Abstract
Cancer-mediated immune dysfunction contributes to tumor progression and correlates with patient outcome. Metastasis to tumor draining lymph nodes (TDLNs) is an important step in breast cancer progression and is used to predict patient outcome and survival. While lymph nodes are important immune organs, the role of immune cells in TDLNs has not been thoroughly investigated. We hypothesized that the host immune response in node negative (NN) patients is more intact and thereby can resist tumor invasion compared to node positive (NP) patients. As such, lymph node metastasis requires breakdown of the host immune response in addition to escape of cancer cells from the tumor. To investigate the immunological differences between NN and NP breast cancer patients, we purified and profiled immune cells from the three major compartments where cancer and immune cells interact: tumor, TDLNs, and peripheral blood. Significant down-regulation of genes associated with immune-related pathways and up-regulation of genes associated with tumor-promoting pathways was consistently observed in NP patients’ TDLNs compared to NN patients. Importantly, these signatures were seen even in NP patients’ tumor-free TDLNs, suggesting that such immune changes are not driven solely by local tumor invasion. Furthermore, similar patterns were also observed in NP patients’ tumor and blood immune cells, suggesting that immunological differences between NN and NP patients are systemic. Together, these findings suggest that alterations in overall immune function may underlie risk for LN metastasis in breast cancer patients.
Keywords: breast cancer, lymph node metastasis, immune profile
Introduction
Tumor modulation of the host immune response is involved in cancer development, progression and metastasis, contributing to tumor escape and failure of therapies. The tumor microenvironment was shown to be polarized towards chronic inflammatory states, leading to impaired immune cell killing of tumor cells and tumor escape 1. CD8+ T-cell infiltration into the tumor is associated with better patient survival 2, while the presence of T-regulatory cells and macrophages predicted worse relapse-free and overall survival 3. Thus, immune status of patients is an important factor in determining tumor progression and clinical outcome.
Immune gene signatures have been observed in whole tumor samples 4, 5. Rarely have immune cells been purified from heterogeneous tumor tissues and profiled independently. In addition, while lymph nodes (LN) are important immune organs, where primary immune responses to antigens are initiated, most studies focus solely on the presence or absence of tumor cells in tumor-draining lymph nodes (TDLN) 6, 7 and ignore their immune status. For tumor cells to survive within TDLNs, impairment of the immune response is likely a critical step in LN invasion and may precede LN metastasis.
Various alterations have been reported in immune cells from TDLNs. We have previously shown that decreased T-cell and dendritic cell proportions in both tumor-invaded and tumor-free TDLNs strongly correlated with worse clinical outcome in breast cancer 8. Quantitative analysis of spatial characteristics of immune cells revealed novel differences in grouping patterns of T and B cells within LNs from healthy vs. breast cancer patients 9. Studies have also shown greater numbers of activated immune cells in metastatic LNs compared to tumor-free LNs 10. In addition, tumor-free LNs were found to be immunologically competent and potentially a site of tumor specific T-cell activation, as evidenced by the presence of greater numbers of mature dendritic cells and cytokine-producing cells 11. Changes in immune cells were also reported in other cancer types. T-regulatory cell populations were higher in regional LNs compared to control LNs in gastric cancer patients 12. Reduced numbers of peritumoural CD8+ T-cells were found in metastatic LNs compared to uninvolved regional nodes in head and neck cancer patients, suggesting a local down-modulation of cellular immunity 13. In addition, development of prostate cancer LN metastasis was shown to correlate with decreased immune LN reactivity 14. The mechanisms by which cancer affects immune populations within TDLNs, especially tumor-free nodes, remain unclear.
In this study, we profiled purified immune cells from breast cancer patients’ TDLNs, as well as from their primary tumor and peripheral blood, to reveal differences in immune-related genes and pathways between node negative (NN) and node positive (NP) patients. We found significant down-regulation of genes associated with immune-related pathways, and up-regulation of tumor-promoting pathways in NP patients’ TDLNs, including NP patients’ tumor-free TDLNs. Similar patterns were observed in NP patients’ immune cells from the primary tumor and peripheral blood, suggesting systemic differences between NP and NN patients in immune system function. These findings confirm the importance of studying immune cell function in the setting of breast cancer, and point to patient-specific differences in immune system function based on LN invasion status.
Materials and Methods
Clinical Samples
A total of 11 primary tumor, 30 lymph nodes and 21 peripheral blood specimens were obtained from 22 breast cancer patients at Stanford Hospital. Patients’ clinico-pathological characteristics are shown in Table 1. Written informed consent was obtained from all patients and the study was approved by Stanford University’s Institutional Review Board.
Table 1.
Patient samples used for microarray analysis
| # of patients | # of arrays | stage | Tumor type | Mean age | ER | PRa | |||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| NN | SLN | 6 | 14 | 0–1: n=6 | DCIS: n=2 | 61.3±10.3 | P: n=7 | P: n=7 | |
|
|
|||||||||
| NSLN | 1 | 1 | 2: n=3 | IDC/DCIS: n=5 | N: n=2 | N: n=2 | |||
|
|
|||||||||
| Primary tumor | 4 | 4 | IDC/LCIS: n=1 | ||||||
|
|
|||||||||
| Peripheral blood | 8 | 8 | ILC/LCIS: n=1 | ||||||
|
| |||||||||
| NP | SLN | T + | 7 | 8 | 2: n=9 | IDC: n=4 | 55.1±13.6 | P: n=11 | P: n=9(+1) |
|
|
|||||||||
| T − | 10 | 16 | 3: n=3 | IDC/DCIS: n=7 | N: n=2 | N: n=3(+1) | |||
|
|
|||||||||
| NSLN | T + | 1 | 1 | 4: n=1 | ILC/LCIS: n=2 | ||||
|
|
|||||||||
| T − | 5 | 22 | |||||||
|
|
|||||||||
| Primary tumor | 7 | 8 | |||||||
|
|
|||||||||
| Peripheral blood | 13 | 13 | |||||||
(+1) refers to a bilateral patient that was PR+ on the right side, and PR-on the left side.
NN = node negative patients; NP = node positive patients; SLN = sentinel lymph nodes; NSLN = non sentinellymph nodes; T+ = tumor-invaded; T− = tumor-free; DCIS = ductal carcinoma in situ; IDC = invasive ductal carcinoma; LCIS = lobular carcinoma in situ; ILC = invasive lobular carcinoma; P=positive; N=negative; n=number of patients.
Isolation of peripheral blood immune cells
Whole blood was collected in heparin tubes and peripheral blood mononuclear cells (PBMCs) were separated by Ficoll-Hypaque density gradient centrifugation (GE Healthcare). Red blood cell (RBC) lysis buffer treatment was used to remove residual RBCs. A total of 1×106 isolated PBMCs were homogenized in TRIzol (Invitrogen) and stored at −80°C.
