Abstract
Long-term therapy with certain drugs, especially P450 inducing agents, confers an increased risk of osteomalacia that is attributed to vitamin D deficiency. Human CYP24A1, CYP3A4 and CYP27B1 catalyze the inactivation and activation of vitamin D and have been implicated in the adverse drug response. In this study, the inducibility of these enzymes and monohydroxylation of 25OHD3 were evaluated following exposure to P450 inducing drugs. With human hepatocytes, treatment with phenobarbital, hyperforin, carbamazepine and rifampin significantly increased the levels of CYP3A4 but not CYP24A1 or CYP27B1 mRNA. In addition, rifampin pretreatment resulted in an 8-fold increase in formation of the major metabolite of 25OHD3, 4β,25(OH)2D3. This inductive effect was blocked by the addition of 6′,7′-dihydroxybergamottin, a selective CYP3A4 inhibitor. With human renal proximal tubular HK-2 cells, treatment with the same inducers did not alter CYP3A4, CYP24A1 or CYP27B1 expression. 24R,25(OH)2D3 was the predominant monohydroxy metabolite produced from 25OHD3, but its formation was unaffected by the inducers. With healthy volunteers, the mean plasma concentration of 4β,25(OH)2D3 was increased 60% (p < 0.01) after short-term rifampin administration. This was accompanied by a statistically significant reduction in plasma 1α,25(OH)2D3 (−10%; p = 0.03), and a non-significant change in 24R,25(OH)2D3 (−8%; p = 0.09) levels. Further analysis revealed a negative correlation between the increase in 4β,25(OH)2D3 and decrease in 1α,25(OH)2D3 levels. Examination of the plasma monohydroxy metabolite/25OHD3 ratios indicated selective induction of the CYP3A4-dependent 4β-hydroxylation pathway of 25OHD3 elimination. These results suggest that induction of hepatic CYP3A4 may be important in the etiology of drug-induced osteomalacia.
Keywords: Cytochrome P450 3A4, Cytochrome P450 24A1, 25-Hydroxyvitamin D3, Pregnane X receptor, Osteomalacia
Introduction
Osteomalacia can be a serious and potentially debilitating side effect from certain drug therapies. It has been associated most strongly with chronic administration of anti-epileptic drugs, such as phenobarbital, carbamazepine, phenytoin and valproic acid (1,2), as well as the antimicrobial drug rifampin (3,4). Although the exact mechanism(s) behind the adverse effect remains unclear, the clinical characteristics of drug-induced osteomalacia resemble that of vitamin D deficiency (5,6). Thus, for some of the drugs implicated, they are thought to induce cytochrome P450-dependent vitamin D catabolism, reduce circulating vitamin D levels and alter the regulation of genes that encode calcium transport proteins in the small intestine (7–9). Decreased absorption of intestinal calcium can trigger compensatory parathyroid hormone-mediated responses that result in bone demineralization in order to maintain systemic calcium homeostasis.
There is a significant body of literature pointing towards the enhanced catabolism of vitamin D by drug treatments linked to osteomalacia. For example, daily administration of rifampin to healthy volunteers (for at least 2 weeks) lowered the plasma concentration of 25-hydroxyvitamin D3 (25OHD3) by 70% (4,10). Similar effects are described for patients receiving phenobarbital and phenytoin for the treatment of epilepsy (11). 25OHD3 is the dominant circulating form of vitamin D. In humans, it is found in plasma at concentrations that are nearly a thousand fold higher than that of the most biologically active vitamin D form, 1α,25-dihydroxyvitamin D3 [1α,25(OH)2D3], which is produced from 25OHD3 mainly in the kidneys. The gene regulatory effects of vitamin D in the body are thought to be mediated predominantly by 1α,25(OH)2D3 (12). Thus, a reduction in the circulating 25OHD3 pool through P450 induction might compromise the synthesis and availability of 1α,25(OH)2D3 to key target organs, such as the small intestine, bone and kidney. P450 induction could also enhance the catabolism of 1α,25(OH)2D3 and more directly affect calcium absorption and renal excretion, particularly if it were to occur within the local environment of target tissues (e.g., intestine and kidney) (8). Interestingly, the data are conflicting with regard to the effect of P450 inducers on plasma 1α,25(OH)2D3 levels, with some investigators reporting a reduction and others reporting no change (10,13–15). Also, there are no data available regarding tissue concentrations of 1α,25(OH)2D3 after treatment with P450 inducers.
The oxidative metabolism of 25OHD3 and 1α,25(OH)2D3 is predominantly catalyzed by a mitochondrial enzyme, CYP24A1 (16,17). These reactions are stereo- and regio-selective, resulting in the respective formation of principally 24R,25-dihydroxyvitamin D3 [24R,25(OH)2D3] and 1α,24R,25-trihydroxyvitamin D3. The primary metabolites can be oxidized further to yield more polar renal excretion products or undergo UDP-glucuronosyltransferase (UGT)-mediated conjugation (18,19). It has been suggested that induction of CYP24A1 in the liver and intestine through activation of human pregnane X receptor (PXR), and enhanced metabolic clearance of 25OHD3 and 1α,25(OH)2D3, is a contributor to drug-induced osteomalacia (7). Indeed, 24R,25(OH)2D3 is a major circulating metabolite of 25OHD3 (20) and thus an enhanced 24R,25(OH)2D3 formation clearance following drug treatment is certainly plausible.
Recently, we reported that both 25OHD3 and 1α,25(OH)2D3 are also substrates for CYP3A4, yielding primary products that are regio- and stereochemically distinct from those produced by CYP24A1 (8,21). CYP3A4 transcription can be greatly enhanced by activation of PXR. Moreover, the major CYP3A4-catalyzed metabolite of 25OHD3, 4β,25-dihydroxyvitamin D3 [4β,25(OH)2D3], also circulates in plasma, and results from two preliminary studies suggest that this metabolic pathway is selectively induced by the P450 inducer, rifampin, both in vitro and in vivo (21,22). In the current report, we describe the results of a more comprehensive investigation of the impact of P450 inducers that are also PXR agonists on the oxidative metabolism of 25OHD3 in healthy volunteers, primary human hepatocytes, and human renal proximal tubular cells (HK-2 cells). We focused on the formation of 24R,25(OH)2D3, 4β,25(OH)2D3, and 1α,25(OH)2D3, in order to distinguish between the competing processes of 25OHD3 elimination and their possible relationship to drug-induced osteomalacia.
Materials and Methods
Materials
Midazolam, 1′-hydroxymidazolam (1′-OH-MDZ), 25OHD3, 24R,25(OH)2D3, 6′7′-dihydroxybergamottin (DHB), 4-phenyl-1,2,4-triazoline-3,5-dione (PTAD) a n d β-glucuronidase from Helix pomatia were purchased from Sigma (St. Louis, MO). 1α,25(OH)2D3 was purchased from Calbiochem (San Diego, CA). 4β,25(OH)2D3 was isolated and purified using HPLC from enzymatic products and its concentration was assigned based on the ultraviolet absorbance and assuming a similar molar absorptivity as that of 1α,25(OH)2D3 (21). Deuterated d6-25OHD3 and d6-1α,25(OH)2D3 were purchased from Medical Isotope Inc. (Pelham, NH). Deuterated d4-midazolam and d4-1′-OH-MDZ were purchased from Cerilliant (Round Rock, TX). William’s E Medium and Dubelcco’s Modified Eagle’s Medium (DMEM) were purchased from Sigma-Aldrich. Human cryopreserved hepatocytes, cryopreserved hepatocyte recovery medium (CHRM), and media supplements were purchased from Invitrogen (Carlsbad, CA). Matrigel was purchased from BD Biosciences (San Jose, CA) and Kreb-Henseleit buffer (KHB) was from Celsis/In Vitro Technologies (Chicago, IL). Nuclease free water, MagMax 96 RNA isolation kit, High Capacity cDNA Transcription Kit, TaqMan primer and probe sets and all TaqMan reagents and consumables were purchased from Applied Biosystems (Foster City, CA).
Metabolism of 25OHD3 and gene expression in primary human hepatocytes
Cryopreserved human hepatocytes were thawed at 37 °C, sedimented at 100 g in CHRM for 10 min and resuspended in plating media (DMEM plus plating supplements: 5% fetal bovine serum, 100 nM dexamethasone, 100 U/mL penicillin and streptomycin, 4 μg/mL insulin, 2 mM GlutaMAX™, 15 mM HEPES, pH 7.4). Viability and density were measured by trypan blue exclusion and 52,000 cells/well were plated onto 96-well collagen I coated plates. Hepatocytes were allowed to attach for 4 to 6 hr, plating media was removed and replaced with maintenance media (William’s Medium E plus maintenance supplements: 100 U/mL penicillin and streptomycin, 6.25 μg/mL insulin, 6.25 μg/mL transferrin, 6.25 ng/mL selenous acid, 1.25 mg/mL bovine serum albumin, 5.35 μg/mL linoleic acid, 2 mM GlutaMAX™, 15 mM HEPES, pH 7.4) containing 0.25 mg/mL Matrigel. After plating and culturing of viable cells for 24 hr, the cells were treated with one of the following P450 inducers: rifampin (10 μM), hyperforin (0.5 μM), phenobarbital (400 μM), carbamazepine (50 μM), levetiracetam (200 μM), or the drug vehicle (0.1% DMSO or ethanol) in a 100 μL solution per well. After 48 hr, the medium was removed and the cells were washed twice with KHB solution. The cells were then incubated with the indicated concentrations of 25OHD3 for various incubation times (t = 2, 4, 8 or 24 hr). In parallel experiments, drug pretreated cells were incubated with midazolam (2 μM) for 30 min for the measurement of CYP3A4 activity. For some of these experiments, the cells were incubated with DHB (20 μM) or vehicle alone (0.1% ethanol) for 4 hr after pretreatment with the P450 inducing drugs. The culture medium was then replaced with fresh medium containing 25OHD3 in the presence or absence of DHB (20 μM) for various incubation times (t = 2, 4 or 24 hr), as indicated. At the end of treatment period, culture medium was collected and pooled for the quantification of monohydroxy metabolites of 25OHD3 by LC-MS/MS. The treated cells were harvested for isolation of mRNA and qRT-PCR analysis.
Evaluation of low dose 25OHD3 metabolism in primary human hepatocytes
Cryopreserved human hepatocytes from three different donors were obtained, prepared and subjected to in vitro cell culture, as described above. After culturing for 24 hr, the cells were treated with rifampin (10 μM) or vehicle (0.1% DMSO) for 48 hr and then incubated with 25OHD3 (50 nM) for various incubation times. In parallel, cells were pretreated with DHB (20 μM) for 4 hr and then co-incubated at the same concentration with 25OHD3. Medium was collected and incubated with β-glucuronidase (1500 U/mL) in 50 mM acetate buffer (pH 5.0) at 37 ° for 2 hr, extracted by liquid-liquid extraction and analyzed for 4β,25(OH)2D3 and 4α,25(OH)2D3 using LC-MS/MS as described previously. Results from a pilot experiment showed that unconjugated 4β,25(OH)2D3 and 4α,25(OH)2D3 concentrations at the end of all incubation periods (2 – 24 hr) were below the limit of quantitation. Thus, the amounts of primary metabolites released after β-glucuronidase treatment represent the levels of the corresponding conjugates in the culture medium.
