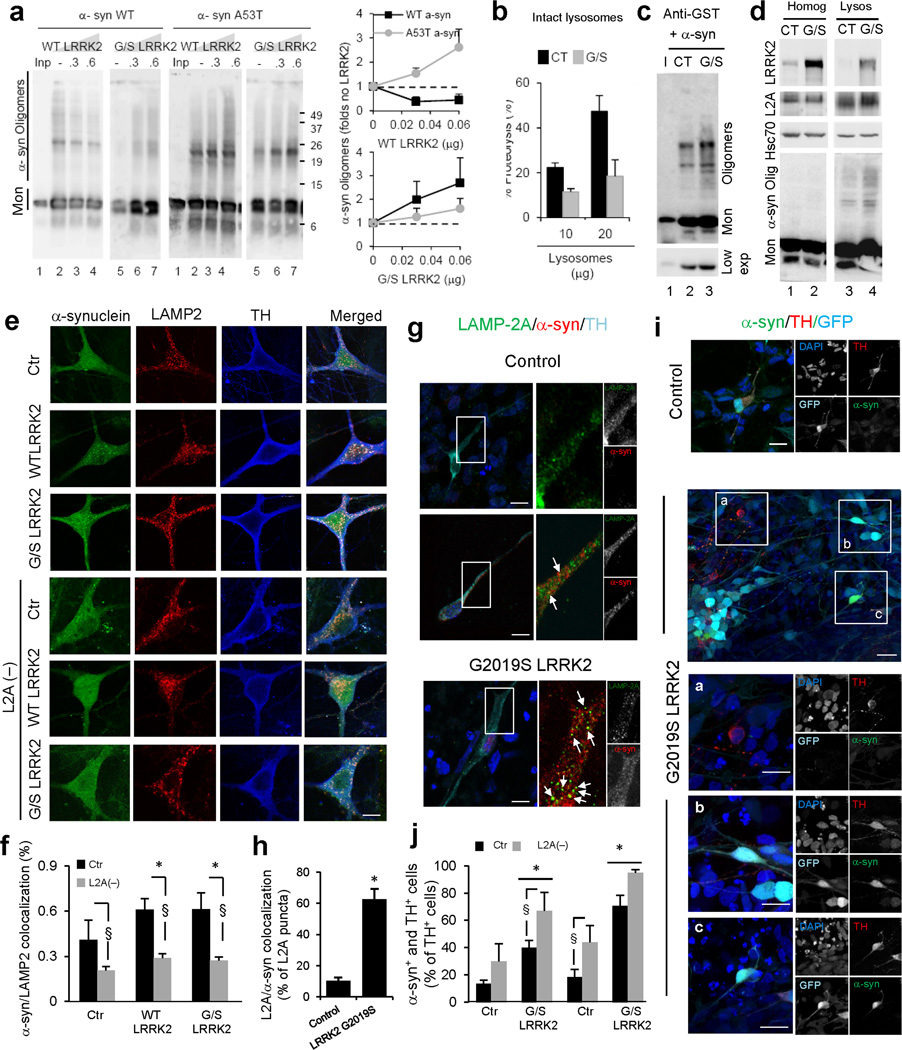
To test this possibility, we analyzed the effect of LRRK2 on the lysosomal association of WT α-syn and a pathogenic α-syn mutant (A53T) that we have previously shown binds LAMP-2A at the lysosomal membrane but does not translocate into the lumen8. WT-LRRK2 competes association of monomeric WT α-syn to lysosomes (as was the case for other CMA substrates) but was less efficient in competing the mutant α-syn variant (Fig. 8a and Supplementary Fig.8a). Increasing concentrations of WT-LRRK2 decreased the presence of WT α-syn oligomers in lysosomes (likely by competing binding when still at the monomeric stage (Supplementary Fig. 8a) whereas combination of WT-LRRK2 with the mutant α-syn protein markedly increased formation of α-syn oligomers in a dose dependent manner (Fig. 8a). LRRK2-induced formation of oligomers of α-syn did not occur when both proteins were incubated alone, indicating that oligomerization required their binding to lysosomal membranes (Supplementary Fig. 8b). These results suggest that the coincidence at the lysosomal membrane of WT-LRRK2 and mutant α-syn is sufficient to aggravate the CMA defect.
Figure 8. Coincidence of LRRK2 and α-syn at lysosomes enhances their toxic effect on CMA.
(a) α-syn immunoblot of rat liver lysosomes incubated with WT or mutant (A53T) α-syn alone (–) or with WT-LRRK2 or G/S MT-LRRK2 (Inp: input). Right: Quantifications of α-syn oligomers expressed as folds levels in samples without LRRK2. (n= 4–5). (b-d) Degradation of radiolabeled proteins (b), binding of GST-α-syn (c) and levels of endogenous α-syn (d) in intact brain lysosomes from control (CT) and G/S LRRK2 transgenic mice. Bottom insert in c shows lower exposure to visualize monomers (Mon) of α-syn. Homog: homogenates; I: input (n=3). (e) Ventral midbrain dopaminergic neuronal cultures from control, or WT or G/S LRRK2 transgenic mice untreated or RNAi for LAMP-2A (L2A(–)). Individual channels and merged images are shown. (f) Colocalization between α-syn and total LAMP-2 from cells in e. (n > 50 TH-positive neurons). Note that levels of total LAMP-2 are not reduced in L2A knock-down cells because of upregulation of LAMP-2B. (g–j) Induced pluripotent stem cells (iPSC) lines from control and G/S LRRK2 mutant PD patients were: (g–h) differentiated for 30 days and immunostained as labeled. (g) Merged channels and magnified green and red channels. Arrowheads: colocalization LAMP-2A/ α-syn. (h) Percentage of LAMP-2A-positive puncta that colocalize with α-syn in TH-positive cells. (n >50) for 2 control and 2 LRRK2 iPSC lines. (i,j) differentiated for 30 or 75 days, and RNAi for LAMP-2A. (i) Immunofluorescence for LAMP-2A and α-syn and GFP expression (light blue). Bottom: Higher magnifications of boxed areas are presented at the bottom. (j) Percentage of TH-positive cells showing cytoplasmic accumulation of α-syn. n≥2 experiments using 2 control and 2 LRRK2 iPSC lines (n > 70 and n >150 TH-positive cells in Ctr and LRRK2, respectively). Scale bars: 10 µm. All values are mean+s.e.m. Differences with WT or control (*) or between the control or knock-down cells (§) are significant for p<0.05). Full-length blots/gels are in Supplementary Figure 12.