Processing and purification of immune and tumor cells from lymph node and primary tumor tissue
Fresh sentinel lymph node, non-sentinel lymph node (SLN and NSLN, respectively) and primary tumor tissues were collected after surgical resection. SLNs were defined by the surgeon based on lymphoscintigraphy and/or gross blue dye inspection. All other LNs not identified by the surgeon as SLNs were considered NSLNs. Fine-needle aspirates were used to collect cells from SLNs. For NSLNs, a small portion of each node was collected and minced to generate single cell suspensions which were filtered through a 70 micron strainer. Primary tumor tissue was collected and the presence of tumor cells in the primary tissue was confirmed by H&E staining from a bisected portion of the received tissue and examined by a pathologists. For grossly tumor involved LNs and primary tumor tissue, minced specimens were enzymatically dissociated with 200units/ml Collagenase-III (Worthington Biochemical Corp.) and 10Kunitz units/ml DNaseI (Sigma) for 30 min-1hr or 1hr–2hr at 37°C, respectively. The digestion process was stopped by addition of M199 10%FBS (Gibco, Invitrogen). Single cell suspensions were generated by filtering cells through a 70 micron strainer followed by a 40 micron strainer (BD Biosciences) followed by RBC lysis buffer treatment.
Purification of immune cells and tumor cells from lymph nodes and primary tumor tissue using flow cytometry
Cells were stained with pan-leukocyte marker CD45-PE-Cy7, fibroblast marker CD140β-PE (both BD Biosciences), epithelial specific antigen ESA-FITC (Biolegend or Biomeda), and a dead cell exclusion marker ViViD (Invitrogen). ESA+CD45-CD140β-tumor cells or ESA-CD45+CD140β-immune cells were sorted using FACSAria (BD Bioscience). Up to 1×106 purified cells (>95%) were homogenized in 1mL TRIzol and stored at −80°C.
Whole genome microarray
Total RNA was extracted using the TRIzoL method and amplified using TrueLabeling-PicoAMPTM kit (QIAGEN), followed by Cy3/Cy5 labeling (GE healthcare). Cy-labeled patient samples were mixed with the same amount of reverse color Cy-labeled universal human reference (UHR) cRNA (Stratagene Corp.) and hybridized to Agilent’s Whole Human Genome Microarray 4×44K. Image files were generated from microarray slides using Agilent Microarray Scanner G2505B. Microarray data is deposited in GEO, accession number: GSE41986.
Microarray analysis
Microarray analysis was performed using R (http://www.r-project.org/) and Bioconductor (http://www.bioconductor.org/). Quantile and LOESS normalization were used to normalize the raw data. Arrays were calibrated to the same scale and variance-stabilizing transformation was applied to the data. The generalized log ratio values for each gene were taken to obtain a relative expression level in samples over the UHR. Probes annotating the same gene whose expression correlated (correlation coefficient > 0.7) were averaged. Probes with no EntrezID, gene ontology (GO) term annotation, or among the 70% probes with the least variance across all microarrays were filtered out. Differentially expressed genes were determined using the Mann-Whitney U test. Differentially expressed genes were filtered for their 1.5 mean fold-change difference between the groups compared. In NN vs. NP patients’ peripheral blood comparison a cutoff of 1.3 was used as a 1.5 cutoff yielded too few genes.
Gene lists were further annotated by association to terms and pathways, using the following tools. Ingenuity pathway analysis (IPA, Ingenuity® Systems) core analysis was performed and canonical pathways were regarded to delineate association of genes with known pathways. Only significant pathways (P-value<0.05) were regarded. DAVID (http://david.abcc.ncifcrf.gov/) functional annotation tool was used to associate gene lists with KEGG pathways. All pathways, including non-significant pathways (trends) were regarded, as KEGG does not yield associated pathways for small gene lists. The hypergeometric test (bioconductor GO and GOstats packages) was used to find the biological process GO terms with a larger than expected (over-representation) subset of differentially expressed genes in their annotation list. Only significant terms (P-value<0.05) with more than two genes (count>2) were regarded. The gene lists examined using these tools included all significantly differentiated genes, prior to the 1.5 fold-change cutoff.
The resulting terms and pathways were further categorized into two main categories: immune-related and other. The immune-related category consisted of terms/pathways including the word “immune”, an immune related population (eg. “T cell”), cytokines (eg. “IL2”), or if genes associated with the term/pathway were immune-related. The “other” category consisted of terms/pathways relating to cell-cycle, DNA-repair, ubiquitin and tumor promoting processes. Tumor promoting terms/pathways were determined according to relevant literature. IPA/KEGG pathways considered as tumor promoting are highlighted and referenced in Supplementary Table 1.
The genes associated with each of the immune-related categorized pathways were enumerated based on the fold-change differences between the expression values of the NN and NP patient groups in each tissue, as follows. Given a gene-list associated with an immune-related pathway that is up-regulated in patient group A (e.g. NN patient’ LNs), the fold-change between the expression value of each gene in that gene-list versus the mean expression value of this gene in the group it was compared to (e.g. NP patients’ LNs) was enumerated for each patient in group A. To emphasize the fold-change differences, each fold-change value was categorized to the following ranges: 0<fold-change<=1.3 → 1; 1.3<fold-change<=2 → 2; fold-change>2 → 4. The fold-change values for all genes in the gene-list were averaged per patient. Statistical differences between the fold-change values in the NN vs. NP groups were computed using Student’s t-test.
Results
Purified immune cells from NP patients’ LNs exhibit lower immune-related signatures compared to NN patients
Microarray gene expression analysis was performed on purified immune cells from TDLNs, including tumor-invaded and tumor-free sentinel LNs (SLN) and tumor-free non-SLN (NSLN), to investigate potential differences in immune cell signatures between NN and NP breast cancer patients (Table 1). To account for alterations in immune cell signatures due solely to tumor invasion, data from NN patients’ LNs were compared separately to each NP patients’ tumor-invaded SLNs, tumor-free SLNs and tumor-free NSLNs (Table 2).
Table 2.
Summary of NN vs. NP patients’ immune cell signatures from the primary tumor, lymph node and peripheral blood comparisons
NN

|
NP

|
|---|---|
|
| |
| Primary tumor | |
|
| |
| Immune | Cell cycle |
| DNA repair | |
| Tumor promoting | |
| Ubiquitination | |
| Immune (B-cell and tolerance related) | |
|
| |
| Lymph nodes | |
|
| |
| Immune | Cell cycle (T+ SLN) |
| DNA repair (T+ SLN) | |
| Immune (B-cell related; T+ SLN) | |
| Tumor promoting (T+/− SLN) | |
|
| |
| Peripheral blood | |
|
| |
| Immune | |
Summary of signatures associated with significantly differentiated genes in comparisons between NN vs. NP patients’ purified immune cells from the primary tumor, lymph nodes and peripheral blood. The left and right hand side columns represent signatures associated with genes up regulated in NN and NP patients, respectively. T+ SLN = tumor-invaded SLN; T−SLN = tumor-free SLN.