Metabolism of 25OHD3 and gene expression in human renal proximal tubular HK-2 cells
The HK-2 cell line was obtained from the American Type Culture Collection (ATCC; Manassas, VA). It retains morphological and biochemical characteristics consistent with those of human renal proximal tubular cells (23). The cells were cultured according to ATCC guidelines in keratinocyte medium supplemented with 5 ng/mL human recombinant epidermal growth factor and 50 μg/mL bovine pituitary extract (Invitrogen). After reaching 80% confluence, the cells were de-attached by 0.25% trypsin solution and then seeded onto 96-well plates (30,000 cells/well) for 24 hr. After culturing for 24 hr, the cells were treated with the panel of inducers: rifampin (10 μM), hyperforin (0.5 μM), phenobarbital (400 μM), carbamazepine (50 μM), and levetiracetam (200 μM) or its vehicle (0.1% v/v) for 48 hr. The medium was removed and the cells were washed with phosphate buffered saline and then harvested for isolation of mRNA for qRT-PCR analysis. In another experiment, the cells were treated with 25OHD3 (2 μM) for 4 hrs after pretreatment with rifampin (10 μM) for 48 hr and the cell media were collected for the measurement of monohydroxy metabolites of 25OHD3 using LC-MS/MS.
Analysis of mRNA levels by qRT-PCR
Immediately following activity assays and saline washing, the cells were lysed and RNA was stabilized with 140 μL of Ambion Lysis/Binding Buffer included in the MagMax 96 RNA Isolation Kit. Cell lysates were either frozen at −70 °C (no longer than one week) or processed immediately. Total RNA was isolated using the MagMax 96 RNA isolation kit according to manufacturer’s protocol, and RNA quantity was assessed using the NanoDrop spectrophotometer. The cDNA was synthesized using the High Capacity cDNA Reverse Transcription kit with a final volume of 20 μL with approximately 100 ng total RNA according to manufacturer’s protocol. After synthesis, the cDNA reactions were diluted to 80 μL total volume with nuclease free water. TaqMan reactions were run on a 7900HT Real-time PCR system in a 384-well optical reaction plate. Each reaction contained 10 μL 2X Gene Expression Master Mix, 5 μL nuclease free water, 1 μL 20X Primer and Probe mix, and 4 μL of cDNA. Probe IDs for the TaqMan assays were as follows: CYP3A4, Hs00604506_m1; CYP24A1, Hs00167999_m1; CYP27A1, Hs01026016_m1; CYP27B1, Hs00168017_m1; CYP2R1, Hs01379776_m1; PXR, Hs01114267_m1; VDR, Hs00172113_m1.
LC-MS/MS analysis of 25OHD3 and its primary monohydroxy metabolites
LC-MS/MS was performed using an Agilent 1200 series HPLC and an Agilent 6410 triple quadrupole tandem mass spectrometer (Palo Alto, CA, USA) equipped with an electrospray ionization source, as described previously (22). Separation was achieved on a Hypersil Gold (2.1 × 100 mm, 1.9 μm) column (Thermo Scientific). Multiple Reaction Monitoring (MRM) of the transitions m/z 574 → 314, 574 → 298, 558 → 298, 564 → 298 and 580 → 314 were employed to detect 1α (4α or 4β),25(OH)2D3, 24R,25(OH)2D3, 25OHD3, d6-25OHD3 and d6-1α,25(OH)2D3, respectively. Because a deuterated form of 4β,25(OH)2D3, 4α,25(OH)2D3 or 24R,25(OH)2D3 was not available, we used d6-1α,25(OH)2D3 as their internal standard. 4β,25(OH)2D3 was used as the standard for generation of the calibration curve of 4α,25(OH)2D3. Calibration curves were constructed by plotting the peak area ratio for each vitamin D3 metabolite and its internal standard, versus the corresponding concentration, and fitting a linear regression equation to the data (22).
LC-MS/MS analysis for the measurement of 1′-OH-MDZ
Midazolam and its major CYP3A-dependent metabolite, 1′-OH-MDZ, were separated by an Agilent Zorbax SB-C18 column (2.1 × 150 mm, 5 μm particle size) on an Agilent 1200 Series LC system. Analytes were eluted under isocratic (45% B) conditions, consisting of 0.1% formic acid (A) and acetonitrile (B), with a flow rate at 0.25 mL/min. Detection was carried out with an Agilent 6410 triple quadrupole LC-MS/MS system. The flow and temperature of the drying gas (N2) were 11 L/min and 300°C, respectively. The nebulizer pressure was 20 psi and the capillary voltage 4500 V for positive ionizations and chamber current was 1.39 μA. The respective MRM for the quantitation of midazolam, 1′-OH-MDZ, d4-midazolam and d4-1′-OH-MDZ were m/z 326 → 291, 342 → 168, 330 → 295 and 346 → 168. Calibration curves were conducted by plotting the peak area ratio for each compound and its internal standard versus the corresponding concentration and fitting with a linear regression equation.
Human subject study procedures
A detailed description of the study population is being reported elsewhere, along with results of the primary study objective – evaluation of sulforaphane (SFN) as an antagonist to PXR activation in vivo. An examination of the effect of rifampin on systemic levels of 25OHD3 and its monohydroxy metabolites represented a secondary use of the samples and was approved by the University of Washington Human Subjects Review Board. Some basic information about the clinical study design is repeated here for clarity. A total of 23 participants were studied, 12 males and 11 females, average age of 23.7 years (range 20 – 34), average body mass index (BMI) of 23.4 kg/m2. Seventeen (74%) of the participants were Caucasian, with others self-identifying as Asian American (5; 22%) or African American (1; 4%). Exclusion criteria for the participants were age < 20 or > 40, BMI over 30 kg/m2, active smoking, use of other prescription medications, and clinical laboratory test values outside the normal range during screening, including tests for liver and kidney function, and hematology. The primary study featured a double-blind crossover design, in which each subject served as his/her own control. There were three treatment periods: rifampin, SFN, and rifampin plus SFN. Treatments with rifampin or SFN (or both) lasted 6 days; the dose of rifampin was 300 mg, given once daily at 6 pm. CYP3A activity was assessed by midazolam clearance (1 mg, PO) on days just prior to the start (baseline) and at the end of each treatment period. Active treatments were separated by a “wash out” period of two weeks to allow cytochrome P450 gene expression to return to baseline. Serial 5 mL blood samples were collected in heparinized Vacutainer tubes between 0 to 6 hrs following midazolam administration, placed on ice and processed within 2 hrs. Plasma was obtained by centrifugation at 4 °C, aliquoted into cryovials, frozen and stored at −80 °C until analyzed. For the purposes of this report, mean data from the three baseline days were compared to data from the day at the end of the rifampin treatment phase. We also compared data from the three baseline days to assess intra-individual variability.
Statistical analysis
Cell culture data are expressed as the mean ± standard error (S.E.) and were compared using one-way ANOVA analysis with Dunnett’s multiple comparison tests. Two-sample, two-tailed, unpaired Students t-tests were performed to examine statistical differences between in vitro treated and control groups. Statistical comparison of the pre- and post-rifampin treatment measurements in vivo were conducted using two-tailed, paired Student t-test. We used GraphPad Prism (v.5, La Jolla, CA) for the statistical analyses. Pharmacokinetic analysis of in vivo midazolam was performed using the Pharsight WinNonlin (Mountain View, CA). Pharmacokinetic estimates from the baseline days and rifampin treatment day were analyzed using Generalized Estimating Equations with treatment period as the independent variable; a p-value less than 0.05 was considered statistically significant.
Results
Induction of CYP3A4, CYP27B1 and CYP24A1 gene expression, and formation of monohydroxy metabolites of 25OHD3, in human hepatocytes treated with PXR agonists
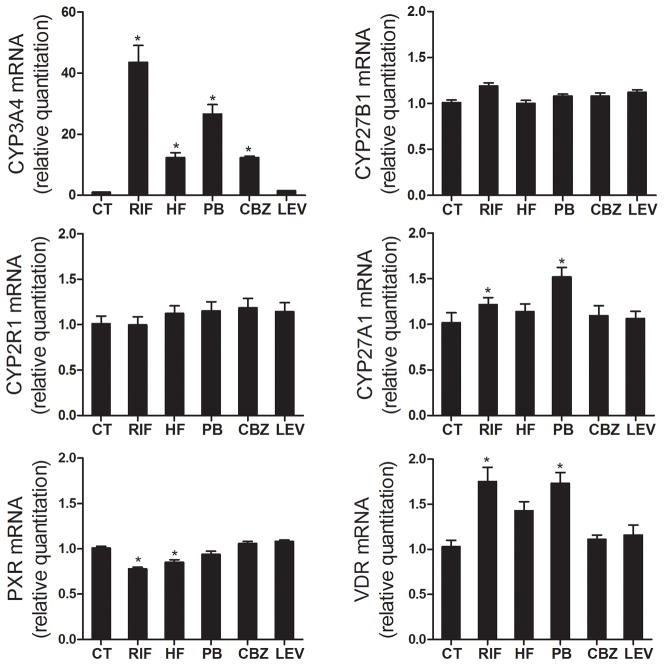
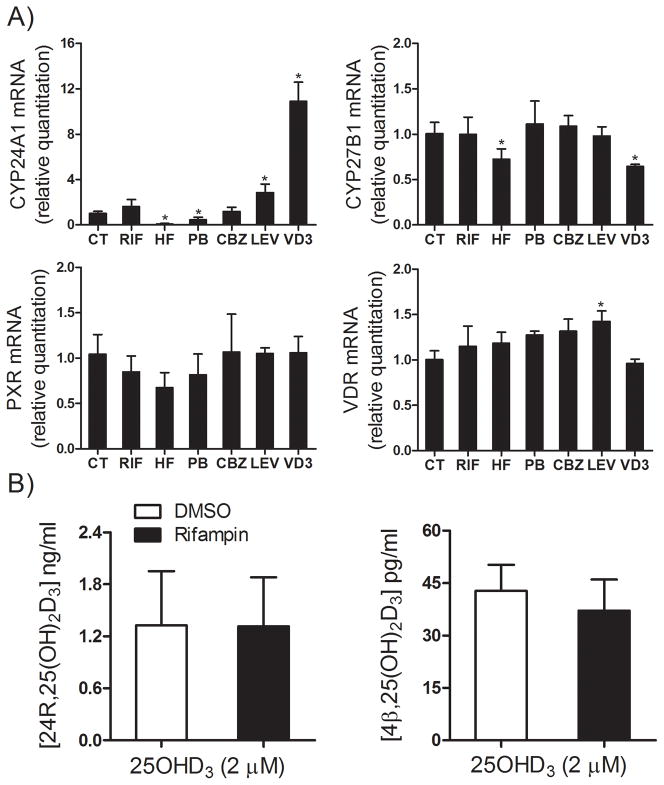
CYP3A4, CYP27B1, and CYP24A1 are all capable of supporting 25OHD3 monohydroxylation, yielding 4β,25(OH)2D3, 1α,25(OH)2D3 and 24R,25(OH)2D3 as the respective predominant metabolite (17,21). CYP2R1 and CYP27A1 are the major 25-hydroxylase, producing 25OHD3 from vitamin D3 mainly in the liver (20). We first investigated the expression of these five genes after treatment with four PXR agonists (rifampin, hyperforin, phenobarbital and carbamazepine) in human hepatocytes obtained from three different donors. Levetiracetam, another anti-epileptic drug that does not activate PXR, was used as a negative control. Following 48 hr of exposure to rifampin, hyperforin, phenobarbital or carbamazepine, the cellular CYP3A4 mRNA content was elevated 43.6-, 12.3-, 26.7-, and 12.3-fold, respectively, while the mRNA levels of CYP27B1 and CYP2R1 were not affected. The level of CYP27A1 mRNA was slightly increased by rifampin and phenobarbital treatment (Figure 1). The basal level of CYP24A1 mRNA in human hepatocytes was not quantifiable (under the initial experimental conditions employed) and was not induced following treatment with PXR agonists. In addition, we also examined relative expression of the nuclear receptors, vitamin D receptor (VDR) and PXR. Rifampin, hyperforin and phenobarbital modestly increased the hepatocellular VDR mRNA content but slightly decreased levels of PXR mRNA. Levetiracetam, as expected, caused no significant changes in the hepatocellular mRNA contents of all five of our target genes. These data suggest selective induction of CYP3A4 25OHD3 monohydroxylase expression, but not CYP24A1 or CYP27B1 gene expression, after treatment with P450 inducers.