G/S-LRRK2, rather than competing binding of the α-syn proteins, preserved or even enhanced binding of monomeric α-syn to the lysosomal membrane (Supplementary Fig. 8a), which probably causes the observed marked increase in formation of oligomers of even the WT form of α-syn at the surface of lysosomes (Fig. 8a). G/S-LRRK2 did not induce oligomerization of α-syn proteins in the absence of lysosomes (Supplementary Fig. 8b). The differences between the effect of WT and G/S-LRRK2 on WT α-syn oligomerization are likely resultant of the different effects that α-syn proteins had on the lysosomal association of WT and G/S-LRRK2. Whereas the combination of WT forms of both proteins did not interfere with lysosomal uptake of LRRK2, combination of WT LRRK2 with mutant α-syn or mutant LRRK2 with any of the α-syn proteins markedly decreased the translocation of LRRK2 into lysosomes (Supplementary Fig. 8c).
Studies in neuronal cell lines co-transfected with WT α-syn and LRRK2 proteins demonstrated significant decreases in the degradation rates of α-syn in the cells co-expressing G/S-LRRK2 (Supplementary Fig. 9). Overexpression of WT-LRRK2 in these cells did not change the early rates of α-syn degradation but delayed the degradation of this protein in the late stages (Supplementary Fig. 9). Further experiments are needed to determine if such two phase kinetics reflect an early compensatory activation of other proteolytic systems in the cells expressing WT-LRRK2 or the amount of time necessary to accumulate enough LRRK2 at the lysosomal membrane to affect α-syn degradation.
To determine if the proposed abnormal oligomerization of α-syn at the lysosomal membrane and the subsequent compromise of CMA also occurred in lysosomes exposed to mutant LRRK2 in vivo, we isolated lysosomes from control and G/S-LRRK2 transgenic mice. As shown in Fig. 8b, intact lysosomes isolated from G/S-LRRK2 mice revealed a significant reduction in their ability to take up proteins via CMA (note that this assay recapitulates binding, uptake and proteolysis but because the proteolytic activity of disrupted lysosomes was comparable in both groups (data not shown), the observed differences are at the level of binding/uptake). We confirmed that in the case of α-syn, this reduced CMA activity favored the formation of the oligomeric complex, similar to what we observed in the in vitro experiments (Fig. 8c). In fact, not only the exogenously added α-syn, but also endogenous α-syn could also be found associated to lysosomes from G/S-LRRK2 mice and it organized there as irreversible oligomeric complexes (Fig. 8d).
We also found a significant increase in the colocalization of α-syn with lysosomal markers (total LAMP2 show in Fig. 8e,f) in neuronal cultures from brains of WT and G/S-LRRK2 transgenic mice when compared to neurons from control mice. This enhanced association of α-syn to lysosomes is a consequence of its delivery to this compartment by CMA because knock-down of LAMP-2A in these neurons reduced colocalization of α-syn with lysosomes (Fig. 8e,f). None of these manipulations changed the already minimal colocalization between LRRK2 and LC3 (Supplementary Fig. 10a). A similar increase in intracellular levels of α-syn and in the association of this protein with lysosomes was observed in primary neuronal cultures from R1441G-LRRK2 (R/G) transgenic mice (Supplementary Fig. 10b), which could be responsible for the reduced CMA activity observed in these cells using the CMA fluorescent reporter (Supplementary Fig. 10c).
Lastly, to determine if a similar association of α-syn with lysosomes is also present in neurons from PD patients with the G/S-LRRK2 mutation, we differentiated dopaminergic neurons from induced pluripotent stem cells (iPSC) from PD patients associated with the LRRK2 G2019S mutation, where we have previously shown that α-syn abnormally accumulates44. After 30 days of differentiation, a time when patients’ iPSC derived neurons do not show overt morphological signs of neurodegeneration44, only 13% of TH-positive neurons displayed detectable amounts of α-syn in control iPSC-derived dopaminergic neurons, and only a small percentage (less than 10%) of these cells containing α-syn showed colocalization between α-syn and the lysosomal marker. In contrast, α-syn was detectable in 40% of the neurons derived from the PD patients and this protein colocalized with LAMP-2A in almost 65% of them (Fig. 8g,h). To address the contribution of changes in CMA activity to the abnormally high levels of α-syn in the patient cells, we knocked-down LAMP-2A in differentiated neurons at 3 weeks and 75 days, (when signs of neurodegeneration are already evident in the patients’ neurons) by lentiviral transduction of the GFP-tagged shRNA against LAMP-2A45. Knock-down of LAMP-2A at 3 weeks of differentiation markedly increased the percentage of α-syn -positive dopaminergic neurons both in control and in PD patient iPSC-derived neurons (Fig. 8i,j). Neurons from cultures infected with a control lentivirus, or including the fraction of neurons in the same cultures that remained untransduced did not show significant changes in their α-syn content (Fig. 8i shows an untransduced neuron). Notably, the increase in cells showing α-syn accumulation was significantly higher in the patient-derived cells when compared with control, suggesting a possible compensatory upregulation of CMA in early pre-symptomatic states (Fig. 8j). The accumulation of α-syn in PD patient-derived cells after CMA blockage was often associated with marked neurite shortening (Fig. 8i). Knock-down of LAMP-2A in cells cultured for 75 days, a condition in which patient-derived cells but not control-derived cells develop evident signs of neurodegeneration44, also increased α-syn content in both groups, but the proportional increase compared to untransduced cells was relatively lower in the patient cells, likely indicating an already compromised CMA of α-syn in these cells (Fig 8i,j).