Comparison of NN SLNs with NP tumor-invaded SLNs yielded 103 significantly differentiated genes (Figure 1A). To further clarify the nature of these genes, they were associated with terms and pathways using three different tools – Gene Ontology biological processes (GO), Ingenuity pathway analysis (IPA) and KEGG pathways using DAVID functional annotation. Genes up-regulated in NN patients’ SLNs were found to be associated with pro-inflammatory immune related pathways such as TREM1 signaling. Genes up-regulated in the NP tumor-invaded SLNs were highly associated with cell-cycle, DNA-repair pathways and tumor-promoting pathways such as angiopoietin signaling 15, in addition to B-cell related genes such as AICDA (activation-induced cytidine deaminase, expressed by germinal center (GC) B-cells) and immunoglobulin genes (Table 3).
Figure 1. Comparison between immune cells from NN and NP patient LNs.
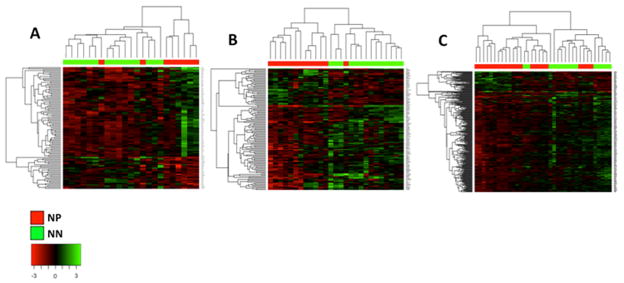
Heatmaps depicting significantly differentiated genes with fold-change differences larger than 1.5 between NN and NP patients’ LNs. All three comparisons show clustering of the NN and NP patient groups. The NN (green) and NP (red) group clusters are shown on the top and the gene clusters are shown on the left. Gene expression values are mean-centered. (A) comparison between immune cells from NN and NP patients’ tumor-invaded SLNs; (B) comparison between immune cells from NN and NP patients’ tumor-free SLNs; (C) comparison between immune cells from NN and NP patients’ tumor-free NSLNs.
Table 3.
Genes and pathways associated with up-regulated genes in NN vs. NP comparisons
| Tissue | Cell type | NN Up-regulated |
NP Up-regulated |
|---|---|---|---|
| Lymph node (NP: T+ SLN, NN: SLN) | Purified immune | TREM1 Signaling (NOD2, TLR5) | Cell Cycle (RAD51, KIF23, PLK4), DNA Repair (RFC2, BRIP1); Angiopoietin Signaling (RASA1, BIRC5); B cell related (AICDA, IGKC, IGKV1-5, IGKV3-20) |
| Lymph node (NP: T-SLN, NN: SLN) | Purified immune | Antigen Presentation (HLA-DQA1, HLA-DQA2); TREM1 Signaling (IL8, PLCG2, TLR7, IL1B, IL6); IL17 Signaling (IL8, PTGS2, IL6) |
Relaxin Signaling (PDE3B, PDE8B, PDE6D) |
| Lymph node (NP: T-NSLN, NN: SLN) | Purified immune | Antigen Presentation (HLA-DOA, HLA-A, HLA-DRB3); Lymphocyte Activation (HLA-DOA, IL23A, IL4, PLCG2, TICAM1); Cytokine-cytokine Receptor Interaction (IL12RB2, IL4, CCR8, TNFRSF21, IL23A, IL3RA) |
Immune signaling related (IL6ST, CD86) |
| Primary tumor | Purified tumor | Innate Immune Response (MIF, IFIH1, NCF2); Inflammatory Response (MIF, CCL19, ACE2, SELE); HIF1a Signaling (MMP12, MMP19) |
|
| Primary tumor | Purified immune | Defense Response (C4B, C8A, IFNA21, CXCL3); NK cell mediated cytotoxicity (FCER1G, ITGAL, RAC3, IFNA21) |
T Cell Receptor Signaling (IL4, BCL10, TNF, CD8B, NFKB1, IFNG, IL2); Cell Cycle (CDK7, CDC26); DNA repair (RAD17, REV3L, RAD23B, GADD45B); IGF-1 Signaling (IGF1, FOXO1, IRS2, SOCS5); mTOR Signaling (MTOR, NRAS, EIF4A2, FNBP1); HMGB1 signaling (HMGB1, TNF, NFKB1, IFNG); Ubiquitin related (UBE2A, UBE4A, UBE3A, UBE2G1) |
| Peripheral blood | Purified immune | Lymphocyte Activation (BLNK, IKZF1, IL7); B cell related (CD22, HLA-DOB, POU2AF1) |
|
| Systemic analysis (Similar expression pattern) | Purified immune | T cell/B cell/Ag presentation related (IGHG1, CR1, HLA-DQA2); | Inflammatory response related (IL10RA, TNF, IL18R1); Protein Ubiquitination (UCHL3, UBE2A, UBE2D2); HIF1a Signaling (SLC2A5, GTF2A1); RhoA Signaling (ACTR2, CFL2); Glucocorticoid Receptor Signaling (GTF2A2, PRKAB2, ANXA1) |
| Systemic analysis (TlowLNhighPBhigh pattern) | Purified immune | Antigen Presentation (HLA-A, HLA-DOB); T/B cell signaling (PCLG2, IFNAR1, IL7) |
Cell Cycle (CDK7, CDC26, CDC23, TOP2B); mTOR Signaling (KRAS, EIF3E, PRKD3); HMGB1 Signaling (HMGB1, KAT2B, KRAS, FNBP1); Breast Cancer Regulation by Stathmin1 Signaling (ROCK1 KRAS, PRKD3) |
| Systemic analysis (ThighLNlowPBlow pattern) | Purified immune | NK/Dendritic Cell Signaling (IFNA21, TYROBP, ITGAL, PLA2G6); HIF1a Signaling (MMP28, EGLN3); Eicosanoid Receptor Signaling (PLA2G6, ALOX12B, PTGDS); Ephrin Receptor Signaling (EFNB3, GRIN2D, LIMK2, RAC3) |
IGF-1 Signaling (IGF1,NOV, IRS2); ILK Signaling (MYC, IRS2) |
Examples of pathways associated with up-regulated genes in the various NN vs. NP patients’ comparisons. The full list of pathways and associated genes is given in Supplementary Table 1 (for the single compartment comparisons) and Supplementary Table 2 (for the systemic comparisons). NN = node negative patients; NP = node positive patients; SLN = sentinel lymph nodes; NSLN = non-sentinel lymph nodes; T+ = tumor-invaded; T−= tumor-free; Systemic = analysis across all primary tumor, LN and peripheral blood tissues.
Comparison of NN SLNs with NP tumor-free SLNs yielded 116 significantly differentiated genes (Figure 1B). The genes up-regulated in NN patients were highly associated with immune cell regulation and signaling pathways such as Antigen presentation in addition to pro-inflammatory immune cell signaling pathways such as TREM1 and IL-17 signaling. In contrast, none of the pathways up regulated in NP patients’ tumor-free SLNs were related to immune cell regulation or signaling, but instead were associated with tumor-promoting pathways, such as relaxin signaling16 and various metabolic and signaling pathways (Table 3).