Figure 1. Expression of CYP3A4, CYP24A1, CYP27A1, CYP27B1, CYP2R1, VDR, and PXR genes in human hepatocytes.
Human hepatocytes from three different donors were treated with 10 μM rifampin (RIF), 400 μM phenobarbital (PB), 0.5 μM hyperforin (HF), 50 μM carbamazepine (CBZ), 200 μM levitiracetam (LEV) or vehicle control (CT, 0.1% v/v) for 48 hr, as indicated. Total RNA from each sample was isolated and the expression of CYP3A4, CYP27A1, CYP27B1, CYP24A1, CYP2R1, VDR, and PXR was determined by qRT-PCR assay. Data represent mean ± S.E. (from four replicate determinations of three different liver donors) of the fold-induction in treated cells, compared to those of vehicle treated cells, after normalization to the 18s ribosomal RNA level. *: p < 0.05 for the inductive effect of PXR agonists, compared to corresponding control group.
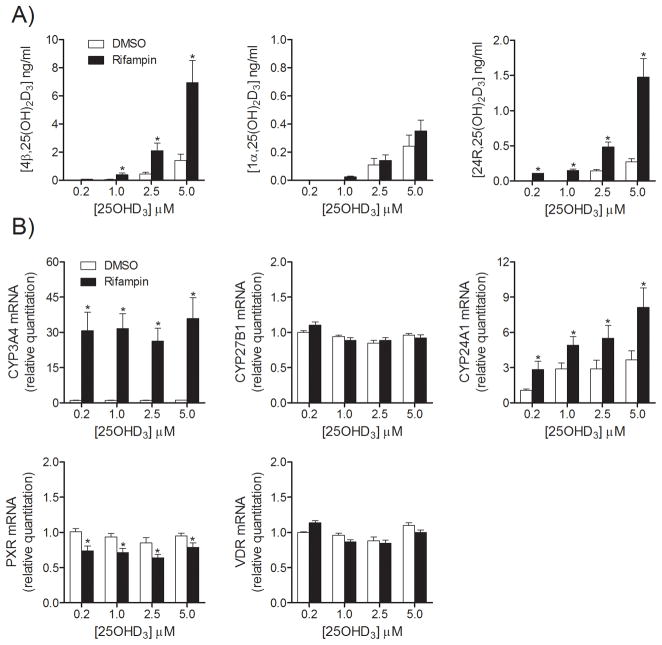
To evaluate which metabolic gene products are involved in 25OHD3 metabolism after treatment with P450-inducing drugs, a two-stage sequential treatment was designed. Rifampin was selected as a prototypical inducer due to its highest inductive capacity. One day after plating, human hepatocytes were pretreated with rifampin for 48 hr to induce target gene expression. After preincubation, the drug was removed by washing and the cells were incubated with 25OHD3 to assess its metabolic fate. First, hepatocytes were incubated with various concentrations of 25OHD3 (0.2 to 5.0 μM) for 4 hr after rifampin pretreatment (Figure 2). Concentrations of three major monohydroxy metabolites of 25OHD3 in culture media were measured using LC-MS/MS. As shown in Figure 2A, all three vitamin D3 metabolites were formed dose-dependently, however, 4β,25(OH)2D3 was the major metabolite at all concentrations tested (>75% of total measured monohydroxy products). 4α,25(OH)2D3 was a minor metabolite, but with a similar formation pattern as that of 4β,25(OH)2D3 (data not shown).
Figure 2. Formation of three 25OHD3 metabolites and expression of VDR, PXR and related genes in human hepatocytes after treatment of 25OHD3 with or without rifampin pretreatment.
After treatment with rifampin (10 μM) for 48 hr, hepatocytes were incubated with various concentrations of 25OHD3 for 4 hr. Media were collected for LC-MS/MS analysis and cell lysates were used for mRNA extraction and RT-PCR. A) Formation of vitamin D metabolites from 25OHD3 metabolism. B) mRNA levels for VDR, PXR and their target genes. Data are represented as mean ± S.E. from three different liver donors. *: p < 0.05 for the effect of rifampin, compared to corresponding control group.
Pretreatment with rifampin greatly enhanced the formation of 4β,25(OH)2D3 (p < 0.05), consistent with induction of CYP3A4 expression in hepatocytes, as reported previously (21). Interestingly, formation of one of the minor metabolites, 24R,25(OH)2D3 (~15%; p < 0.05) was also enhanced by rifampin pretreatment, whereas there was no effect on 1α,25(OH)2D3 formation (~5%; NS). This result prompted parallel measurements of mRNA levels for the three 25OHD3 metabolizing enzymes (CYP3A4, CYP24A1 and CYP27B1) following the different pretreatment conditions and after the different 25OHD3 doses. As shown in Figure 2B, CYP3A4 mRNA content was increased approximately 30-fold following rifampin pretreatment and 25OHD3 (0.2 – 5 μM, 4 hr) did not have a post-treatment synergetic effect on CYP3A4 expression. The basal level of CYP24A1 mRNA (cycle threshold Ct = 35 ± 2) was very low but detectable in this experiment, and was increased slightly (p < 0.05) after rifampin pretreatment. Post-treatment incubation with 25OHD3 increased CYP24A1 mRNA modestly (compared to CYP3A4 mRNA). In contrast, the mRNA level of CYP27B1 was not affected by rifampin pretreatment or the post- treatment period with 25OHD3. Finally, mRNA levels for the two nuclear receptors, VDR and PXR, were relatively unaffected by sequential rifampin and 25OHD3 treatments, although a ~10% reduction in PXR mRNA content after rifampin was significant for all 25OHD3 doses (Figure 2B).
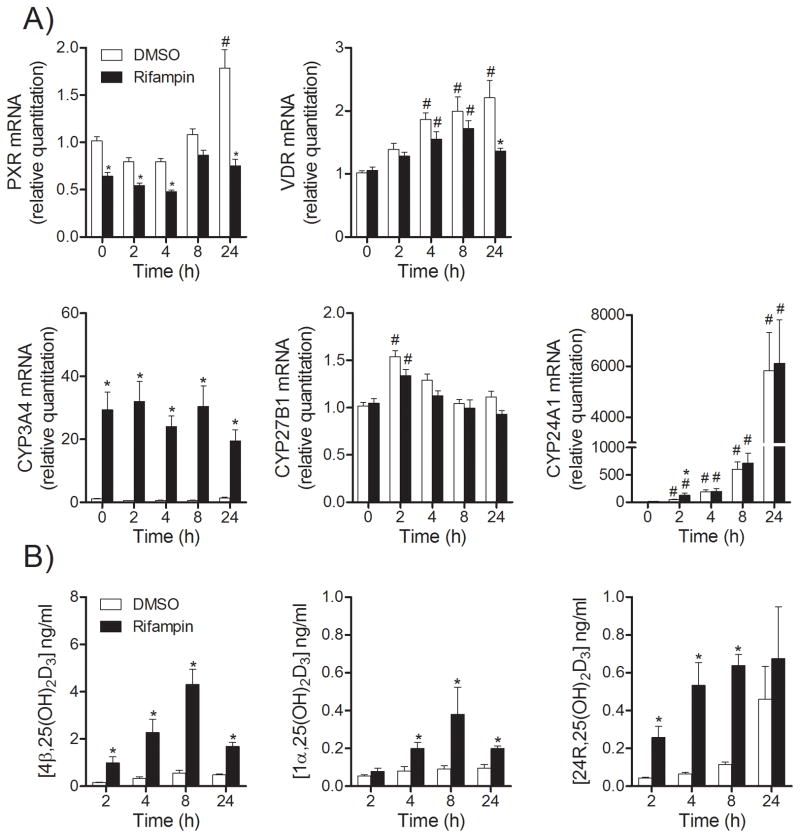
Incubation of hepatocytes with micromolar concentrations of 25OHD3 may induce gene expression (e.g., CYP24A1) in a time-dependent manner by activating a VDR-mediated transcription pathway. Therefore, we measured the mRNA levels of CYP3A4, CYP27B1 and CYP24A1 at different periods of 25OHD3 incubation time after preincubation with rifampin, in parallel with the measurement of 4β,25(OH)2D3, 1α,25(OH)2D3 and 24R,25(OH)2D3 formation. Hepatocytes were first pretreated with rifampin and then incubated with 25OHD3 (2 μM) for an additional period of time (t = 0, 2, 4, 8, and 24 hr). As shown in Figure 3, pretreatment with rifampin prior to 25OHD3 incubation slightly decreased PXR, but not VDR mRNA levels; without rifampin pretreatment, the mRNA levels of PXR and VDR were modestly increased (~2 fold over 24 hr). Cellular CYP3A4 mRNA content was increased substantially (~30-fold) by rifampin pretreatment and once again the post-incubation with 25OHD3 did not enhance this effect (Figure 3A). The formation of 4β,25(OH)2D3 peaked at 8 hr and declined by 24 hr. Pretreatment with rifampin enhanced the formation of 4β,25(OH)2D3 at all-time points (Figure 3B). The basal CYP24A1 mRNA level was very low, but detectable, and treatment with rifampin only increased it slightly (see t = 0 hr with 25OHD3). Surprisingly, post-treatment incubation with 25OHD3 out to 24 hr resulted in a ~6,000 fold increased CYP24A1 expression in both rifampin and vehicle pretreated groups, with little difference between the two pre-treatment groups (t = 4, 8 and 24 hr). The formation of 24R,25(OH)2D3 also increased in a time-dependent manner, suggesting that activation of VDR might be responsible for the time-dependent induction of CYP24A1 after 2 to 24 hr of incubation with 25OHD3 (Figure 3A). Rifampin pretreatment greatly enhanced 24R,25(OH)2D3 formation within the first 8 hr of 25OHD3 incubation time, including the t = 0 time point, but this effect diminished after incubation for 24 hr (Figure 3B). Interestingly, rifampin pretreatment did not enhance CYP27B1 expression, but modestly increased 1α,25(OH)2D3 production (p < 0.05), particularly over 4 to 24 hr of 25OHD3 incubation. Together, these data suggested a change in the relative contribution of enzymes (e.g., CYP3A4 and CYP24A1) to 24R- and 1α-monohydroxy metabolite formation over the 25OHD3 incubation time.