Overall, our findings support the conclusion that LRRK2 at lysosomes not only blocks the degradation of α-syn by CMA, but also that the presence of mutations in only one of these proteins is sufficient to elicit the CMA toxic effect of the other. We propose that the enhanced binding of LRRK2 to the lysosomal membrane in the presence of CMA substrates may serve to self-perpetuate the negative effect of LRRK2 on CMA. The observed LRRK2-mediated inhibition at the level of translocation but not binding is particularly detrimental for pathogenic proteins that tend to oligomerize into toxic species, as shown here for α-syn (see model in Supplementary Fig. 11).
Discussion
We have found that the PD associated protein LRRK2 can undergo degradation in lysosomes via CMA and that this degradation is compromised for the most common LRRK2 mutant (G2019S) and additional pathogenic mutations of this protein. LRRK2 is the first CMA substrate identified for which binding to the lysosomal membrane is enhanced by other CMA substrates, and this enhanced binding inhibits the assembly of the CMA translocation complex at the lysosomal membrane. Cells respond to this blockage by increasing levels of LAMP-2A, consistent with the high levels of LAMP-2A that we observed in brains of PD patients with the G2019S LRRK2 mutation. Our studies in neuronal cell lines, primary neuronal mouse cultures, lysosomes from different LRRK2 transgenic mouse models and in iPSC-derived dopaminergic neurons from PD patients with the G/S-LRRK2 mutation, support the conclusion that LRRK2-mediated blockage of LAMP-2A multimerization leads to accumulation of other CMA substrates, including α-syn, at the surface of the lysosomal membrane by increasing the time that substrate proteins remain bound to the membrane prior to translocation. Thus, two dominant mutations that cause PD converge at the same step, leading to blockage of degradation of CMA substrates, which can contribute to the accumulation of neuronal α-syn widely thought to underlie PD.
A surprising finding of our study is that the toxic effect of LRRK2 on CMA was manifest not only for mutant forms of LRRK2 but also for WT-LRRK2 both in vitro and in intact cells when present at high concentrations, highlighting the need for a tight control of intracellular LRRK2 levels. This low threshold of tolerance for LRRK2 levels could result, in part, from its peculiar interplay with CMA. LRRK2 is, in all aspects, a rather unconventional CMA substrate, which rather than competing for lysosomal membrane binding with other CMA substrates, displays an unusually enhanced membrane binding. Further analysis of the peculiarities of LRRK2 in the context of CMA may reveal the existence of a different subgroup of CMA substrates depending on this cooperative binding for their internalization into lysosomes. As for most intracellular proteins, multiple pathways contribute to the degradation of LRRK2, making it likely that posttranslational modifications in LRRK2 determine its degradative fate. Ubiquitinization of LRRK2 by CHIP and its interaction with hsp90 have been shown to determine its degradation by the proteasomal system27. Whether specific modifications in LRRK2 modulate its CMA targeting requires future investigation. Functional association of LRRK2 with other cellular membranes has been shown to depend on its GTPase activity and phosphorylation status46. While we did not find LRRK2 binding to the lysosomal membrane to be dependent on GTP, changes in the kinase activity of LRRK2 may modulate its binding to the lysosomal compartment. Indeed, we found that changes in the kinase activity of the protein may have a different impact on its lysosomal binding depending on whether it is in the wild-type or mutant background and in the presence of CMA substrates at the membrane. Thus, a kinase-dead form of the protein displays reduced binding to lysosomes, but its binding is still enhanced in the presence of CMA substrates, indicating that kinase activity per se is not required for lysosomal binding once CMA substrate are added. In contrast, modulation of the kinase activity in the background of a mutation in LRRK2 (i.e. G/S-LRRK2 shown in Fig. 5b) had a marked impact on its binding to lysosomes in the presence of substrate proteins. It is possible that binding of LRRK2 to its substrates or changes in its conformation in the presence of the inhibitors may modify its capability to interact with CMA substrates at the lysosomal membrane. Alternatively, LRRK2 could phosphorylate unidentified components at the lysosomal membrane and regulate its association to this organelle. A third possibility is that inhibition of the kinase activity may promote delivery of the no longer active protein for degradation and that the observed differences between the WT and G/S mutant in response to the treatment with kinase inhibitors reflects, the lower efficiency of uptake of the mutant protein and consequently higher membrane-bound levels.
The inhibitory effect of LRRK2 on CMA could be due to direct interaction with LAMP-2A, as mutant forms of LRRK2 engage in a more stable binding with LAMP-2A. Alternatively, LRRK2 may abnormally bind to CMA-substrates forming oligomeric structures that compromise the lateral mobility of LAMP-2A along the lysosomal membrane to form the translocation complex or the additional lysosomal binding of LRRK2 observed in the presence of a-syn could occur through oligomerization with molecules of this protein already bound to the receptor (Supplementary Fig. 11). Association of LRRK2 could thus prevent α-syn translocation, since only single unfolded proteins can be transported through CMA. Notably, we have not found among the described pathogenic mutations in LRRK2 any that affect the possible CMA targeting motifs. It is possible that, as described for α-syn24, mutations in the motifs may not lead to disease because they cannot be targeted to lysosomes by hsc70, preventing thus the CMA blockage.
The increase in levels of LAMP-2A in cells expressing LRRK2 and in brains from LRRK2 PD patients contrasts with the previously described decrease in levels of this protein in brains of idiopathic PD patients26. These differences may highlight the distinctive characteristics of the LRRK2-mediated blockage of CMA. Our previous data suggest that pathogenic forms of α-syn that bind tightly to the lysosomal membrane prevent further binding of other CMA substrates. In contrast, LRRK2, once bound to the lysosomal membrane, prevents LAMP-2A multimerization, and consequently, translocation of substrate proteins, but does not interfere with their binding to this membrane. It is possible that the higher levels of LAMP-2A observed in the G2019S LRRK2 mutant PD brains result from a chronic decrease in the turnover of this receptor that we have previously shown is inhibited upon binding of substrates to LAMP-2A47. Alternatively, the increase in LAMP-2A in these patients could be a compensatory response to the CMA blockage, as previously described in the brain of chemically and genetically-induced PD mouse models35. This last possibility is also supported by the higher impact that the knock-down of LAMP-2A has in the PD patient iPSC-derived neuronal cells before signs of neurodegeneration become manifest, suggesting an initial compensatory upregulation of CMA. In this respect, the use of the iPSC-derived neuronal cells is particularly advantageous as it not only allows for the study of the process in the relevant cell type expressing endogenous levels of the mutant protein, but in addition, it permits following the process of neurodegeneration from an initially healthy differentiated neuron. Our analysis of the iPSC-derived dopaminergic neurons from PD patients with the G/S-LRRK2 mutation indicates that alterations in CMA are an early event, detectable before overt neurodegeneration and the previously described compromise of macroautophagy in these cells44.