Comparison of NN patients’ SLNs with NP patients’ tumor-free NSLNs yielded 219 significantly differentiated genes (Figure 1C). The genes up regulated in the NN SLNs were highly associated with immune cell regulation/signaling pathways such as antigen processing and presentation, lymphocyte activation and cytokine-cytokine receptor interaction. Only a negligible number of genes up regulated in tumor-free NSLNs from NP patients were related to immune cell signaling (Table 3).
The significantly differentiated genes associated with immune related pathways were used to enumerate the fold-change differences between NN and NP patients in each LN compartment examined, as described in the methods section (Figure 2A). The immune related genes had significantly higher fold-change values in NN patients compared to NP patients in all LN compartments. These results point to down-regulation of immune-related pathways in NP patients’ immune cells. Given that immune cells from NP patients were found to up-regulate genes associated with other, non-immune-related pathways such as cell cycle, DNA repair and tumor promoting processes, these results suggest a deviation in the processes in which NP patients’ lymphatic immune cells participate, which may be directly tumor-derived in tumor-invaded LNs or indirectly in tumor-free LNs.
Figure 2. Fold-change differences between genes associated with immune-related pathways in NN and NP patients.
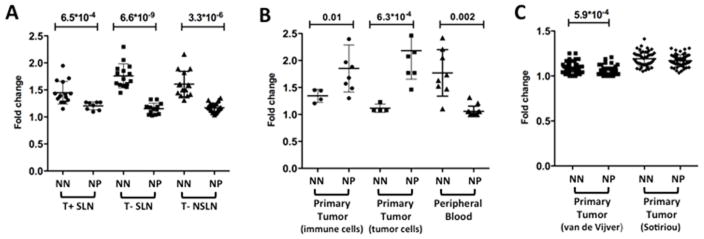
Patient specific fold change differences between genes associated with immune-related pathways resulting from NN vs. NP patient comparison in each of the comparisons performed. (A) NN vs. NP patients’ purified immune cell genes from tumor-invaded SLNs (T+SLN), tumor-free SLN (T-SLN) and tumor-free NSLNs (T-NSLN). (B) NN vs. NP patients’ purified immune and tumor cells from the primary tumor and purified immune cells from the peripheral blood. (C) NN vs. NP patients’ heterogeneous cell population from the primary tumor in two publically available datasets – van de Vijver et al. 4 (295 patients: 151 NN, 144 NP) and Sotiriou et al. 5 (99 patients: 46 NN, 53 NP). The numbers above the comparison represent significant P-values obtained in Student’s t-test. NN = Node negative patients; NP = Node positive patients. Similar results were obtained using the gene expression values rather than the fold change differences (Supplementary Figure 1).
Differences in immune cell signatures observed between NN and NP patients’ primary tumor and peripheral blood
Microarray analysis was also performed with purified immune cells and purified tumor cells from patients’ tumor compartment (Table 1). Comparison of immune cells from NN with NP patients’ primary tumor yielded 1267 significantly differentiated genes (Supplementary Figure 2A). Genes up-regulated in both NN and NP patients were associated with immune cell regulation/signaling pathways such as Defense response and Natural killer cell mediated cytotoxicity in NN patients and T cell receptor signaling in NP patients. However, genes up-regulated in NP patients were also associated with cell-cycle, DNA-repair and tumor-promoting pathways such as IGF-1 17, mTOR 18 and HMGB1 signaling 19, consistent with the pathways observed in NP patients’ LNs, in addition to ubiquitin related pathways (Table 3). Fold-change values for genes associated with immune related pathways were significantly higher in NP patients compared to NN patients (P-value = 0.01, Figure 2B). However, a closer look at these pathways reveals up-regulation of processes such as T cell anergy and regulation of tolerance. In addition, many B-cell and GC B-cell related pathways (e.g. B-cell differentiation, isotype switching, immunoglobulin secretion) were up regulated in NP patients in addition to other tumor-promoting pathways, consistent with our observations in NP patients’ T+ SLNs.
Purified tumor cells from patients’ primary tumor were examined. The comparison between NN and NP patients yielded 71 differentially expressed genes, which separated the NN and NP patient groups (Supplementary Figure 2B). The genes up-regulated in NP patients were associated with immune cell regulation and signaling pathways such as Inflammatory Response and Innate Immune Response in addition to tumor-promoting pathways such as HIF1a 20 signaling (Table 3). Indeed, fold-change values for immune related genes were significantly higher in NP patients compared to NN patients (P-value = 6.3*10−4, Figure 2B). However, the scores were computed based on immune-related genes that were up-regulated in NP patients only. Similar to the purified immune cell signatures from the same compartment, a close examination of these genes shows that they are mainly involved in inflammatory processes, consistent with prior observations that neoplastic microenvironments favor chronic inflammatory states 1.
The tumor and immune cells in the NP patients’ primary tumor compartment were involved in similar immune-related inflammatory processes and in addition the immune cells were involved in tumor promoting pathways and additional pathways consistent with our observations in NP patients’ LNs. These results suggest immune-tumor cell cross talk which may result in pro-tumor regulation of immune cells in NP patients’ primary tumor compartment.
Microarray analysis was also performed with purified immune cells from patients’ peripheral blood (Table 1). Comparison of NN with NP patients’ peripheral blood immune cells yielded 96 significantly differentiated genes (Supplementary Figure 2C). Genes up-regulated in NN patients were highly associated with immune regulation/signaling pathways such as Lymphocyte activation and B cell related pathways, consistent with some of our observations in the LN compartments. Few genes were up regulated in NP patients’ peripheral blood immune cells, none of which were immune cell signaling related (Supplementary Table 1). Similar to the LN compartments, the fold-change values for immune-related genes were significantly higher in NN patients compared to NP patients (P-value = 0.0002, Figure 2B).
Overall, purified immune cell signatures in NN and NP patients’ peripheral blood were consistent with the signatures observed in the LN compartments, whereas immune cell signatures from the primary tumor were different than the LN and peripheral blood compartments.
Profiles of heterogeneous tumor tissue cells could not distinguish between NN and NP patients
We next examined whether the signatures observed in NN and NP patients could be observed in gene expression studies of non-purified breast tumor tissues, consisting largely of tumor cells but also tumor-infiltrating immune cells and microenvironmental cells such as fibroblasts 1. We examined two large publically available datasets - Van de Vijver et al.4 and Sotiriou et al.5. In an NN vs. NP patient comparison using each of these datasets, none of the differentially expressed genes had a fold-change larger than 1.5 nor clearly separated the NN and NP groups (Supplementary Figure 3). Indeed, the fold-change values for immune-related genes were similar in both datasets (Figure 2C). The NN group in the Van De Vijver et al. dataset 4 was significantly higher compared to the NP group, however the actual fold-change values were very similar between the groups, with mean fold-change values that are very close to 1 in both groups (1.032 and 1.049 in the NN and NP groups, respectively). Thus, the significant P-value in this case does not indicate biological significance and results from the low standard deviations in the NN and NP groups (0.05 and 0.04, respectively).