Figure 3. Time-dependent formation of 25OHD3 metabolites and related gene expression after rifampin treatment.
After treated with rifampin (10 μM) for 48 hr, hepatocytes were incubated with 25OHD3 (2 μM) for different periods of incubation time, as indicated. After incubation, media were collected for LC-MS/MS analysis and the total mRNA were extracted for RT-PCR analysis. A) mRNA levels for VDR, PXR and their targeted genes. Data are represented as mean ± S.E. from three different liver donors. B) Formation of vitamin D metabolites. *: p < 0.05 for the effect of rifampin, compared to corresponding control group. #: p < 0.05 for the effect of time, compared to corresponding control group (t = 0).
Effects of CYP3A4 inhibition by DHB on the formation of three major monohydroxy metabolites of 25OHD3 in human hepatocytes
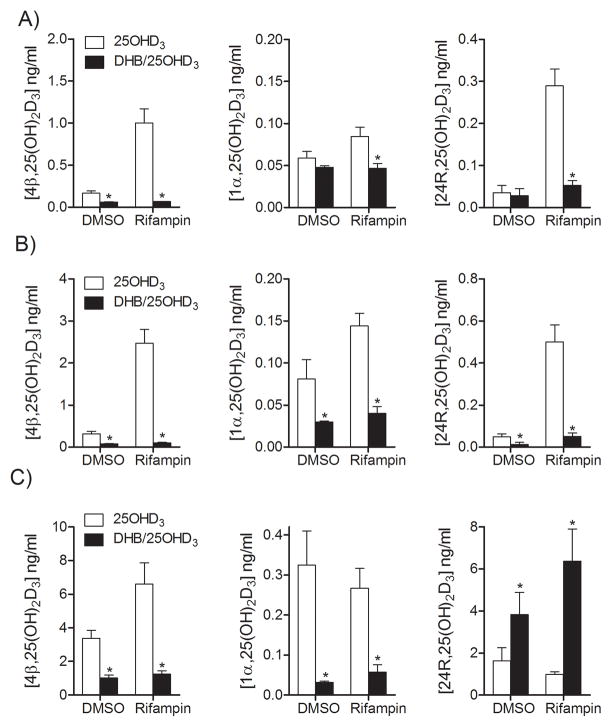
In order to elucidate the specific contribution of CYP3A4, CYP24A1 and CYP27B1 to the rifampin-enhanced and 25OHD3-enhanced formation of the three major 25OHD3 metabolites, a selective CYP3A4 inhibitor, DHB, was added to the cell culture system after rifampin pretreatment. The addition of 20 μM DHB greatly reduced CYP3A4 activity in both basal and rifampin-induced conditions (Supplemental Figure 1). This same treatment did not inhibit recombinant CYP24A1 activity in vitro (24). After rifampin pretreatment, hepatocytes were incubated with DHB for 4 hr and throughout the period of 25OHD3 treatment (t = 2, 4 and 24 hr) (Figure 4). DHB treatment caused a 63% and 93% (control and rifampin-induced conditions), 74% and 96%, and 70% and 81% reduction in 4β,25(OH)2D3 levels after dosing 25OHD3 for 2, 4, and 24 hr, respectively. Moreover, DHB inhibited control and rifampin-induced 1α,25(OH)2D3 formation by 19% and 45%, 63% and 72%, and 90% and 79% after dosing 25OHD3 for 2, 4, and 24 hr, respectively. Interestingly, DHB treatment also reduced the formation of 24R,25(OH)2D3 in drug vehicle control and rifampin pretreated cells at the early stage of 25OHD3 incubation (2 hr: 20% and 82%; 4 hr: 76% and 90%); whereas it greatly enhanced the production of 24R,25(OH)2D3 after 24 hr. These results suggested that CYP3A4 was the predominant enzyme catalyzing the formation of 24R,25(OH)2D3 in hepatocytes during the early incubation time (e.g., 2 and 4 hr). However, after 24 hr, up-regulation of what appeared to be VDR-mediated CYP24A1 expression generated an alternative pathway for 24R,25(OH)2D3 formation, and inhibition of CYP3A4 activity by DHB switched the metabolism of 25OHD3 from mainly 4β-hydroxylation (CYP3A4-mediated) to 24R-hydroxylation (CYP24A1-mediated). The data also suggested that CYP3A4 contributed significantly to 1α,25(OH)2D3 formation, in both the control and rifampin-induced states, particularly after 4 hr and 24 hr of 25OHD3 incubation.
Figure 4. Effects of DHB on the formation of 25OHD3 metabolites in hepatocytes.
After treatment with rifampin (10 μM) for 48 hr, hepatocytes were pre-incubated with DHB (20 μM) for 4 hr, followed by incubation with 25OHD3 and DHB mixture under the certain conditions, as indicated: A) incubation with 25OHD3 (2 μM) and DHB for 2 hr; B) incubation with 25OHD3 (2 μM) and DHB for 4 hr; C) 25OHD3 (5 μM) and DHB for 24 hr. Media were collected for LC-MS/MS analysis. Data are represented as mean ± S.E. from three different liver donors.
Because CYP3A4 appeared to be the major enzyme responsible for 25OHD3 hydroxylation under both basal and rifampin-induced conditions (in the absence of DHB), we tested the inductive capacity of other PXR agonists and the inhibitory effects of DHB on CYP3A4 activity and formation of the three monohydroxy metabolites of 25OHD3 (Table 1). Hepatocytes were incubated with 25OHD3 in the presence or absence of DHB for 4 hr after rifampin pretreatment. Consistent with the mRNA results shown in Figure 1, CYP3A4 activity was increased 4- to 7-fold, with the following potency order, e.g. rifampin > phenobarbital > hyperforin > carbamazepine. Again, 4β,25(OH)2D3 was the major metabolite produced from 25OHD3 and its formation was induced by all four PXR agonists and inhibited by DHB. 24R,25(OH)2D3 was a minor metabolite and its formation was also induced by PXR agonists and inhibited by DHB to an undetectable level. This effect was consistent with a significant CYP3A4 contribution to its formation after a short (4 hr) period of incubation with 25OHD3.
Table 1. Effect of PXR agonists on CYP3A4 activity and formation of three monohydroxy metabolites of 25OHD3 in cultured human hepatocytes.
Human hepatocytes were pretreated with 10 μM rifampin, 400 μM phenobarbital, 0.5 μM hyperforin, 50 μM carbamazepine, 200 μM levitiracetam or vehicle for 48 hr.
| CYP3A4 activity a | Concentrations of three major monohydroxy metabolites of 25OHD3 in culture media (ng/mL)b | |||||
|---|---|---|---|---|---|---|
| 4β,25(OH)2D3 | 1α,25(OH)2D3 | 24R,25(OH)2D3 | ||||
| (−) DHB | (+) DHB | (−) DHB | (+) DHB | (−) DHB | ||
| Control | 53.1 (17.3) | 0.65 (0.16) | 0.19 (0.08)# | 0.10 (0.00) | 0.09 (0.02) | 0.02 (0.02) |
| Rifampin | 341.5 (74.5)* | 2.95 (0.55)* | 0.20 (0.03)# | 0.12 (0.01) | 0.09 (0.01) | 0.39 (0.10)* |
| Hyperforin | 248.4 (71.4)* | 1.81 (0.50)* | 0.28 (0.13)# | 0.11 (0.02) | 0.07 (0.01) | 0.19 (0.07)* |
| Phenobarbital | 287.0 (78.1)* | 2.10 (0.83)* | 0.16 (0.00)# | 0.09 (0.02) | 0.07 (0.01) | 0.17 (0.09)* |
| Carbamazepine | 216.7 (65.4)* | 1.52 (0.56)* | 0.27 (0.01)# | 0.09 (0.01) | 0.11 (0.02) | 0.10 (0.06)* |
| Levitiracetam | 64.3 (20.3) | 0.56 (0.20) | 0.27 (0.11)# | 0.10 (0.01) | 0.08 (0.01) | N.D. |
At the end of preincubation, cells were incubated with 2 μM midazolam for 30 min. CYP3A4 activity was calculated as the rate of 1′-hydroxylation of midazolam (pmol/mL/hr).
In a parallel experiment, cells were washed with phosphate buffer saline to remove the pretreatment drugs, incubated with 20 μM DHB or vehicle for 4 hrs to block CYP3A4 activity, and then treated with 2 μM 25OHD3 with/without DHB in fresh medium for 4 hr. After incubation, the media were collected for LC-MS/MS analysis.
Data represent mean (S.E.) from three different liver donors (n = 3).
p < 0.05 for the effect of agonists, compared to corresponding control group.
p < 0.05 for the effect of DHB, compared to corresponding control or agonist pretreated groups.