CMA is a common target for three of the four mutations analyzed in this study (see summary in Supplementary Fig. 11c). Although there are some discrete differences among the three mutants that have a toxic effect on CMA, all of them have also revealed themselves as poor substrates for this pathway when compared to the WT protein. Their reduced lysosomal uptake and augmented binding to the lysosomal membrane in the presence of CMA substrates, are likely important determinants of their inhibitory effect on this pathway. Notably, the mutant that did not inhibit CMA is the one that revealed a much more pronounced swelling of the lysosomal endocytic system, a phenotype recently described in flies expressing a LRRK2 homolog34. Future studies with additional mutant variants (as they are still being identified) may help to separate them into groups depending on the primary target of their toxicity in the lysosomal system, and thus help in a more efficient design of therapeutic interventions.
As both LRRK2 and α-syn are degraded by CMA, and pathogenic mutant forms of both impair CMA, we suggest that alterations in this autophagic pathway could play an important role in the pathogenesis of PD7, 8. The manners by which mutations in LRRK2 and α-syn affect CMA differ in important aspects. First, whereas mutant α-syn proteins have a higher lysosome-binding affinity and low rates of lysosomal translocation compared to the wild-type protein8, the mutant LRRK2 analyzed in this study shows a more discrete, yet significant, decrease in its lysosomal uptake compared to the wild-type protein, and does not display abnormally enhanced binding to the lysosomal membrane when presented alone to these organelles. Importantly, however, both wild-type and even more so mutant LRRK2, display a marked increase in their association with lysosomes when in the presence of other CMA substrate proteins, which was not seen for α-syn. This enhanced binding of LRRK2 can inhibit the CMA transporter, and we show here that this blockage promotes the formation of α-syn oligomers at the lysosomal membrane, even for the wild-type form of α-syn, which is further toxic for CMA activity and could underlie the formation of Lewy body pathology in LRRK2 mutant PD patients. Considering the toxic effect on CMA described in this study for the most common pathogenic mutant forms of LRRK2, interventions aimed at enhancing CMA activity or at preventing its decrease in pathologies and with age may prove to be valuable in the treatment of PD and other age-related disorders resulting from alterations in cellular proteostasis.
Online Methods
Animals, cells and human brain tissue
Adult (4 months old) male Wistar rats and male (3–5 months old) mice BAC transgenic for WT, G2019S and R1441G LRRK2 or null for LRRK2 generated as described before48 were from Jackson Laboratory and were utilized under animal study protocols approved by the Institutional Animal Care and Use Committee of Albert Einstein College of Medicine and Columbia University. Rats (3/cage) and mice (5/cage) were maintained in a 12h light/dark cycle. Where indicated, rodents were starved for 48 hours to maximally activate CMA. SH-SY5Y cells were from the ATCC. HEK293 cells expressing WT or the various LRRK2 mutations under the control of tetracycline were generated as described before31. Plasmids for myc- and GFP-tagged full length or truncated LRRK2 (RCK, ROC, K, COR) were obtained from Addgene40, 41 Cells were maintained in Dulbecco's modified Eagle's medium (Sigma, St. Louis, MO) in the presence of 10% fetal bovine serum or newborn calf serum (Sigma), 50 μg/ml penicillin, and 50 μg/ml streptomycin at 37°C with 5% CO2. Ventral midbrain dopaminergic neurons were derived from postnatal day 1 pups and were prepared as described49. Previously generated lines of iPSC SP-11.1 and SP-17.2 (from control) and SP-05.1 and SP-12.3 (from familial PD patients with the LRRK2 G2019S mutation) were used and culture and differentiation were carried out as described44, following a protocol approved by the Spanish competent authorities (Commission on Guarantees concerning the Donation and Use of Human Tissues and Cells of the Carlos III Health Institute). Frozen and formalin fixed brain tissue from the dorsal motor nucleus of the vagus nerve (DMV) was obtained from the New York Brain Bank at Columbia University, and deidentified, under a protocol approved by the Columbia University Institutional Review Board. Brain tissue was available from two cases with known LRRK2 G2019S mutations (case 1: female, 80-years-old; case 2: male, 60-years-old) and two approximately age matched normal controls, with normal neuropathological findings (control 1: female, 66-years-old; control 2: female, 83-years-old)50. The DMV was the only available region from the original brain samples know to be targeted in PD.
Chemicals
Sources of chemicals and antibodies were as described before20, 21. 3-methyladenine (3MA), ammonium chloride and MG-132 were from Sigma, lactacystin was from Enzo Life Sciences, and leupeptin was from Calbiochem. The antibody against the cytosolic tail of rat and mouse LAMP-2A was prepared in our laboratory20 and the antibodies (clone, dilution) against total LAMP-2 (H4B4 (human, 1:3000) and GL27B (mouse, 1:100-500), LAMP-1 (H4A3 (human, 1:100-3000)) and anti-myc (9E10, 1:1000) were from the Developmental Hybridoma Bank (University of Iowa, Iowa City, Iowa). The antibody against mouse LAMP-1 (Ly1C6, 1:1000) was from Stressgen, against human LAMP-2A (18528, 1:200-500) from Abcam, against LRRK2 from the Michael J Fox Foundation (C5, 1:100-1000), Epitomics (3515-1, 1:50-1000), Everest (EB06550, 1:100-1000) and Abcam (AB38003, 1:100-1000), against actin (S12-I, 1:10000) and hsp90 (16F1, 1:10000) from Stressgen, against GAPDH (AB8245, 1:3000) and cylophilin A (AB41684, 1:1000) from Abcam, against tyrosine hydroxylase from Millipore (MAB318, 1:200), against flotillin-1 (18/f-1, 1:3000) and a-syn (42/a-s, 1:500) from BD Biosciences, against RNase A (100-4188, 1:3000) from Rockland Immunochemicals, against hsc70 (13D3, 1:5000) from Novus, against GFP from Roche (7.1, 1:1000) and Aves Labs (1020; 1:1000), against GST (1H14L28, 1:1000) and V5 (R960-25, 1:1000) from Invitrogen, against MAP2 from Sigma (AP-20, 1:250), and against LC3 (2775S, 1:100-1000) from Cell Signaling. Lysotracker and the purified WT and various mutant LRRK2 proteins were from Invitrogen. WT and mutant α-syn were purified as described before7. Cyclophilin A was from Abcam. GAPDH, RNaseA, ovalbumin, GTP and the kinase inhibitor Su 6656 were from Sigma.