These results demonstrate that purified tumor and immune cell signatures can better distinguish between NN and NP patients and reveal cell-type specific mechanisms compared to heterogeneous non-purified samples from tumor tissue.
Differences between NN and NP patients’ purified immune cell signatures are systemic
To further clarify the relationships between the pathways observed in the primary tumor, LNs and peripheral blood, we examined gene expression patterns amongst these three compartments. For this purpose, the significantly up-regulated genes in the NN and NP patient groups resulting from each NN vs. NP comparisons described above (i.e., LNs, primary tumor and peripheral blood purified immune cell comparisons) were gathered and their mean gene expression values in each NN and NP patient groups were evaluated for each compartment. That is, significantly differentiated genes resulting from a comparison in a certain compartment (e.g. NN vs. NP in the LNs) were examined in the other compartments (e.g. NN vs. NP in the peripheral blood and primary tumor) as well. IPA was used to associate the genes following various expression patterns across the compartments with corresponding pathways. The genes up-regulated in NP patients’ tumor-invaded and tumor-free SLNs (vs. NN patients’ SLNs) were combined as similar results were obtained using either tumor-invaded or tumor-free SLNs only (data not shown).
The first pattern examined was that of similar expression across all three compartments. The genes following this pattern were selected based on having a mean fold-change smaller than 1.5 across the three compartments. 74 genes up regulated in the NN group matched this pattern, including immune-related genes associated with B/T cells and antigen presentation. The 264 genes up regulated in the NP group with this trend were associated with inflammatory immune response, protein ubiquitination and tumor-promoting signaling pathways such as HIF1a 20, RhoA 21 and Glucocorticoid Receptor 22. These results are similar to our observations in the LN compartments, suggesting that some signatures observed in the LN compartments are also observed systemically (Table 3).
We next examined patterns with differences in gene expression values across the compartments. Amongst all patterns examined, the patterns representing differences between the primary tumor compartment and the other compartments – LNs and peripheral blood – were the most interesting, including the TlowLNhighPBhigh and ThighLNlowPBlow patterns (T=tumor, LN=lymph nodes, PB=peripheral blood). Additional patterns examined included ascending and descending gradient patterns across the compartments (e.g. ThighLNmedPBlow) in addition to all combinations of different expression patterns between all three compartments (e.g. ThighLNlowPBhigh, TlowLNhighPBlow, etc., Supplementary Table 2). The TlowLNhighPBhigh pattern included genes with a mean fold-change larger than 2 between the primary tumor compartment and the LNs or the peripheral blood compartments, where the gene expression in the primary tumor compartment was lower compared to the other compartments. In the NN group 63 genes followed this trend, mainly immune-related genes in T/B cell signaling and antigen presentation pathways. In the NP group, the 265 genes following this trend associated with cell-cycle and tumor-promoting signaling pathways such as mTOR 18, HMGB119 and Breast Cancer Regulation by Stathmin1 23 (Table 3).
The opposite pattern, ThighLNlowPBlow pattern, denoting higher gene expression in the primary tumor compartment (with a mean fold-change larger than 2) compared to the LNs and peripheral blood compartments, was also examined. The 184 genes up-regulated in NN patients following this pattern were associated with Natural Killer and Dendritic cell related pathways as well as tumor-promoting signaling pathways such as HIF1a 20, Eicosanoid Receptor 24 and Ephrin Receptor 25. In the NP group, the 33 genes following this pattern associated with tumor-promoting signaling pathways such as IGF-1 17 and ILK 26 (Table 3).
These results show similarities between the LNs and peripheral blood compartments and differences between these compartments and the primary tumor compartment in both NN and NP patients. The pathways associated with the genes following these patterns were consistent with the pathways observed in the LN comparisons between NN and NP patients, including up-regulation of immune-related pathways in NN patients, and cell cycle and tumor-promoting pathways in NP patients. In the primary tumor compartment both NN and NP patients’ signatures associated with tumor-promoting pathways, however NP patients’ signatures was also associated with tumor promoting pathways. Overall, the expression pattern analysis across the tumor, LNs and peripheral blood compartments suggests that the differences in purified immune cell signatures observed between NN and NP patients are systemic. NN patients up-regulate diverse immune-related signatures including antigen presentation while NP patients up-regulated genes are associated with inflammatory immune pathways, consistent with previous studies 1, and tumor-promoting pathways.
Discussion
The traditional temporal view of cancer metastasis is that once cancer cells acquire the ability to exit the primary tumor, they invade TDLNs, then distant organs. As such, LN invasion is inevitable over time. This view ignores the role of the host immune response, and that LNs are important immune organs that should resist tumor invasion. Recent data show that tumor cells disseminate early, before tumors become clinically detectable, but immunosurveillance limits metastatic outgrowth 27. Epidemiological analyses of breast cancer patients indicated that metastasis might be initiated already 5–7 years before diagnosis of the primary tumor 28, yet only a subset of patients develop clinically evident relapse. Under this new view, the host immune response plays an important role in suppressing micrometastases and determines if patients develop clinically evident metastasis in LNs or distant organs. In this study, we hypothesized that NN patients’ immune population within the LNs is more resistant to tumor-mediated immune modulation, and thus tumor invasion, compared to NP patients. We have found that NP patients’ immune-related signatures were found to be down-regulated, while other pathways such as cell cycle, DNA repair, ubiquitin and tumor-promoting signaling were up regulated compared to NN patients. These differences observed in the purified immune cells are likely present in patients prior to metastasis, as they were also observed in NN patients’ immune cells from non-invaded LNs as well as systemically. In addition, these differences were not observed in bulk heterogeneous tumor samples, illustrating the power of analyzing purified cell populations rather than heterogeneous tissue samples, and emphasizing the important functional status that immune cells play in determining LN metastasis.
NP patients’ immune-related signatures were found to be down regulated compared to NN patients in all LN types examined. These signatures included antigen presentation, leukocyte activation, cytokine mediated signaling and production and additional immune-related signaling pathways, thus suggesting that NP patients’ immune cell functions are reduced or dysfunctional compared to NN patients. Previous studies have reported changes in immune cell population and/or function in the setting of cancer. Breast cancer cells have been reported to inhibit antigen processing and presenting function of dendritic cells 29. In addition, decreased dendritic cell numbers were found in tumor-involved LNs as opposed to tumor-free LNs 8 and lower abundance of CD38+ activated lymphocytes was observed in metastatic LNs 14.