N.D.: the analyte was not detectable
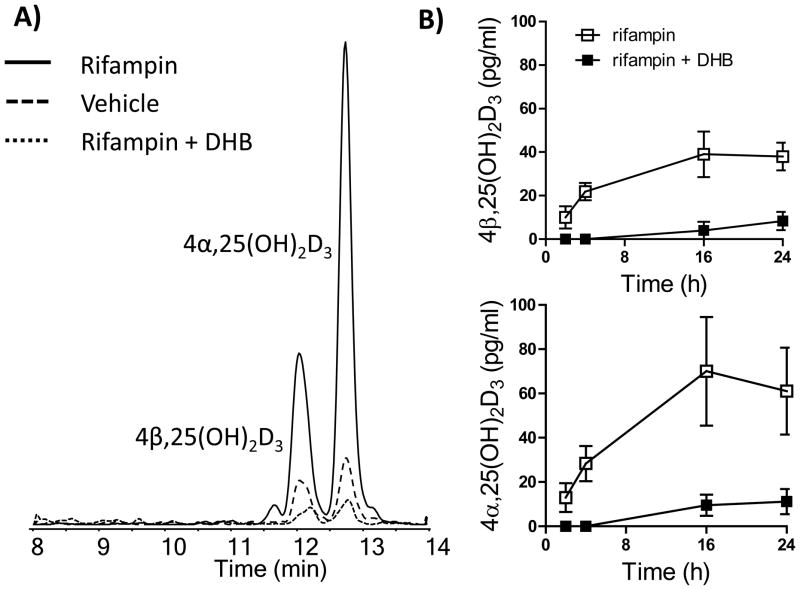
In order to determine whether CYP3A4-dependent 4-hydroxylation of 25OHD3 also occurs in cultured hepatocytes when more physiological concentrations of 25OHD3 are introduced, hepatocytes were incubated with 50 nM 25OHD3 in the presence or absence of DHB after rifampin pretreatment (Figure 5). In agreement with the results from Figure 2, unconjugated 4β,25(OH)2D3 and 4α,25(OH)2D3 were not detectable in hepatocytes treated with 50 nM of 25OHD3. Considering the possibility that rapid sequential Phase II conjugation could mask the formation of the primary oxidation products, the collected cell culture media were incubated with β-glucuronidase and then analyzed by LC-MS/MS. As seen in Figure 5A, significant amounts of 4β,25(OH)2D3 and 4α,25(OH)2D3 were detected in those β-glucuronidase treated samples, indicating that 4β,25(OH)2D3 and 4α,25(OH)2D3 were indeed formed when the hepatocytes were incubated with 50 nM 25OHD3. Moreover, rifampin pretreatment greatly enhanced the formation of the conjugated metabolites (Figure 5A), whereas co-treatment of 25OHD3 with DHB suppressed their formation below that produced by control cells (Figure 5A). Looking at the time-course of secondary metabolite formation (Figure 5B), the conjugates of both 4α and 4β-hydroxy metabolites appeared rapidly after initiation of 25OHD3 incubation with rifampin pretreated cells, reaching a peak at 16 hr and declining slightly 8 hr later. Interestingly, the peak 4α,25(OH)2D3 conjugate concentration exceeded that of the 4β,25(OH)2D3 conjugate by about 79%. Again, DHB blocked the inductive effect of rifampin yielding peak levels of 4β,25(OH)2D3 and 4α,25(OH)2D3 conjugates that were lower than 20 pg/ml (Figure 5B). Preliminary experimental results suggests that the conjugates of 4β,25(OH)2D3 and 4α,25(OH)2D3 are glucuronidation and not sulfation products. As expected, conjugates of 24R,25(OH)2D3 and 1α,25(OH)2D3 were not detected under the same 25OHD3 incubation conditions. Overall, these results strongly support a conclusion that 4-hydroxylation of 25OHD3 is a major pathway of 25OHD3 metabolism under physiological conditions in hepatocytes.
Figure 5. Formation of 4β,25(OH)2D3 and 4α,25(OH)2D3 conjugates in human hepatocytes.
A) Representative chromatograms of 4β,25(OH)2D3 and 4α,25(OH)2D3; B) Time-dependent formation of 4β,25(OH)2D3 and 4α,25(OH)2D3 in hepatocytes after β-glucuronidase treatment. Hepatocytes (n = 3) were pretreated with 10 μM rifampin for 48 hr and then treated with 50 nM 25OHD3 with various incubation times. In parallel, cells were pre-incubated with 20 μM DHB for 4 hr and then co-incubated with 25OHD3. Medium was collected and incubated with β-glucuronidase, extracted and analyzed as described previously. The released amounts of 4β,25(OH)2D3 and 4α,25(OH)2D3 represent the levels of the corresponding conjugates in the culture medium. Both primary oxidation products were undetectable following incubation of 50 nM 25OHD3 without β-glucuronidase treatment of the culture media.
Induction of CYP3A4, CYP27B1 and CYP24A1 gene expression by PXR agonists in human renal HK-2 cells
To determine whether PXR agonists induce gene CYP3A4 or CYP24A1 in human kidney epithelial cells, we investigated the inductive effects of rifampin, phenobarbital, hyperforin and carbamazepine on the expression of CYP3A4, CYP27B1 and CYP24A1 in human HK-2 cells. 1α,25(OH)2D3 (0.5 nM) was used as a positive control for the induction of CYP24A1 via a VDR-mediated pathway. Similar to what is seen in normal human kidney (9), the HK-2 cells contained an extremely low level of mRNA for the nuclear receptor PXR (Ct = 39 ± 1). Also, the basal mRNA levels for CYP24A1 (Ct = 33 ± 2) and CYP27B1 (Ct = 33 ± 1) were higher than that for CYP3A4 (Ct > 39). As shown in Figure 6A, after exposure to PXR agonists, CYP27B1 mRNA content was unchanged by the various pretreatments, except for a slight decrease in 1α,25(OH)2D3 and hyperforin-treated groups. CYP3A4 mRNA content was below limits of detection in both untreated and treated HK-2 cells and thus not quantifiable. In contrast, CYP24A1 mRNA content was significantly increased by 1α,25(OH)2D3 and levetiracetam, but modestly decreased by hyperforin and phenobarbital (p < 0.05). These results suggest that a VDR, rather than PXR-mediated transcriptional activation was the primary mechanism of CYP24A1 induction in human HK-2 cells, and that it was relatively unaffected by treatment with PXR agonists. This conclusion was also supported by an examination of 25OHD3 metabolite formation. As shown in Figure 6B, 24R,25(OH)2D3 was the predominant product in HK-2 cells after exposure to 25OHD3, and its formation was not inducible by the PXR agonist rifampin. In contrast, the concentration of 1α,25(OH)2D3 was below the limit of detection under all conditions. Formation of 4β,25(OH)2D3 was detectable in HK-2 cells incubated with 25OHD3, but at concentrations of approximately 0.04 ng/ml, compared to ~1.34 ng/ml for 24R,25(OH)2D3. Rifampin pretreatment had no effect on 4β,25(OH)2D3 formation. Because CYP3A4 mRNA was undetectable in HK-2 cells, production of picomolar levels of 4β,25(OH)2D3 might be due to the expression of CYP3A5 in the kidney cells, which was not tested.
Figure 6. Effect of PXR agonists on expression of VDR, PXR and their target genes, and formation of 24R,25(OH)2D3 in HK2 cells.
A) HK2 cells were treated with 10 μM rifampin (RIF), 400 μM phenobarbital (PB), 0.5 μM hyperforin (HF), 50 μM carbamazepine (CBZ), 200 μM levitiracetam (LEV) or vehicle (CT, 0.1% v/v) for 48 hr. 1α,25(OH)2D3 (VD3, 0.5 nM) was used as a positive control for CYP24A1 induction and incubated for 24 hr, as previously described (24). Total RNA was isolated and the expression of CYP3A4, CYP27B1, CYP24A1, VDR, and PXR was determined by qRT-PCR assay. B) Formation of 24R,25(OH)2D3 and 4β,25(OH)2D3. HK2 cells were pretreated with 10 μM rifampin for 48 hr. Cells were washed with PBS twice and then treated with 2 μM 25OHD3 for 4 hr. Media were collected for LC-MS/MS analysis.
Effect of rifampin on 25OHD3 metabolism in healthy volunteers
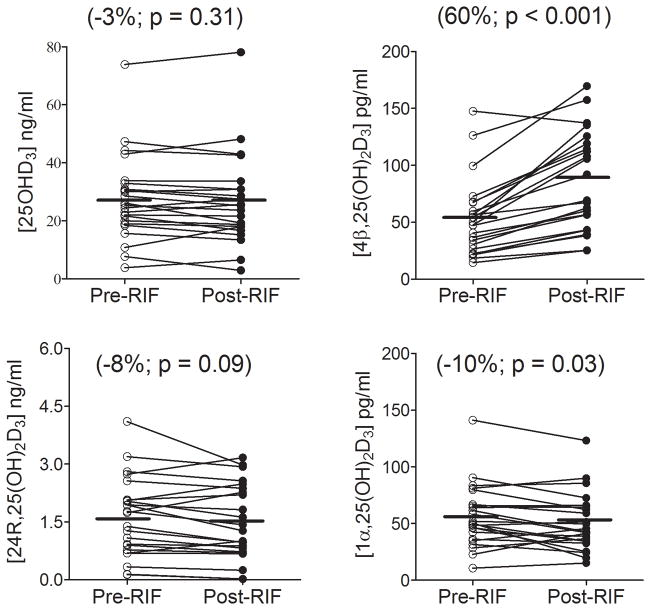
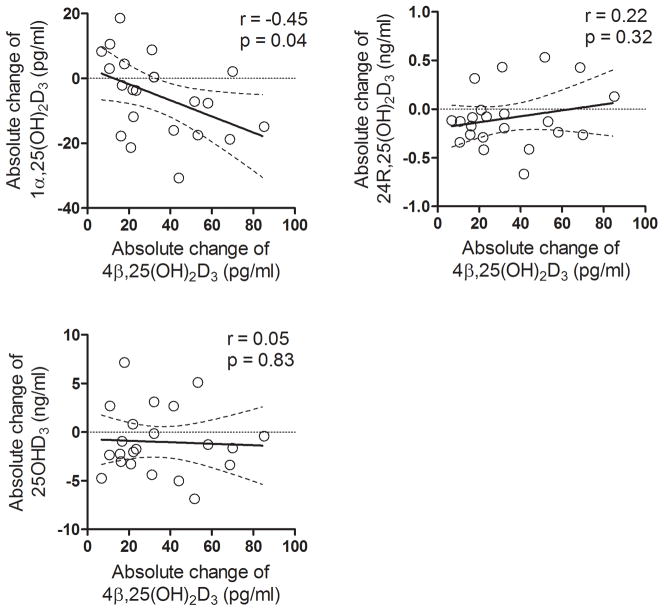
The plasma concentrations of 25OHD3 and its monohydroxy metabolites were measured before and after 6 days of rifampin (300 mg, PO) treatment. As shown in Figure 7, there was considerable inter-subject variability in the levels of all four vitamin D species under both baseline and rifampin treatment conditions. The measured values appeared to be normally distributed; hence, no data transformations were conducted for statistical comparisons. Paired comparisons showed a mean reduction of 10% (p < 0.03) in the plasma 1α,25(OH)2D3 concentration following rifampin treatment. There was a similar magnitude mean reduction in the 24R,25(OH)2D3 level following rifampin treatment, although this failed to reach statistical significance (p = 0.09). The mean change in the concentration of 25OHD3 was also not significant following rifampin treatment, although 16 out of 23 subjects showed a decline in concentration, compared to six showing an increase and one with no change. In contrast to these observations, the plasma level of 4β,25(OH)2D3 was increased by 63% (p < 0.001), on average, following rifampin treatment. There was some inter-individual variability in the magnitude of the response to the P450 inducer. However, all but one subject recorded an increase over baseline. Among the 22 of 23 subjects with increased levels of 4β,25(OH)2D3, there was a significant correlation between the absolute change in plasma 4β,25(OH)2D3 concentration following rifampin treatment and the absolute change in the plasma 1α,25(OH)2D3 concentration (r = −0.45, p < 0.05), but not with the absolute changes in 25OHD3 and 24R,25(OH)2D3 (Figure 8).