Lysosomal isolation and lysosomal subfractionation
Lysosomes with high activity for CMA were isolated by centrifugation of a light mitochondrial-lysosomal fraction from rodent liver or brain in a discontinuous metrizamide density gradient21,29. Preparations with more than 10% broken lysosomes at the moment of the isolation, measured by β-hexosaminidase latency51, were discarded. Where indicated, lysosomal membrane proteins were removed by incubation with the indicated concentrations of trypsin at room temperature.
Intracellular protein turnover
To measure degradation of long-lived proteins, confluent cells were labeled with [3H]leucine (2 μCi/ml) for 48 h at 37°C and then extensively washed and maintained in complete (10% FBS) or serum-deprived media containing an excess of unlabeled leucine (2.8 mM), to prevent reutilization of radiolabeled leucine45. Aliquots of the media taken at different times were precipitated with TCA and proteolysis was measured as the percentage of the initial acid-insoluble radioactivity (protein) transformed into acid-soluble radioactivity (amino acids and small peptides) at the end of the incubation. Total radioactivity incorporated into cellular proteins was determined as the amount of acid-precipitable radioactivity in labeled cells immediately after washing29. Cell viability was routinely monitored in parallel cells at the end of the experiment and we did not find significant differences in viability between control cells and those expressing LRRK2 proteins during the duration of the assay.
Degradation of substrate proteins by intact lysosomes in vitro
Lysosomes isolated from rat livers were incubated with radiolabeled proteins in 3-(N-morpholino) propanesulfonic acid (MOPS) buffer (10 mM MOPS, pH 7.4, 0.3 M sucrose, 1 mM dithiothreitol, and 5.4 µM cysteine) for 30 min at 37°C29. Reactions were stopped with 20% TCA and filtered through a Millipore Multiscreen Assay System (Millipore, Billerica, MA) and detected in a liquid scintillation analyzer (PerkinElmer Wallac, Gaithersburg, MD). Proteolysis was measured as the percentage of the initial acid-insoluble radioactivity transformed into acid-soluble radioactivity as described above.
Binding and uptake of CMA substrate proteins by isolated lysosomes
Freshly isolated intact lysosomes were incubated with the substrate protein in MOPS buffer at 37°C for 20 min29. Where indicated, lysosomes were preincubated with a cocktail of protease inhibitors for 10 min at 0°C as described before29. Lysosomes were collected by centrifugation, washed with MOPS buffer, and subjected to SDS-PAGE and immunoblotted for the utilized substrate protein. Binding was calculated from the densitometric analysis as the amount of substrate protein bound to the lysosomal membrane in the absence of protease inhibitors. Uptake was calculated by subtracting the amount of protein associated with lysosomes in the presence (protein bound to the lysosomal membrane and taken up by lysosomes) and absence (protein bound to the lysosomal membrane) of protease inhibitors.
Measurement of CMA activity in intact cells
HEK293 cell lines and primary mouse neurons in culture (ventral midbrain dopaminergic neurons at DIV1) were transduced with lentivirus carrying the CMA reporter KFERQ-PA-mCherry133. Cells were photoactivated in the presence of oxyrase and lactic acid 48 h post-infection (in non-neuronal cells) or 30min to 5 h post-infection (in neuronal cells) by a 405 nm light emitting diode (LED: Norlux) for 9 min (non neuronal) or 3 min (neuronal) with the intensity of 3.5mA (current constant). Levels of red fluorescence were checked under the microscope 1 hour post-photoactivation to confirm photoactivation and compare levels of expression of the reporter. We did not find visible differences in expression levels of the reporter between cells expressing or not LRRK2 proteins. After 16 h (non neuronal) or 0.5–6 h (neuronal), cells were fixed with 4% paraformaldehyde and images were acquired with an Axiovert 200 fluorescence microscope (Carl Zeiss Ltd., Thornwood, NY), equipped with an apotome or in a Leica Multiphoton microscope (in neuronal cell studies). Images were prepared using Adobe Photoshop 6.0 software (Adobe Systems Inc., Mountain View, CA). CMA activity is determined in this system as a change in the fluorescent pattern from diffuse (cytosolic) to punctate (lysosomal). The number of fluorescent puncta per cell was quantified using Image J software (NIH) in individual frames after thresholding.
LAMP-2A dynamics
Multimerization of LAMP-2A at the lysosomal membrane was studied on 3–12% NativePAGE Bis Tris Gels (Invitrogen) after solubilization in 1% octylglucoside (in 20 mM MOPS and 150 mM NaCl buffer)21. Lysosomal membrane microdomains were isolated after detergent solubilization through floatation in a sucrose density gradient as described previously23.