In addition to down-regulation of immune-related signatures, NP patients’ immune cells up-regulated genes associated with tumor promoting pathways such as relaxin, HIF1a and Angiopoietin signaling as well as several metabolic processes. Studies have shown that immune cells may be involved in tumor-promoting pathways such as angiogenesis and metastasis. Elevated relaxin serum levels positively correlated with breast cancer mestastasis and relaxin-mediated signaling was shown to enhance the migratory activity of leukocytes into primary tumors 30. The development of hypoxic regions is an indicator of poor prognosis in many tumors; HIF1a, the direct effector of hypoxia, was found to be involved in recruitment of myeloid cells to promote neovascularization in glioblastoma 31. Targeted deletion of HIF1a in macrophages resulted in reduced breast tumor growth 32. Myeloid cells were shown to have an important role in regulating angiogenesis in tumors 33. Moreover, immune cell metabolic changes caused by hypoxia and growth factors were linked to immune recognition and the nature of the immune response 34, where increased lipid metabolism in dendritic cells was found to impair presentation of tumor antigens 35 and inherited defects in purine metabolite degradation was found to be involved with T and B-cell immune deficiency 36.
A unique immune signature was seen in NP patients’ immune cells from tumor-invaded SLNs and primary tumor compartments, largely consisting of up-regulated B-cell/GC B-cell genes. In addition, NP patients’ immune cells showed up-regulation of cell-cycle and DNA-repair pathways which may also be GC-related, as GC B-cells undergo proliferation and somatic hypermutation involving DNA repair mechanisms 37. Ectopic GCs were observed in primary breast tumors, where they were found to produce tumor antigen-specific antibodies 38. B cells were suggested to be tumor promoting and associated with chronic inflammation 1, 39. Indeed, ectopic GCs are the hallmark of autoimmune diseases, which are chronic and inflammatory by nature 40. On the other hand, large numbers of peritumoral B cells in LN metastases were associated with favorable outcome in head and neck cancer patients 13. According to our data, it is possible that tumor-invaded TDLN and tumor infiltrating B cells involved in cell cycle and/or DNA repair pathways may promote tumor invasion and/or progression in NP patients.
Analysis of gene expression patterns across the three compartments – primary tumor, LNs and peripheral blood – revealed that the signatures observed in the LNs, i.e. down regulation of immune-related signatures and up-regulation of tumor-promoting pathways in immune cells from NP versus NN patients, are also observed systemically. Interestingly, the LN and peripheral blood compartments emerged as having similar signatures in both NN and NP patients, where in NP patients these signatures were consistent with up-regulation of cell cycle and tumor-promoting pathways and down regulation of immune-related pathways. Thus, patterns observed in the peripheral blood compartment echo the patterns observed in TDLNs. As false negative diagnosis rates of TDLN biopsies determining tumor positivity may reach 43% 41, immune cell signatures within TDLNs may be used to add sensitivity and specificity. Furthermore, similarities in LN and peripheral blood immune signatures suggest that immune analysis of blood samples may serve as a replacement, or at least adjunct, to TDLN biopsy.
The genes up regulated in immune cells from the tumor compartment were associated with tumor-promoting pathways in both NN and NP patients, possibly due to the close proximity of the tumor-infiltrating immune cells to the primary tumor cells and the tumor-influenced microenvironment in both patient types. In contrast to the LN and peripheral blood compartments, NP patients’ purified immune and tumor cells from the primary tumor showed a significantly higher association with immune-related pathways compared to NN patients. Examination of the genes involved in these pathways showed that they are mainly inflammatory-related, thus suggesting tumor-promoting tumor-immune cell cross-talk. NP patients’ immune cell up regulated genes were also involved with tumor-promoting, cell cycle and DNA repair pathways, similar to the observations in the LN and peripheral blood compartments, further suggesting that NP patients’ tumor infiltrating immune cells are more involved in tumor-promoting processes compared to NN patients. Studies report correlations between immune cell infiltration into tumors and prognosis, depending on the infiltrating immune cell composition and the nature of the immune response. CD8+ T cell infiltration tends to correlate with good prognosis, whereas macrophage and T-regulatory cell and infiltration correlate with worse prognosis, suggesting that the identity of the infiltrating immune cells is crucial in dictating tumor-immune interplay 2,3,42,43. Further studies are needed to determine the composition of immune cells in NN and NP patients and their correlation to good and poor prognosis. We note that comparison of immune cell signatures between different compartments should be treated with caution, as the heterogeneity of immune cell populations differs between the compartments as well as between different patients. Nevertheless, the focus of this study was to examine the global state of the immune system in patient groups with and without LN metastasis rather than differences between specific immune cell types.
Overall, significant differences were found in purified immune cell gene expression signatures from TDLNs between NN and NP breast cancer patients. In comparison to NN patients, NP patients show significant down regulation of immune-related pathways such as antigen presentation, and up regulation of cell cycle and tumor-promoting pathways. Similar patterns were also observed systemically in the peripheral blood compartment and to some extent also in the primary tumor compartment. The immunosurveillence theory states that cancer cells arise but few develop into clinically evident tumors due at least in part to the immune status of the individual 1. Tumor dissemination to TDLNs can potentially occur in all breast cancer patients, as a natural part of tumorigenesis. However, while in some patients establishment of metastasisin the TDLNs becomes clinically evident (NP patients), in others they do not (NN patients). Our findings suggest that the key difference lies in NN patients’ immune responses being more intact and thus capable of responding to and eradicating tumor metastases in TDLNs, while NP patients’ immune cells are modulated to other, tumor-promoting processes. Understanding the mechanisms underlying the differences in immune cell functions between NN and NP patients will lead to the development of therapeutic strategies to restore and enhance the immune response against tumors in NP patients.
Supplementary Material
Novelty
It is increasingly clear that the host immune response plays an important role in modulating cancer progression. However, rarely are immune cells purified and profiled independently from tumor samples or tumor draining lymph nodes for detailed analyses of immune signatures. The novelty of this paper is that it describes the characteristics of purified immune cells from major compartments where cancer and immune cells interact: tumor, tumor draining lymph nodes (tumor-invaded or tumor-free), and peripheral blood.
Impact
Purified immune cell signatures enable the identification of differences in genes and pathways between immune cells from node-negative and node-positive breast cancer patients otherwise not detected in heterogeneous tumor tissues. These differences, which are shown to be local and systemic using various bioinformatics methods, suggest that node-positive patients’ immune-related signatures are down-regulated, and tumor-promoting signatures are up-regulated compared to node-negative patients. The differences between these patient groups will help unravel the mechanisms underlying immune dysfunction in the setting of breast cancer and metastasis.
Acknowledgments
The authors would like to thank Dr. Andrea K. Miyahira for critical reading of the manuscript.
Financial support
This work was supported by DoD Era of Hope Scholar Award BC051650.
Abbreviations used
- DC
dendritic cell
- GC
germinal center
- GO
gene ontology
- IPA
ingenuity pathway analysis
- LN
lymph node
- NN
node negative
- NP
node positive
- NSLN
non-sentinel lymph nodes
- PB
Peripheral blood
- SLN
sentinel lymph node
- T
Primary tumor
- TDLN
tumor draining lymph nodes
Footnotes
Conflict of interest
The authors declare no financial or commercial conflict of interest.