Figure 7. Effect of short-term rifampin treatment on the plasma concentrations of vitamin D metabolites.
Plasma samples were collected before (pre-RIF) and after (post-RIF) rifampin administration (300 mg, PO, 6 days). Paired comparisons were conducted to show the direct effect of rifampin on the levels of four metabolites in each individual. Statistical analysis was conducted using a paired t-test; a p value less than 0.05 indicates significance.
Figure 8. Correlation between the absolute change in plasma 4β,25(OH)2D3 concentration with absolute change in plasma 1α,25(OH)2D3, 24R,25(OH)2D3, and 25OHD3 levels.
The absolute concentration change was calculated as the post-rifampin minus pre-rifampin concentrations. Sample correlation coefficients and corresponding p-values were calculated; the null hypothesis that the population correlation is zero was tested with a t-test, under the assumption that the population was bivariate and normally distributed.
Under certain steady-state conditions, the metabolite to parent plasma concentration ratio is equivalent to the ratio of the metabolite formation clearance to the metabolite elimination clearance (25). As seen in Table 2, there was no significant change in the ratios of 1α,25(OH)2D3/25OHD3 and 24R,25(OH)2D3/25OHD3 following rifampin treatment. In contrast, the 4β,25(OH)2D3/25OHD3 ratio was increased by 60% (p < 0.01), on average, following treatment with rifampin. The design of the primary SFN interaction study provided repeated measurements (n = 3) of the vitamin D species under un-induced, baseline conditions. This permitted an assessment of intra-individual variation over the two-week study period. As seen in Supplemental Figure 2, there was no statistically significant differences in the measured concentrations of 25OHD3, 1α,25(OH)2D3, 24R,25(OH)2D3 and 4β,25(OH)2D3 over the three baseline periods.
Table 2. Changes in the concentration ratio for three monohydroxy metabolites of 25OHD3 after oral treatment with rifampin.
Plasma samples were collected before and after orally dosing rifampin for six days in 23 subjects. Individual mean ratios at baseline (pre-RIF) and after rifampin treatment (post-RIF) are shown: 4β,25(OH)2D3/25OHD3 ratio (indicator of CYP3A4 activity); 24R,25(OH)2D3/25OHD3 ratio (indicator of CYP24A1 activity) and 1α,25(OH)2D3/25OHD3 ratio (indicator of CYP27B1 activity). Data are shown as mean ± S.E. (min; max) from 23 subjects.
| Plasma ratios of dihydroxyvitamin D3 to parent compound 25OHD3 (× 102) | Fold change of ratio (post-RIF/pre-RIF) | ||
|---|---|---|---|
| Pre-RIF | Post-RIF | ||
| 1α,25(OH)2D3 | 0.24 ± 0.16 (0.10; 0.90) | 0.25 ± 0.25 (0.08; 1.25) | 1.04 ± 0.55 (0.51; 3.38) |
| 4β,25(OH)2D3 | 0.20 ± 0.07 (0.10; 0.38) | 0.37 ± 0.18* (0.18; 0.87) | 1.85 ± 0.58* (0.88; 3.58) |
| 24R,25(OH)2D3 | 5.83 ± 2.78 (1.75; 14.02) | 5.55 ± 3.49 (0.30; 13.71) | 0.88 ± 0.34 (0.08; 1.77) |
p < 0.05 for the effect of rifampin, compared to corresponding control group.
Correlation between 4β,25(OH)2D3 formation and midazolam elimination in hepatocytes and in vivo
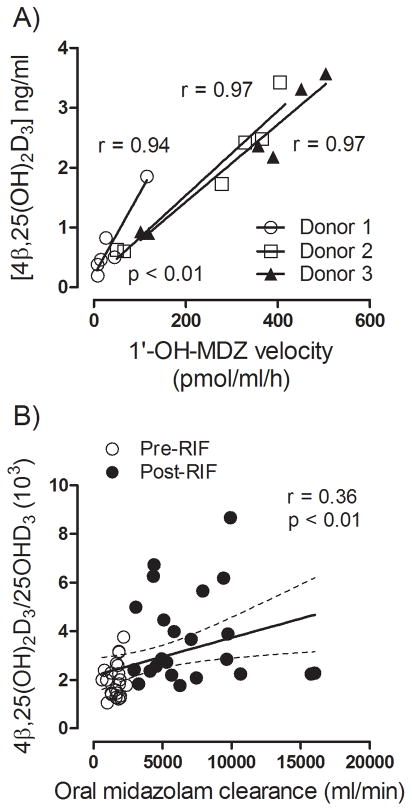
We showed previously that hepatic formation of 4β,25(OH)2D3 by human liver microsomes is correlated with CYP3A4-dependent midazolam hydroxylation activity (21). Here we examined whether formation of 4β,25(OH)2D3 is correlated with CYP3A4 activity in human hepatocytes and in vivo. Hepatocytes from three donors were pre-treated with PXR agonists to induce various degrees of CYP3A4 activity. The rate of 1′-hydroxylation of midazolam formation was used as the index of CYP3A4 activity. As shown in Figure 9A, the rates of 4β,25(OH)2D3 formation in hepatocytes were strongly correlated with the rate of 1′-OH-MDZ formation (r > 0.9, p < 0.01). Moreover, in vivo, oral midazolam clearance was significantly correlated with the 4β,25(OH)2D3/25OHD3 plasma concentration ratio (r = 0.36, p < 0.01) (Figure 9B), although the correlation coefficient was not as strong as that obtained in hepatocytes. This could be due to the substantial first-pass intestinal metabolism of midazolam that occurs in vivo (26,27), a phenomenon that might not be as relevant for endogenous 25OHD3 metabolism. It could also be the consequence of a parallel increase in the clearance of 4β,25(OH)2D3 through glucuronidation.
Figure 9. Correlation between 4β,25(OH)2D3 formation and midazolam elimination in both human hepatocytes and healthy volunteers.
A). After treated with rifampin (10 μM), phenobarbital (400 μM), hyperforin (0.5 μM), carbamazepine (50 μM), or vehicles (0.1% v/v) for 48 hr, human hepatocytes (from three different liver donors) were incubated with either midazolam (2 μM) for 30 min or 25OHD3 (2 μM) for 4 hr. The rate of 1′-hydroxylation of midazolam was correlated with the concentration of 4β,25(OH)2D3 using linear regression. B). Correlation between the ratio of plasma 4β,25(OH)2D3/25OHD3 and oral midazolam clearance for pre-RIF and post-RIF treatment periods. Open circle: constitutive condition; closed circle: rifampin-induced condition. Oral midazolam clearance was estimated using the Pharsight WinNonlin. Correlation coefficients and corresponding p-values were calculated according to the null hypothesis.
Discussion
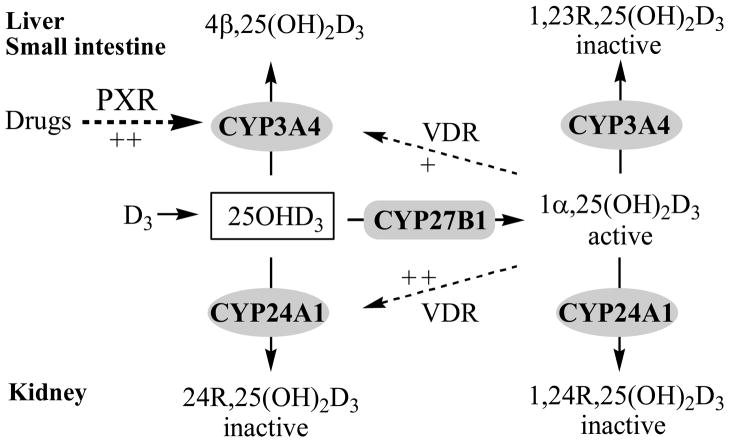
Long-term therapy with certain antiepileptic drugs and the antimicrobial agent rifampin, is associated with an increased risk of osteomalacia (28–31). Dysregulation of the bioactivation and degradation of 25OHD3 and 1α,25(OH)2D3 by P450 induction has been proposed, based on the fact that many of the drugs implicated in this adverse drug reaction (ADR) are known or thought to alter the expression and activity of vitamin D biotransformation enzymes (Scheme 1). In this context, the induction of CYP3A4-dependent vitamin D catabolism as a mechanism behind the ADR has significant appeal, because basal specific content of the enzyme in the liver and small intestine is high and it is highly inducible through activation of PXR [liver and intestine; (32,33)] or the constitutive androstane receptor [CAR; liver; (34,35)].
Scheme 1. Proposed mechanism for the tissue-specific induction of CYP3A4 and CYP24A1 gene expression on regulation of 25OHD3 and 1α,25(OH)2D3 catabolism in humans.
Both CYP3A4 and CYP24A1 can catalyze 25OHD3 and 1α,25(OH)2D3 hydroxylation. Under the constitutive conditions in the liver, CYP3A4 is the most abundant enzyme, catalyzing 4β-hydroxylation of 25OHD3 and 23R-hydroxylation of 1α,25(OH)2D3. While under the constitutive conditions in the kidney, CYP24A1 is the most abundant enzyme, catalyzing 24R-hydroxylation of 25OHD3 and 1α,25(OH)2D3. In the liver and small intestine, drugs (PXR agonists, e.g. rifampin) induce CYP3A4, but not CYP24A1 gene expression via a PXR pathway. In the kidney, neither CYP3A4 nor CYP24A1 is significantly induced by PXR activation; however CYP24A1 can be induced by vitamin D metabolites [e.g. 1α,25(OH)2D3] via a VDR pathway. Relative induction capability: ++ > +
Previously we reported that induction of 1α,25(OH)2D3 catabolism by CYP3A4, particularly within the proximal intestinal epithelium, might lead to a reduction in cellular VDR activation and a reduction in the expression of proteins involved in calcium absorption (8,24). In the current investigation, we sought to determine whether treatment with drugs that activate PXR alters the hepatic metabolism of 25OHD3, and, specifically, whether they induce the formation of CYP3A4-dependent 25OHD3 metabolites. Our results show that the overwhelming effect of the PXR agonists was to enhance the formation of 4β,25(OH)2D3 in vitro (rifampin, phenobarbital, hyperforin and carbamazepine) and in vivo (rifampin), with little to no induction of 24R,25(OH)2D3 and 1α,25(OH)2D3 formation. In cultured human hepatocytes, 4β,25(OH)2D3 was the predominant oxidative metabolite formed and its formation was attributed almost exclusively to CYP3A4 (Table 1 and Figure 4). The same findings held true when human hepatocytes were incubated with a physiologically relevant concentration of 25OHD3, although in this case rapid secondary conjugation of the primary oxidation products needed to be taken into account (Figure 5). It is worth speculating on whether the conjugates of 4α,25(OH)2D3 and 4β,25(OH)2D3 are also found in blood at concentrations that exceed that of the primary metabolites. Further characterization of conjugate formation and their preferred route of hepatic secretion (basolateral or canalicular) is warranted to more firmly establish the importance of 4-hydroxylation to the metabolic clearance of 25OHD3.