Immunofluorescence and immunohistochemistry
Non neuronal cells: Cells grown on coverslips were fixed with 4% paraformaldehyde (PFA) or methanol, blocked, and then incubated with the primary and corresponding fluorophore-conjugated secondary antibodies23. Mounting medium contained DAPI (4',6– diamidino-2-phenylindole) to highlight the cellular nucleus. Neuronal cultures: Ventral midbrain dopaminergic neuron cultures were derived from postnatal day 1 pups. Neurons were transduced into dopaminergic neurons at DIV1 and fixed at DIV7 with 4% PFA with 4% glucose in PBS for 2 minutes at room temperature and 10 min with −20°C methanol. Cultures were blocked with 10% donkey serum with 0.2% saponin for 1 hour before immunostaining. iPSC-derived neurons: iPSC-derived cells were fixed with 4% PFA in PBS at 4°C for 10 min and blocked in 0.3% of Triton-X100 with 3% donkey serum for 2 h. Images were taken using a Leica SP5 confocal microscope. For quantification analyses, a minimum of 3 fields per condition were randomly selected and counted. Data points represent the average of at least two independent experiments. Human brains: Formalin fixed brain tissues were paraffin embedded and sectioned for immunohistochemistry analysis. Brains were chilled 2– 4 h post mortem and frozen 6–30 h post mortem. The two LRKK2 G2019S brains were rated at a grade of neuropathological Braak & Braak staging of 4/6. Tissue blocks of the medulla were sectioned at 7µm thickness. Antigen retrieval was performed with Trilogy (Cell Marque) in a vegetable steamer for 40 minutes. Immunostaining for the desired proteins followed the standard procedures35. Sections were imaged and neurons were selected randomly and fields of 75 × 75μm were photographed in a confocal fluorescent microscope (Leica). The micrographs were analyzed for the number of fluorescent puncta per neuronal cell using the Image J software (NIH) after thresholding. Colocalization was quantified using the JACop-pluggin in the same software and Costes’ automatic thresholding to calculate the Pearson’s coefficient.
RNA interference
For downregulation of LAMP-2A, we used previously described lentiviral vectors carrying shRNA against LAMP-2A37,45 packed in 293T cells and purified by ultra-centrifugation as previously described37, 45. Expression titers were 5×108 to 1×109 transducing units×ml−1. For transduction: 1) cultured non-neuronal cells and ventral midbrain dopaminergic neurons at DIV1p were incubated with the packed virus for 12h and the placed in fresh media; 2) iPSC were grown on coverslips and incubated after 3 or 9 weeks of differentiation with 1 μl of concentrated lentivirus in a final volume of 1 ml for 24 h at 37°C, and with an additional 1ml of fresh media for 48 h. GFP positive cells started to appear 3 days after infection. Cells were fixed and analyzed 7–10 days after infection.
LC3 Flux
Macroautophagy activity was quantified as the degradation of the autophagosome-associated protein LC3. Cells were untreated or treated with 20 mM NH4Cl and 100 µM leupeptin for 6 hrs followed by immunoblot against LC3. Flux was calculated as the ratio of treated to untreated lines after normalization to actin52.
General procedures
Protein concentration was determined using the Lowry method utilizing bovine serum album as a standard53. Cells were solubilized on ice with RIPA buffer (1% Triton-X 100, 1% sodium deoxycholate, 0.1% SDS, 0.15 M NaCl, 0.01 M sodium phosphate, pH 7.2). Immunoblotting was performed after transferring SDS-PAGE gels on nitrocellulose membranes54. These proteins were visualized by chemiluminescence using peroxidase conjugated secondary antibodies in a LAS-3000 Imaging System (Fujifilm). Image J. Software (NIH) was used for densitometric quantification. Co-immunoprecipitation was performed after solubilization of cellular extracts in a low astringency buffer as described before8. Immunoblots shown in the figures are representative of at least 3 different experiments.
Statistical analysis
All numerical results are reported as mean + S.E., and represent data from a minimum of three independent experiments unless otherwise stated. Statistical significance of difference between groups was determined in instances of single comparisons by the two-tailed unpaired Student’s t-test of the means. In instances of multiple means comparisons, we used one-way analysis of variance (ANOVA) followed by the Bonferroni post-hoc test to determine statistical significance. Statistic analysis was performed in all the assays and significant differences are noted in the graphical representations.
Supplementary Material
Acknowledgements
We thank the Michael J. Fox Foundation for kindly providing some of the LRRK2 antibodies, C. Calatayud for help with neuronal differentiation of iPSC, D. Arduino and B. Caballero for their help with the generation of the LRRK2 knockdown cells, J.P. Vonsattel and R. Alcalay for assistance with human tissue and A. Diaz and B. Patel for technical assistance in several aspects of this work. This work was supported by grants from the US National Institutes of Health National Institute on Aging (AG031782 and AG038072 (A.M.C.) and AG08702 (Alzheimer's Disease Research Center at Columbia University), MINECO (SAF2012-33526, PLE2009-0144 and ACI2010-1117 (A.R.), RyC-2008-02772 and BFU2010-21823 (A.C.), the Beatrice and Roy Backus Foundation , the Rainwaters Foundation and a Robert and Renee Belfer gift (A.M.C.), the JPB Foundation (D.S.), the Parkinson's Disease Foundation (D.S. and S.-H.K.), Fondazione Guido Berlucchi (A.C.), a CIBERNED Cooperative Project (A.R.), a US National Institute of Neurological Disorders and Stroke Udall Center of Excellence (A.M.C. and D.S.) and a Hirschl/Weill-Caulier Career Scientist Award (A.M.C.). S.J.O. was supported by US National Institutes of Health National Institute on Aging training grant T32AG023475 and S.-H.K. by an American Academy of Neurology Research Fellowship.
Abbreviations
- α-syn
alpha synuclein
- CMA
chaperone-mediated autophagy
- hsc
heat shock cognate protein
- hsp
heat shock protein
- iPSC
induced pluripotent stem cells
- LAMP
lysosome-associated membrane protein
- LRRK2
leucine-rich repeat kinase 2
- PD
Parkinson’s disease
Footnotes
Conflict of interest: Authors declare no conflict of interest
Author Contributions
SJO performed most of the experiments, analyzed data, and prepared a draft of the manuscript; SHK analyzed the human brain samples and conducted some of the experiments on mouse neuronal cultures; IT performed some studies in isolated rat liver and mouse, pull-down experiments and some analysis of the kinase inhibitors; EA performed studies in neuronal cultures; HK assisted with the CMA reporter analysis; IF-C, AC and AR were responsible for all the studies with differentiated iPSC; EC and LSH provided the brain samples; WD provided the LRRK2 expressing cells and advice on some aspects of the project; DS critically discussed the results, directed experiments in neuronal culture and human brain and edited and reviewed the final version of the manuscript; AMC directed the study, designed most of the experiments, and edited and reviewed the final version of the manuscript.