References
- 1.DeNardo DG, Coussens LM. Inflammation and breast cancer. Balancing immune response: crosstalk between adaptive and innate immune cells during breast cancer progression. Breast Cancer Res. 2007;9:212. doi: 10.1186/bcr1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mahmoud SM, Paish EC, Powe DG, Macmillan RD, Grainge MJ, Lee AH, Ellis IO, Green AR. Tumor-Infiltrating CD8+ Lymphocytes Predict Clinical Outcome in Breast Cancer. J Clin Oncol. 2011;29:1949–55. doi: 10.1200/JCO.2010.30.5037. [DOI] [PubMed] [Google Scholar]
- 3.Mahmoud SM, Paish EC, Powe DG, Macmillan RD, Lee AH, Ellis IO, Green AR. An evaluation of the clinical significance of FOXP3+ infiltrating cells in human breast cancer. Breast Cancer Res Treat. 2011;127:99–108. doi: 10.1007/s10549-010-0987-8. [DOI] [PubMed] [Google Scholar]
- 4.van de Vijver MJHY, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, Parrish M, Atsma D, Witteveen A, Glas A, Delahaye L, van der Velde T, Bartelink H, Rodenhuis S, Rutgers ET, Friend SH, Bernards R. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 5.Sotiriou C, Neo SY, McShane LM, Korn EL, Long PM, Jazaeri A, Martiat P, Fox SB, Harris AL, Liu ET. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci U S A. 2003;100:10393–8. doi: 10.1073/pnas.1732912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hernandez-Aya LF, Chavez-Macgregor M, Lei X, Meric-Bernstam F, Buchholz TA, Hsu L, Sahin AA, Do KA, Valero V, Hortobagyi GN, Gonzalez-Angulo AM. Nodal status and clinical outcomes in a large cohort of patients with triple-negative breast cancer. J Clin Oncol. 2011;29:2628–34. doi: 10.1200/JCO.2010.32.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Torrenga H, Fabry H, van der Sijp JR, van Diest PJ, Pijpers R, Meijer S. Omitting axillary lymph node dissection in sentinel node negative breast cancer patients is safe: a long term follow-up analysis. J Surg Oncol. 2004;88:4–7. doi: 10.1002/jso.20101. discussion 7–8. [DOI] [PubMed] [Google Scholar]
- 8.Kohrt HE, Nouri N, Nowels K, Johnson D, Holmes S, Lee PP. Profile of immune cells in axillary lymph nodes predicts disease-free survival in breast cancer. PLoS Med. 2005;2:e284. doi: 10.1371/journal.pmed.0020284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Setiadi AF, Ray NC, Kohrt HE, Kapelner A, Carcamo-Cavazos V, Levic EB, Yadegarynia S, van der Loos CM, Schwartz EJ, Holmes S, Lee PP. Quantitative, architectural analysis of immune cell subsets in tumor-draining lymph nodes from breast cancer patients and healthy lymph nodes. PLoS One. 2010;5:e12420. doi: 10.1371/journal.pone.0012420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Afrimzon E, Deutsch A, Shafran Y, Zurgil N, Sandbank J, Pappo I, Deutsch M. Intracellular esterase activity in living cells may distinguish between metastatic and tumor-free lymph nodes. Clin Exp Metastasis. 2008;25:213–24. doi: 10.1007/s10585-007-9135-1. [DOI] [PubMed] [Google Scholar]
- 11.Poindexter NJ, Sahin A, Hunt KK, Grimm EA. Analysis of dendritic cells in tumor-free and tumor-containing sentinel lymph nodes from patients with breast cancer. Breast Cancer Res. 2004;6:R408–15. doi: 10.1186/bcr808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawaida H, Kono K, Takahashi A, Sugai H, Mimura K, Miyagawa N, Omata H, Ooi A, Fujii H. Distribution of CD4+CD25high regulatory T-cells in tumor-draining lymph nodes in patients with gastric cancer. J Surg Res. 2005;124:151–7. doi: 10.1016/j.jss.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 13.Pretscher D, Distel LV, Grabenbauer GG, Wittlinger M, Buettner M, Niedobitek G. Distribution of immune cells in head and neck cancer: CD8+ T-cells and CD20+ B-cells in metastatic lymph nodes are associated with favourable outcome in patients with oro- and hypopharyngeal carcinoma. BMC Cancer. 2009;9:292. doi: 10.1186/1471-2407-9-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gannon PO, Alam Fahmy M, Begin LR, Djoukhadjian A, Filali-Mouhim A, Lapointe R, Mes-Masson AM, Saad F. Presence of prostate cancer metastasis correlates with lower lymph node reactivity. Prostate. 2006;66:1710–20. doi: 10.1002/pros.20466. [DOI] [PubMed] [Google Scholar]
- 15.Saharinen P, Eklund L, Pulkki K, Bono P, Alitalo K. VEGF and angiopoietin signaling in tumor angiogenesis and metastasis. Trends Mol Med. 2011;17:347–62. doi: 10.1016/j.molmed.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 16.Binder C, Simon A, Binder L, Hagemann T, Schulz M, Emons G, Trumper L, Einspanier A. Elevated concentrations of serum relaxin are associated with metastatic disease in breast cancer patients. Breast Cancer Res Treat. 2004;87:157–66. doi: 10.1023/B:BREA.0000041622.30169.16. [DOI] [PubMed] [Google Scholar]
- 17.Walsh LA, Damjanovski S. IGF-1 increases invasive potential of MCF 7 breast cancer cells and induces activation of latent TGF-beta1 resulting in epithelial to mesenchymal transition. Cell Commun Signal. 2011;9:10. doi: 10.1186/1478-811X-9-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petroulakis E, Mamane Y, Le Bacquer O, Shahbazian D, Sonenberg N. mTOR signaling: implications for cancer and anticancer therapy. Br J Cancer. 2007;96 (Suppl):R11–5. [PubMed] [Google Scholar]
- 19.Campana L, Bosurgi L, Rovere-Querini P. HMGB1: a two-headed signal regulating tumor progression and immunity. Curr Opin Immunol. 2008;20:518–23. doi: 10.1016/j.coi.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 20.Chiavarina B, Whitaker-Menezes D, Migneco G, Martinez-Outschoorn UE, Pavlides S, Howell A, Tanowitz HB, Casimiro MC, Wang C, Pestell RG, Grieshaber P, Caro J, et al. HIF1-alpha functions as a tumor promoter in cancer associated fibroblasts, and as a tumor suppressor in breast cancer cells: Autophagy drives compartment-specific oncogenesis. Cell Cycle. 2010;9:3534–51. doi: 10.4161/cc.9.17.12908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma L, Liu YP, Zhang XH, Xing LX, Wang JL, Geng CZ. Effect of RhoA signaling transduction on expression of Ezrin in breast cancer cell lines. Ai Zheng. 2009;28:108–11. [PubMed] [Google Scholar]