An analysis of hepatocyte mRNA under the different pretreatment conditions also indicated that there was a selective response to the PXR agonists, in that the transcription of CYP3A4 was significantly enhanced, with little if any change in CYP2R1, CYP27B1 and CYP24A1 transcription. In experiments with cultured human kidney epithelial cells, we found prominent expression of CYP24A1 under basal conditions, but it was not induced by the PXR agonists tested (Figure 6), which is consistent with a low level of PXR expression in human kidney (9,32). These in vitro findings were confirmed in vivo, where we observed selectiveinduction of 4β,25(OH)2D3 formation following six days of rifampin treatment (Table 2 and Figure 7), and no change in the formation clearance to the 24R,25(OH)2D3 and 1α,25(OH)2D3 metabolites. Although we did not attempt to measure conjugates of 4β,25(OH)2D3 and 4α,25(OH)2D3 in plasma, it is possible that they may circulate in blood, given that they form readily in human hepatocytes, and that they are also sensitive biomarkers of PXR activation and CYP3A4 induction. Overall, the only mechanistically significant change in the metabolism of 25OHD3 following treatment with rifampin, phenobarbital, hyperforin and carbamazepine in vitro, and rifampin in vivo, was an induction of CYP3A4-dependent 4-hydroxylation.
Results from our experiments with human hepatocytes also revealed some interesting cross-talk between PXR and VDR. When the metabolism of 25OHD3 was studied between 2 – 24 hr, it appeared that initial formation of the minor 24R,25(OH)2D3 metabolite, was mediated largely by CYP3A4, in both the basal and rifampin-induced states. DHB co-treatment largely abolished its formation during the first 4 hrs of 25OHD3 exposure (Figure 4A and 4B). However, over a longer time period (4 – 24 hr), any contribution from CYP3A4 appeared to be overwhelmed by production of 24R,25(OH)2D3 from CYP24A1 (Figure 4C), which was clearly induced following sustained exposure to 25OHD3 (Figure 3). While it is possible that this effect represented a response to 1α,25(OH)2D3 that was being produced by CYP3A4 (Figure 4A–C), it most likely was elicited directly by 25OHD3 (present initially at micromolar concentrations), given that DHB treatment reduced 1α,25(OH)2D3 production at all-time points but enhanced 24R,25(OH)2D3 production at the later time points. This observation is somewhat surprising in that the expression of VDR in human liver appears to be much lower than that found in the kidney and small intestine (8,9). In fact, some have suggested that its presence in the liver is restricted largely to Kupffer and biliary epithelial cells and that it is essentially absent from hepatocytes (36). It is possible that response to vitamin D exposure might be due to the presence of Kupffer or biliary epithelial cells in these commercially available human hepatocytes. However, it is also possible that a low level of VDR expression in primary cultured hepatocytes is sufficient to support the induction of CYP24A1 in response to vitamin D exposure. Whether this occurs in vivo in the intact liver is not clear and merits further investigation.
The opportunistic clinical study design we employed in our investigation does not permit us to say anything definitive about whether the induction of 25OHD3 4-hydroxylation could contribute to a reduction in VDR activation in the small intestine (or in other tissues) and result in drug-induced osteomalacia. Such a causal link would require an assessment of biomarkers of calcium and phosphate homeostasis following treatment with PXR agonists and intervention with compounds like DHB that selectively block CYP3A4 function. However, we observed a significant, albeit weak, inverse correlation between the change in 4β,25(OH)2D3 concentration and the change in the 1α,25(OH)2D3 concentration in subjects treated short-term with rifampin. This observation is in agreement with results from our previous pilot study conducted in six subjects after seven days of oral rifampin administration, where we observed a strong correlation between the absolute concentration change of 4β,25(OH)2D3 and 1α,25(OH)2D3 (21). Although in both studies, there was no indication that the formation clearance for 1α,25(OH)2D3 was altered by rifampin (Table 2), the increased formation of 4β,25(OH)2D3 appears to have resulted in metabolic switching, where a greater fraction of the 25OHD3 pool was directed away from 1α,25(OH)2D3 formation. Confirmation of this trend will require an evaluation of 25OHD3 products in plasma after longer-term treatments with PXR agonists, given the relatively long half-life (~10 days) of the precursor molecule, 25OHD3 (37).
Although CYP24A1-mediated monohydroxylation is clearly an important pathway for 25OHD3 and 1α,25(OH)2D3 elimination from circulation (19,38), particularly under constitutive conditions, our data suggest that it is not induced to any significant extent by the drugs associated with osteomalacia. Moreover, as expected from our gene expression work, the kidney is not a major site of CYP3A4 expression and thus does not contribute to 4β,25(OH)2D3 formation. This suggests that while 25OHD3 homeostasis is regulated by renal CYP24A1 (and factors that control its expression), there may be parallel regulation in the liver and possibly small intestine through the 4-hydroxylation pathway. Specifically, the high capacity of 4-hydroxylation in the liver and small intestine, as a consequence of a high CYP3A4 enzyme content (39,40), may represent a quantitatively important pathway of 25OHD3 elimination from blood, in addition to renal CYP24A1-catalyed 24R-hydroxylation. Moreover, unlike CYP24A1, hepatic and intestinal CYP3A4 is highly inducible by PXR (and CAR) agonists.
The fact that 24R,25(OH)2D3 levels in blood are relatively high, in comparison to 1α,25(OH)2D3 and 4β,25(OH)2D3, illustrates its importance in 25OHD3 homeostasis. However, a low systemic 4β,25(OH)2D3 plasma concentration, relative to 24R,25(OH)2D3 concentration, may simply reflect a quantitative difference in the clearance of the respective metabolites. Information from the literature indicates that 24R,25(OH)2D3 undergoes further oxidation to a C- 23 acid product that is excreted into the urine, but it can also undergo glucuronidation (41). Although we have not rigorously evaluated the metabolic clearance of the newly characterized 4β,25(OH)2D3 metabolite (21), results from Figure 5 and some preliminary experiments suggest that glucuronidation of 4β,25(OH)2D3 and 4α,25(OH)2D3 is highly facile and, thus, may contribute to a high clearance of these primary 25OHD3 metabolite in vivo.
In conclusion, CYP3A4 catalyzes both 1α,25(OH)2D3 and 25OHD3 hydroxylation reactions. PXR activation regulates CYP3A4, but not CYP24A1, expression in human hepatocytes and in vivo, as well as the hepatic formation of specific monohydroxy metabolites of 25OHD3. Long-term treatment with certain drugs which are PXR ligands could up-regulate CYP3A4 expression, enhance 4-hydroxylation of 25OHD3 and eventually decrease circulating levels of 25OHD3. The current study supports an important role of PXR in vitamin D and bone homeostasis. In addition, it establishes a potential therapeutic target for clinical treatment or prevention of drug-induced osteomalacia by modulation of CYP3A4-mediated catabolic pathway in the liver and the small intestine.
Supplementary Material
Acknowledgments
This work was supported in part by grants from the National Institutes of Health: R01 GM063666 (K.E.T.), R01 GM079280 (D.L.E.), a Clinical and Translational Science Award, UL1 RR025014, NIEHS Center Grant, P30 ES07033 (Y.S.L, D.L.E, J.W.L., D.D.S., and K.E.T.) and T32 ES07032 (E.J.P.). The authors would like to thank Lisa Levy and Julia Tracy for their assistance in the conduct of the clinical study and Amgen Inc. for laboratory resources.
Abbreviations
- ADR
adverse drug reaction
- CAR
constitutive androstane receptor
- CYP24A1
cytochrome P450 24A1
- CYP27A1
cytochrome P450 27A1
- CYP3A4
cytochrome P450 3A4
- CYP27B1
cytochrome P450 27B1
- CYP2R1
cytochrome P450 2R1
- DHB
6′,7′-dihydroxybergamottin
- MRM
multiple reaction monitoring
- 1α,25(OH)2D3
1α,25-dihydroxyvitamin D3
- 4α,25(OH)2D3
4α,25-dihydroxyvitamin D3
- 4β,25(OH)2D3
4β,25-dihydroxyvitamin D3
- 24R, 25(OH)2D3
24R,25-dihydroxyvitamin D3
- 25OHD3
25-hydroxyvitamin D3
- 1′-OH-M D Z
1′-hydroxymidazolam
- PTAD
4-phenyl-1,2,4-triazoline-3,5-dione
- PXR
pregnane X receptor
- SFN
sulforaphane
- UGT
UDP-glucuronosyltransferase
- VDR
vitamin D receptor
Footnotes
All authors state that they have no conflicts of interest.
Authors’ roles: Study design – ZW, YSL, DLE, KET; Data acquisition – ZW, YSL, LJD, EJP; Data analysis – ZW, LJD, KET; Drafting manuscript – ZW, DLE, JWL, DDS, CLD, MSC, KET; All authors approved the final version of the submitted manuscript.