References
- 1.Spillantini M, et al. Alpha-synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 2.West AB, et al. Parkinson's disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc. Natl. Acad. Sci. U S A. 2005;102:16842–16847. doi: 10.1073/pnas.0507360102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 4.Anglade P, et al. Apoptosis and autophagy in nigral neurons of patients with Parkinson's disease. Histol Histopathol. 1997;12:25–31. [PubMed] [Google Scholar]
- 5.Zhu JH, Guo F, Shelburne J, Watkins S, Chu CT. Localization of phosphorylated ERK/MAP kinases to mitochondria and autophagosomes in Lewy body diseases. Brain Pathol. 2003;13:473–481. doi: 10.1111/j.1750-3639.2003.tb00478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stefanis L, Larsen K, Rideout H, Sulzer D, Greene L. Expression of A53T mutant but not wild-type alpha-synuclein in PC12 cells induces alterations of the ubiquitin-dependent degradation system, loss of dopamine release, and autophagic cell death. J. Neurosci. 2001;21:9549–9560. doi: 10.1523/JNEUROSCI.21-24-09549.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martinez-Vicente M, et al. Dopamine-modified alpha-synuclein blocks chaperone-mediated autophagy. J Clin Invest. 2008;118:777–788. doi: 10.1172/JCI32806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science. 2004;305:1292–1295. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- 9.Mizushima N, Levine B, Cuervo A, Klionsky D. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gomez-Suaga P, et al. Leucine-rich repeat kinase 2 regulates autophagy through a calcium-dependent pathway involving NAADP. Hum. Mol. Genet. 2011 doi: 10.1093/hmg/ddr481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biskup S, et al. Localization of LRRK2 to membranous and vesicular structures in mammalian brain. Ann Neurol. 2006;60:557–569. doi: 10.1002/ana.21019. [DOI] [PubMed] [Google Scholar]
- 12.Alegre-Abarrategui J, et al. LRRK2 regulates autophagic activity and localizes to specific membrane microdomains in a novel human genomic reporter cellular model. Hum. Mol. Genet. 2009;18:4022–4034. doi: 10.1093/hmg/ddp346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramonet D, et al. Dopaminergic neuronal loss, reduced neurite complexity and autophagic abnormalities in transgenic mice expressing G2019S mutant LRRK2. PLoS One. 2011;6:e18568. doi: 10.1371/journal.pone.0018568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plowey ED, Cherra SJ, 3rd, Liu YJ, Chu CT. Role of autophagy in G2019S-LRRK2-associated neurite shortening in differentiated SH-SY5Y cells. J Neurochem. 2008;105:1048–1056. doi: 10.1111/j.1471-4159.2008.05217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tong Y, et al. Loss of leucine-rich repeat kinase 2 causes impairment of protein degradation pathways, accumulation of alpha-synuclein, and apoptotic cell death in aged mice. Proc. Nat.l Acad. Sci. U S A. 2010;107:9879–9884. doi: 10.1073/pnas.1004676107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orenstein SJ, Cuervo AM. Chaperone-mediated autophagy: Molecular mechanisms and physiological relevance. Semin Cell Dev Biol. 2010;21:719–726. doi: 10.1016/j.semcdb.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cuervo AM. Chaperone-mediated autophagy: selectivity pays off. Trends Endocrin Met. 2010;21:142–150. doi: 10.1016/j.tem.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiang HL, Dice JF. Peptide sequences that target proteins for enhanced degradation during serum withdrawal. The Journal of biological chemistry. 1988;263:6797–6805. [PubMed] [Google Scholar]
- 19.Chiang HL, Terlecky SR, Plant CP, Dice JF. A role for a 70-kilodalton heat shock protein in lysosomal degradation of intracellular proteins. Science. 1989;246:382–385. doi: 10.1126/science.2799391. [DOI] [PubMed] [Google Scholar]
- 20.Cuervo AM, Dice J. A receptor for the selective uptake and degradation of proteins by lysosomes. Science. 1996;273:501–503. doi: 10.1126/science.273.5274.501. [DOI] [PubMed] [Google Scholar]
- 21.Bandyopadhyay U, Kaushik S, Vartikovsky L, Cuervo AM. Dynamic organization of the receptor for chaperone-mediated autophagy at the lysosomal membrane. Mol. Cell Biol. 2008;28:5747–5763. doi: 10.1128/MCB.02070-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cuervo AM, Mann L, Bonten E, d'Azzo A, Dice J. Cathepsin A regulates chaperone-mediated autophagy through cleavage of the lysosomal receptor. EMBO J. 2003;22:12–19. doi: 10.1093/emboj/cdg002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaushik S, Massey AC, Cuervo AM. Lysosome membrane lipid microdomains: novel regulators of chaperone-mediated autophagy. EMBO J. 2006;25:3921–3933. doi: 10.1038/sj.emboj.7601283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vogiatzi T, Xilouri M, Vekrellis K, Stefanis L. Wild type alpha-synuclein is degraded by chaperone-mediated autophagy and macroautophagy in neuronal cells. J. Biol. Chem. 2008;283:23542–23556. doi: 10.1074/jbc.M801992200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kabuta T, Furuta A, Aoki S, Furuta K, Wada K. Aberrant interaction between Parkinson disease-associated mutant UCH-L1 and the lysosomal receptor for chaperone-mediated autophagy. J. Biol. Chem. 2008;283:23731–23738. doi: 10.1074/jbc.M801918200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alvarez-Erviti L, et al. Chaperone-mediated autophagy markers in Parkinson disease brains. Arch Neurol. 2010;67:1464–1472. doi: 10.1001/archneurol.2010.198. [DOI] [PubMed] [Google Scholar]