- 22.Moutsatsou P, Papavassiliou AG. The glucocorticoid receptor signalling in breast cancer. J Cell Mol Med. 2008;12:145–63. doi: 10.1111/j.1582-4934.2007.00177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alli E, Yang JM, Ford JM, Hait WN. Reversal of stathmin-mediated resistance to paclitaxel and vinblastine in human breast carcinoma cells. Mol Pharmacol. 2007;71:1233–40. doi: 10.1124/mol.106.029702. [DOI] [PubMed] [Google Scholar]
- 24.Thomas W, Caiazza F, Harvey BJ. Estrogen, phospholipase A and breast cancer. Front Biosci. 2008;13:2604–13. doi: 10.2741/2869. [DOI] [PubMed] [Google Scholar]
- 25.Kumar SR, Singh J, Xia G, Krasnoperov V, Hassanieh L, Ley EJ, Scehnet J, Kumar NG, Hawes D, Press MF, Weaver FA, Gill PS. Receptor tyrosine kinase EphB4 is a survival factor in breast cancer. Am J Pathol. 2006;169:279–93. doi: 10.2353/ajpath.2006.050889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hannigan G, Troussard AA, Dedhar S. Integrin-linked kinase: a cancer therapeutic target unique among its ILK. Nat Rev Cancer. 2005;5:51–63. doi: 10.1038/nrc1524. [DOI] [PubMed] [Google Scholar]
- 27.Husemann Y, Geigl JB, Schubert F, Musiani P, Meyer M, Burghart E, Forni G, Eils R, Fehm T, Riethmuller G, Klein CA. Systemic spread is an early step in breast cancer. Cancer Cell. 2008;13:58–68. doi: 10.1016/j.ccr.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 28.Engel J, Eckel R, Kerr J, Schmidt M, Furstenberger G, Richter R, Sauer H, Senn HJ, Holzel D. The process of metastasisation for breast cancer. Eur J Cancer. 2003;39:1794–806. doi: 10.1016/s0959-8049(03)00422-2. [DOI] [PubMed] [Google Scholar]
- 29.Tourkova IL, Shurin GV, Ferrone S, Shurin MR. Interferon regulatory factor 8 mediates tumor-induced inhibition of antigen processing and presentation by dendritic cells. Cancer Immunol Immunother. 2009;58:567–74. doi: 10.1007/s00262-008-0579-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Figueiredo KA, Mui AL, Nelson CC, Cox ME. Relaxin stimulates leukocyte adhesion and migration through a relaxin receptor LGR7-dependent mechanism. J Biol Chem. 2006;281:3030–9. doi: 10.1074/jbc.M506665200. [DOI] [PubMed] [Google Scholar]
- 31.Du R, Lu KV, Petritsch C, Liu P, Ganss R, Passegue E, Song H, Vandenberg S, Johnson RS, Werb Z, Bergers G. HIF1alpha induces the recruitment of bone marrow-derived vascular modulatory cells to regulate tumor angiogenesis and invasion. Cancer Cell. 2008;13:206–20. doi: 10.1016/j.ccr.2008.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doedens AL, Stockmann C, Rubinstein MP, Liao D, Zhang N, DeNardo DG, Coussens LM, Karin M, Goldrath AW, Johnson RS. Macrophage expression of hypoxia-inducible factor-1 alpha suppresses T-cell function and promotes tumor progression. Cancer Res. 2010;70:7465–75. doi: 10.1158/0008-5472.CAN-10-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murdoch C, Muthana M, Coffelt SB, Lewis CE. The role of myeloid cells in the promotion of tumour angiogenesis. Nat Rev Cancer. 2008;8:618–31. doi: 10.1038/nrc2444. [DOI] [PubMed] [Google Scholar]
- 34.Newell MK, Villalobos-Menuey E, Schweitzer SC, Harper ME, Camley RE. Cellular metabolism as a basis for immune privilege. J Immune Based Ther Vaccines. 2006;4:1. doi: 10.1186/1476-8518-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zitvogel L, Kroemer G. Targeting dendritic cell metabolism in cancer. Nat Med. 2010;16:858–9. doi: 10.1038/nm0810-858. [DOI] [PubMed] [Google Scholar]
- 36.Grunebaum E, Chung CT, Dadi H, Kim P, Brigida I, Ferrua F, Cicalese MP, Aiuti A, Roifman CM. Purine metabolism, immune reconstitution, and abdominal adipose tumor after gene therapy for adenosine deaminase deficiency. J Allergy Clin Immunol. 2011;127:1417–9. e3. doi: 10.1016/j.jaci.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 37.Odegard VH, Schatz DG. Targeting of somatic hypermutation. Nat Rev Immunol. 2006;6:573–83. doi: 10.1038/nri1896. [DOI] [PubMed] [Google Scholar]
- 38.Coronella JA, Spier C, Welch M, Trevor KT, Stopeck AT, Villar H, Hersh EM. Antigen-driven oligoclonal expansion of tumor-infiltrating B cells in infiltrating ductal carcinoma of the breast. J Immunol. 2002;169:1829–36. doi: 10.4049/jimmunol.169.4.1829. [DOI] [PubMed] [Google Scholar]
- 39.de Visser KE, Korets LV, Coussens LM. De novo carcinogenesis promoted by chronic inflammation is B lymphocyte dependent. Cancer Cell. 2005;7:411–23. doi: 10.1016/j.ccr.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 40.William J, Euler C, Christensen S, Shlomchik MJ. Evolution of autoantibody responses via somatic hypermutation outside of germinal centers. Science. 2002;297:2066–70. doi: 10.1126/science.1073924. [DOI] [PubMed] [Google Scholar]
- 41.Fujii T, Yanagita Y, Kinoshita T, Fujisawa T, Hirakata T, Yamaki S, Matsumoto A, Uchida N, Iijima M, Kuwano H. Accuracy of intraoperative macroscopic diagnosis of sentinel node metastases in breast cancer: is accurate prediction possible? Tumori. 2011;97:62–5. doi: 10.1177/030089161109700112. [DOI] [PubMed] [Google Scholar]
- 42.Ghebeh H, Barhoush E, Tulbah A, Elkum N, Al-Tweigeri T, Dermime S. FOXP3+ Tregs and B7-H1+/PD-1+ T lymphocytes co-infiltrate the tumor tissues of high-risk breast cancer patients: Implication for immunotherapy. BMC Cancer. 2008;8:57. doi: 10.1186/1471-2407-8-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mukhtar RA, Nseyo O, Campbell MJ, Esserman LJ. Tumor-associated macrophages in breast cancer as potential biomarkers for new treatments and diagnostics. Expert Rev Mol Diagn. 2011;11:91–100. doi: 10.1586/erm.10.97. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