References
- 1.Pack AM, Gidal B, Vazquez B. Bone disease associated with antiepileptic drugs. Cleve Clin J Med. 2004;71(Suppl 2):S42–8. doi: 10.3949/ccjm.71.suppl_2.s42. [DOI] [PubMed] [Google Scholar]
- 2.Pack AM. Treatment of epilepsy to optimize bone health. Curr Treat Options Neurol. 2011;13 (4):346–54. doi: 10.1007/s11940-011-0133-x. [DOI] [PubMed] [Google Scholar]
- 3.Shah SC, Sharma RK, Hemangini, Chitle AR. Rifampicin induced osteomalacia. Tubercle. 1981;62(3):207–9. doi: 10.1016/0041-3879(81)90008-8. [DOI] [PubMed] [Google Scholar]
- 4.Brodie MJ, Boobis AR, Hillyard CJ, Abeyasekera G, Stevenson JC, MacIntyre I, Park BK. Effect of rifampicin and isoniazid on vitamin D metabolism. Clin Pharmacol Ther. 1982;32(4):525–30. doi: 10.1038/clpt.1982.197. [DOI] [PubMed] [Google Scholar]
- 5.Peterson P, Gray P, Tolman KG. Calcium balance in drug-induced osteomalacia: response to vitamin D. Clin Pharmacol Ther. 1976;19(1):63–7. doi: 10.1002/cpt197619163. [DOI] [PubMed] [Google Scholar]
- 6.Bhan A, Rao AD, Rao DS. Osteomalacia as a result of vitamin D deficiency. Endocrinol Metab Clin North Am. 2010;39(2):321–31. doi: 10.1016/j.ecl.2010.02.001. table of contents. [DOI] [PubMed] [Google Scholar]
- 7.Pascussi JM, Robert A, Nguyen M, Walrant-Debray O, Garabedian M, Martin P, Pineau T, Saric J, Navarro F, Maurel P, Vilarem MJ. Possible involvement of pregnane X receptor-enhanced CYP24 expression in drug-induced osteomalacia. J Clin Invest. 2005;115(1):177–86. doi: 10.1172/JCI21867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu Y, Hashizume T, Shuhart MC, Davis CL, Nelson WL, Sakaki T, Kalhorn TF, Watkins PB, Schuetz EG, Thummel KE. Intestinal and hepatic CYP3A4 catalyze hydroxylation of 1α,25-dihydroxyvitamin D3: implications for drug-induced osteomalacia. Mol Pharmacol. 2006;69(1):56–65. doi: 10.1124/mol.105.017392. [DOI] [PubMed] [Google Scholar]
- 9.Zhou C, Assem M, Tay JC, Watkins PB, Blumberg B, Schuetz EG, Thummel KE. Steroid and xenobiotic receptor and vitamin D receptor crosstalk mediates CYP24 expression and drug-induced osteomalacia. J Clin Invest. 2006;116(6):1703–12. doi: 10.1172/JCI27793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brodie MJ, Boobis AR, Dollery CT, Hillyard CJ, Brown DJ, MacIntyre I, Park BK. Rifampicin and vitamin D metabolism. Clin Pharmacol Ther. 1980;27(6):810–4. doi: 10.1038/clpt.1980.115. [DOI] [PubMed] [Google Scholar]
- 11.Lee RH, Lyles KW, Colon-Emeric C. A review of the effect of anticonvulsant medications on bone mineral density and fracture risk. Am J Geriatr Pharmacother. 2010;8(1):34–46. doi: 10.1016/j.amjopharm.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeLuca HF. Evolution of our understanding of vitamin D. Nutr Rev. 2008;66(10 Suppl 2):S73–87. doi: 10.1111/j.1753-4887.2008.00105.x. [DOI] [PubMed] [Google Scholar]
- 13.Burt R, Freston JW, Tolman KG. The influence of phenobarbital on biotransformation of 25-hydroxycholecalciferol. J Clin Pharmacol. 1976;16(8–9):393–8. doi: 10.1002/j.1552-4604.1976.tb02413.x. [DOI] [PubMed] [Google Scholar]
- 14.Eastwood JB, de Wardener HE, Gray RW, Lemann JL., Jr Normal plasma-1,25-(OH)2-vitamin-D concentrations in nutritional osteomalacia. Lancet. 1979;1(8131):1377–8. doi: 10.1016/s0140-6736(79)92012-9. [DOI] [PubMed] [Google Scholar]
- 15.Pack AM, Morrell MJ. Epilepsy and bone health in adults. Epilepsy Behav. 2004;5(Suppl 2):S24–9. doi: 10.1016/j.yebeh.2003.11.029. [DOI] [PubMed] [Google Scholar]
- 16.Sakaki T, Sawada N, Komai K, Shiozawa S, Yamada S, Yamamoto K, Ohyama Y, Inouye K. Dual metabolic pathway of 25-hydroxyvitamin D3 catalyzed by human CYP24. Eur J Biochem. 2000;267(20):6158–65. doi: 10.1046/j.1432-1327.2000.01680.x. [DOI] [PubMed] [Google Scholar]
- 17.Prosser DE, Jones G. Enzymes involved in the activation and inactivation of vitamin D. Trends Biochem Sci. 2004;29(12):664–673. doi: 10.1016/j.tibs.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 18.Hashizume T, Xu Y, Mohutsky MA, Alberts J, Hadden C, Kalhorn TF, Isoherranen N, Shuhart MC, Thummel KE. Identification of human UDP-glucuronosyltransferases catalyzing hepatic 1alpha,25-dihydroxyvitamin D3 conjugation. Biochem Pharmacol. 2008;75(5):1240–50. doi: 10.1016/j.bcp.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christakos S, Ajibade DV, Dhawan P, Fechner AJ, Mady LJ. Vitamin D: Metabolism. Endocrinol Metab Clin North Am. 2010;39(2):243–253. doi: 10.1016/j.ecl.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohyama Y, Yamasaki T. Eight cytochrome P450s catalyze vitamin D metabolism. Front Biosci. 2004;9:3007–18. doi: 10.2741/1455. [DOI] [PubMed] [Google Scholar]
- 21.Wang Z, Lin YS, Zheng XE, Senn T, Hashizume T, Scian M, Dickmann LJ, Nelson SD, Baillie TA, Hebert MF, Blough D, Davis CL, Thummel KE. An Inducible Cytochrome P450 3A4-Dependent Vitamin D Catabolic Pathway. Mol Pharmacol. 2012;81(4):498–509. doi: 10.1124/mol.111.076356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Z, Senn T, Kalhorn T, Zheng XE, Zheng S, Davis CL, Hebert MF, Lin YS, Thummel KE. Simultaneous measurement of plasma vitamin D(3) metabolites, including 4beta,25-dihydroxyvitamin D(3), using liquid chromatography-tandem mass spectrometry. Anal Biochem. 2011;418(1):126–33. doi: 10.1016/j.ab.2011.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ryan MJ, Johnson G, Kirk J, Fuerstenberg SM, Zager RA, Torok-Storb B. HK-2: an immortalized proximal tubule epithelial cell line from normal adult human kidney. Kidney Int. 1994;45(1):48–57. doi: 10.1038/ki.1994.6. [DOI] [PubMed] [Google Scholar]
- 24.Zheng XE, Wang Z, Liao MZ, Lin YS, Shuhart MC, Schuetz EG, Thummel KE. Human PXR-mediated induction of intestinal CYP3A4 attenuates 1alpha,25-dihydroxyvitamin D(3) function in human colon adenocarcinoma LS180 cells. Biochem Pharmacol. 2012;84(3):391–401. doi: 10.1016/j.bcp.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Houston JB. Drug metabolite kinetics. Pharmacol Ther. 1981;15(3):521–52. doi: 10.1016/0163-7258(81)90056-5. [DOI] [PubMed] [Google Scholar]
- 26.Paine MF, Shen DD, Kunze KL, Perkins JD, Marsh CL, McVicar JP, Barr DM, Gillies BS, Thummel KE. First-pass metabolism of midazolam by the human intestine. Clin Pharmacol Ther. 1996;60(1):14–24. doi: 10.1016/S0009-9236(96)90162-9. [DOI] [PubMed] [Google Scholar]
- 27.Thummel KE, O’Shea D, Paine MF, Shen DD, Kunze KL, Perkins JD, Wilkinson GR. Oral first-pass elimination of midazolam involves both gastrointestinal and hepatic CYP3A-mediated metabolism. Clin Pharmacol Ther. 1996;59(5):491–502. doi: 10.1016/S0009-9236(96)90177-0. [DOI] [PubMed] [Google Scholar]
- 28.Karaaslan Y, Haznedaroglu S, Ozturk M. Osteomalacia associated with carbamazepine/valproate. Ann Pharmacother. 2000;34(2):264–5. doi: 10.1345/aph.19099. [DOI] [PubMed] [Google Scholar]
- 29.Andress DL, Ozuna J, Tirschwell D, Grande L, Johnson M, Jacobson AF, Spain W. Antiepileptic drug-induced bone loss in young male patients who have seizures. Arch Neurol. 2002;59(5):781–6. doi: 10.1001/archneur.59.5.781. [DOI] [PubMed] [Google Scholar]
- 30.Petty SJ, O’Brien TJ, Wark JD. Anti-epileptic medication and bone health. Osteoporos Int. 2007;18(2):129–42. doi: 10.1007/s00198-006-0185-z. [DOI] [PubMed] [Google Scholar]
- 31.Carbone LD, Johnson KC, Robbins J, Larson JC, Curb JD, Watson K, Gass M, Lacroix AZ. Antiepileptic drug use, falls, fractures, and BMD in postmenopausal women: findings from the women’s health initiative (WHI) J Bone Miner Res. 2010;25(4):873–81. doi: 10.1359/jbmr.091027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lehmann JM, McKee DD, Watson MA, Willson TM, Moore JT, Kliewer SA. The human orphan nuclear receptor PXR is activated by compounds that regulate CYP3A4 gene expression and cause drug interactions. J Clin Invest. 1998;102(5):1016–23. doi: 10.1172/JCI3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng J, Ma X, Gonzalez FJ. Pregnane X receptor- and CYP3A4-humanized mouse models and their applications. Br J Pharmacol. 2011;163(3):461–8. doi: 10.1111/j.1476-5381.2010.01129.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goodwin B, Hodgson E, D’Costa DJ, Robertson GR, Liddle C. Transcriptional regulation of the human CYP3A4 gene by the constitutive androstane receptor. Mol Pharmacol. 2002;62(2):359–65. doi: 10.1124/mol.62.2.359. [DOI] [PubMed] [Google Scholar]
- 35.Burk O, Koch I, Raucy J, Hustert E, Eichelbaum M, Brockmoller J, Zanger UM, Wojnowski L. The induction of cytochrome P450 3A5 (CYP3A5) in the human liver and intestine is mediated by the xenobiotic sensors pregnane X receptor (PXR) and constitutively activated receptor (CAR) J Biol Chem. 2004;279(37):38379–85. doi: 10.1074/jbc.M404949200. [DOI] [PubMed] [Google Scholar]
- 36.Gascon-Barre M, Demers C, Mirshahi A, Neron S, Zalzal S, Nanci A. The normal liver harbors the vitamin D nuclear receptor in nonparenchymal and biliary epithelial cells. Hepatology. 2003;37(5):1034–42. doi: 10.1053/jhep.2003.50176. [DOI] [PubMed] [Google Scholar]
- 37.Vicchio D, Yergey A, O’Brien K, Allen L, Ray R, Holick M. Quantification and kinetics of 25-hydroxyvitaminD3 by isotope dilution liquid chromatography/thermospray mass spectrometry. Biol Mass Spectrom. 1993;22(1):53–8. doi: 10.1002/bms.1200220107. [DOI] [PubMed] [Google Scholar]
- 38.Schuster I. Cytochromes P450 are essential players in the vitamin D signaling system. Biochim Biophys Acta. 2011;1814(1):186–199. doi: 10.1016/j.bbapap.2010.06.022. [DOI] [PubMed] [Google Scholar]
- 39.Paine MF, Khalighi M, Fisher JM, Shen DD, Kunze KL, Marsh CL, Perkins JD, Thummel KE. Characterization of interintestinal and intraintestinal variations in human CYP3A-dependent metabolism. J Pharmacol Exp Ther. 1997;283(3):1552–62. [PubMed] [Google Scholar]
- 40.von Richter O, Burk O, Fromm MF, Thon KP, Eichelbaum M, Kivisto KT. Cytochrome P450 3A4 and P-glycoprotein expression in human small intestinal enterocytes and hepatocytes: a comparative analysis in paired tissue specimens. Clin Pharmacol Ther. 2004;75(3):172–83. doi: 10.1016/j.clpt.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 41.Higashi T, Horike M, Kikuchi R, Shimada K. In vitro and in vivo glucuronidation of 24,25-dihydroxyvitamin D3. Steroids. 1999;64(10):715–25. doi: 10.1016/s0039-128x(99)00057-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.