- 27.Ko HS, et al. CHIP regulates leucine-rich repeat kinase-2 ubiquitination, degradation, and toxicity. Proc. Natl. Acad Sci. U S A. 2009;106:2897–2902. doi: 10.1073/pnas.0810123106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dice J. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem Sci. 1990;15:305–309. doi: 10.1016/0968-0004(90)90019-8. [DOI] [PubMed] [Google Scholar]
- 29.Kaushik S, Cuervo AM. Methods to monitor chaperone-mediated autophagy. Methods Enzymol. 2009;452:297–324. doi: 10.1016/S0076-6879(08)03619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cuervo AM, Terlecky SR, Dice JF, Knecht E. Selective binding and uptake of ribonuclease A and glyceraldehyde-3-phosphate dehydrogenase by isolated rat liver lysosomes. The Journal of biological chemistry. 1994;269:26374–26380. [PubMed] [Google Scholar]
- 31.Kett LR, et al. LRRK2 Parkinson disease mutations enhance its microtubule association. Hum. Mol. Genet. 2012;21:890–899. doi: 10.1093/hmg/ddr526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanida I, Minematsu-Ikeguchi N, Ueno T, Kominami E. Lysosomal Turnover, but Not a Cellular Level, of Endogenous LC3 is a Marker for Autophagy. Autophagy. 2005;1:84–91. doi: 10.4161/auto.1.2.1697. [DOI] [PubMed] [Google Scholar]
- 33.Koga H, Martinez-Vicente M, Macian F, Verkhusha VV, Cuervo AM. A photoconvertible fluorescent reporter to track chaperone-mediated autophagy. Nat Commun. 2011;2:386. doi: 10.1038/ncomms1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dodson MW, Zhang T, Jiang C, Chen S, Guo M. Roles of the Drosophila LRRK2 homolog in Rab7-dependent lysosomal positioning. Hum. Mol. Genet. 2011 doi: 10.1093/hmg/ddr573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mak SK, McCormack AL, Manning-Bog AB, Cuervo AM, Di Monte DA. Lysosomal degradation of alpha-synuclein in vivo. J. Biol. Chem. 2010;285:13621–13629. doi: 10.1074/jbc.M109.074617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang C, Cuervo AM. Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nat Med. 2008;14:959–965. doi: 10.1038/nm.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Massey AC, Kaushik S, Sovak G, Kiffin R, Cuervo AM. Consequences of the selective blockage of chaperone-mediated autophagy. Proc Nat Acad Sci USA. 2006;103:5905–5910. doi: 10.1073/pnas.0507436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blake RA, et al. SU6656, a selective src family kinase inhibitor, used to probe growth factor signaling. Mol. Cell. Biol. 2000;20:9018–9027. doi: 10.1128/mcb.20.23.9018-9027.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y, et al. Tau fragmentation, aggregation and clearance: the dual role of lysosomal processing. Hum. Mol. Genet. 2009;18:4153–4170. doi: 10.1093/hmg/ddp367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Greggio E, et al. The Parkinson disease-associated leucine-rich repeat kinase 2 (LRRK2) is a dimer that undergoes intramolecular autophosphorylation. J. Biol. Chem. 2008;283:16906–16914. doi: 10.1074/jbc.M708718200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Greggio E, et al. Kinase activity is required for the toxic effects of mutant LRRK2/dardarin. Neurobiol. Dis. 2006;23:329–341. doi: 10.1016/j.nbd.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 42.Sulzer D, Surmeier DJ. Neuronal vulnerability, pathogenesis, and Parkinson's disease. Mov. Disord. 2012 doi: 10.1002/mds.25187. [DOI] [PubMed] [Google Scholar]
- 43.Sen S, Webber PJ, West AB. Dependence of leucine-rich repeat kinase 2 (LRRK2) kinase activity on dimerization. J. Biol. Chem. 2009;284:36346–36356. doi: 10.1074/jbc.M109.025437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanchez-Danes A, et al. Disease-specific phenotypes in dopamine neurons from human iPS-based models of genetic and sporadic Parkinson's disease. EMBO Mo.l Med. 2012;4:380–395. doi: 10.1002/emmm.201200215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kon M, et al. Chaperone-mediated autophagy is required for tumor growth. Sci. Transl. Med. 2011;3:109ra117. doi: 10.1126/scitranslmed.3003182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berger Z, Smith KA, Lavoie MJ. Membrane localization of LRRK2 is associated with increased formation of the highly active LRRK2 dimer and changes in its phosphorylation. Biochemistry. 2010;49:5511–5523. doi: 10.1021/bi100157u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cuervo A, Dice J. Regulation of lamp2a levels in the lysosomal membrane. Traffic. 2000;1:570–583. doi: 10.1034/j.1600-0854.2000.010707.x. [DOI] [PubMed] [Google Scholar]
- 48.Li Y, et al. Mutant LRRK2(R1441G) BAC transgenic mice recapitulate cardinal features of Parkinson's disease. Nat. Neurosci. 2009;12:826–828. doi: 10.1038/nn.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Petersen A, et al. Expanded CAG repeats in exon 1 of the Huntington's disease gene stimulate dopamine-mediated striatal neuron autophagy and degeneration. Hum. Mol. Genet. 2001;10:1243–1254. doi: 10.1093/hmg/10.12.1243. [DOI] [PubMed] [Google Scholar]
- 50.Poulopoulos M, et al. Clinical and Pathological Characteristics of LRRK2 G2019S Patients with PD. J Mol Neurosci. 2011 doi: 10.1007/s12031-011-9696-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Storrie B, Madden E, et al. Isolation of subcellular organelles. Meth Enzymol. 1990;182:203–225. doi: 10.1016/0076-6879(90)82018-w. [DOI] [PubMed] [Google Scholar]
- 52.Rubinsztein DC, et al. In search of an "autophagomometer". Autophagy. 2009;5:585–589. doi: 10.4161/auto.5.5.8823. [DOI] [PubMed] [Google Scholar]
- 53.Lowry CC, Kraeft NH, Hughes FA., Jr Blastomycosis of the lung. Am J Surg. 1951;81:676–679. doi: 10.1016/0002-9610(51)90161-4. [DOI] [PubMed] [Google Scholar]
- 54.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proceedings of the National Academy of Sciences of the United States of America. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.