Abstract
MicroRNAs and cancer stem cells have emerged as critical players in glioblastoma, one of the deadliest human cancers. In this study, we investigated the expression and function of microRNA-10b in glioblastoma cells and stem cells. An analysis of The Cancer Genome Atlas data revealed a correlation between high miR-10b levels and poor prognosis in glioblastoma patients. We measured the levels of miR-10b and found that it is upregulated in human glioblastoma tissues, glioblastoma cell and stem cell lines as compared to normal human tissues or astrocytes. Inhibition of miR-10b with a specific antagomir inhibited the proliferation of glioblastoma established and stem cell lines. Inhibition of miR-10b strongly reduced cell invasion and migration in glioblastoma cell and stem cell lines while overexpression of miR-10b induced cell migration and invasion. We also investigated several predicted targets of miR-10b but could not verify any of them experimentally. Additionally, miR-10b inhibition significantly decreased the in vivo growth of stem cell-derived orthotopic GBM xenografts. Altogether, our findings confirm the oncogenic effects of miR-10b in GBM cells and show for the first time a role of this microRNA in GBM stem cells. Targeting miR-10b might therefore inhibit glioblastoma stem cells, which are thought to be at the origin of glioblastoma and to contribute its recurrence and resistance to therapy.
Keywords: MicroRNA-10b, Glioblastoma, Glioblastoma stem cells, Migration, Invasion
Introduction
Gliomas are extremely aggressive brain tumors that account for the majority of deaths due to primary brain neoplasms. Grade IV glioma, glioblastoma (GBM) is the most frequently diagnosed glioma and one of the least successfully treated human tumors [1]. Despite recent therapeutic advances, GBM treatment combining surgical resection, chemotherapy and radiotherapy remains ineffective and is associated with an average life expectancy of 12-15 months [2]. The origin of GBM is largely unknown but there is an increasing speculation that they arise from a subset of cells, called GBM stem cells, that putatively consist of transformed neural stem cells [3–5]. Accumulating evidence suggests that these stem cells are responsible for GBM initiation, growth, and resistance to therapy [6]. Migration and invasion are hallmarks of cancer progression including in GBM. GBMs rarely metastasize beyond the central nervous system, but these highly infiltrative cancers often invade into normal adjacent brain tissues limiting surgical resection, a context where glioblastoma stem cells display a very aggressive phenotype [7].
MicroRNAs (miRNAs) are small non-coding RNAs that are frequently deregulated in many cancers, acting either as tumor suppressors by targeting oncogenes or as oncogenes by targeting tumor suppressors [8, 9]. miRNAs modulate protein expression by binding to the 3′-untranslated region (3′-UTR) of mRNA and promoting its degradation or inhibiting its transcription [10]. miRNAs play critical roles in multiple biological processes characteristic of glioblastoma including cell growth, migration, invasion, angiogenesis and stem cell behavior [11–14]. A few studies have identified miR-10b as a tumor promoter that is positively associated with malignancy and grade of various cancers [15]. Indeed, miR-10b is the most significantly upregulated miRNA in metastatic breast cancer cell lines, breast cancer samples [16], human pancreatic adenocarcinomas [17] and GBM [18]. In human GBM, miR-10b levels correlate with tumor grade, invasiveness, and the tumor invasive factors urokinase plasminogen activator receptor uPAR and RhoC [19, 20]. However a role for miR-10b in GBM stem cells has not been described before. Targeting oncogenic miR-NAs might represent a novel therapeutic approach for cancer treatment. Various types of antisense-based miRNAs inhibitors, including antagomirs, locked nucleic acid oligonucleotides, 2′-O-modified oligonucleotides, anti-miR poly(l-Lysine)-based nanoparticles have been successfully used for oncogenic miRNAs inhibition both in vitro and in vivo [19, 21–24], but to our knowledge, no such study has been carried out in GSCs.
In the present work, we screened human GBM specimens, GBM cell and stem cell lines (GSCs) for miR-10b levels by quantitative RT-PCR. We found a high expression of miR-10b in GBM tissues as compared to normal brain and higher miR-10b expression in GSCs as compared to established GBM cell lines and normal human astrocytes. We then investigated the effects of miR-10b inhibition on GBM malignancy, in vitro and in vivo. Our data showed that miR-10b inhibition significantly reduced cell proliferation, migration and invasion in GSCs as compared to GBM cells, or normal astrocytes. We also found that targeting of miR-10b in GSCs significantly decreased in vivo tumor growth. We therefore propose miR-10b as an attractive therapeutic target in GBM cells in general and GSCs in particular.
Materials and methods
Cells, reagents and tissue culture
Human GBM tissues, GBM cell lines, primary GBM cells and GBM stem cell lines (GSCs) were used for this study. GBM and normal brain tissues were obtained from consenting patients undergoing surgery at the University of Virginia Health System. All specimens were handled in compliance with the policies of the UVA Health System Institutional Review Board. GSCs 0308, 0822 and 1228 were a kind gift from Dr. H. Fine (NIH, Bethesda, MA). GSCs were grown in neurobasal media with N2 and B27 supplements (0.5× each), and supplemented with human recombinant EGF, bFGF (50 ng/mL each). Primary GBM cells (GBM8 and GBM10) were isolated from surgical specimens of patients who underwent surgical treatment at the Mayo Clinic and who consented to the use of the tissue for research. The primary cells were propagated in animals via heterotopic implantation in the flanks of immunodeficient mice and subsequently were grown in α-MEM with 2.5 % FBS. U1242 cells were a kind gift from Dr. I. Hussaini (University of Virginia, Charlottesville, VA), and were grown in α-MEM supplemented with l-Glutamine. Astrocytes and GBM cell lines A172, U87, U373, SNB19, T98G, SF767 cells were obtained from American Type Culture Collection (Manassas, VA). Astrocytes and A172 cells were grown in DMEM supplemented with 10 % FBS. U87 cells were grown in Eagle's MEM supplemented with 1 mmol/L sodium pyruvate, 0.15 % sodium bicarbonate, 0.1 mol/L nonessential aminoacids and 10 % FBS. U373 cells were grown in DMEM (1 mg/L glucose with l-glutamine) supplemented with HEPES buffer, and 10 % FBS. T98G cells were grown in MEM supplemented with 1 mmol/L sodium pyruvate, 0.1 mol/L nonessential amino acids and 10 % FBS. SNB19 cells were grown in DMEMF12 and supplemented with 0.1 mol/L nonessential amino acids and 10 % FBS. SF767 cells were grown in MEM supplemented with Earl's salts and l-Glutamine. All cells were grown at 37 °C in 5 % CO2 in the presence of penicillin and streptomycin.
Except for stem cell media and supplements, all cell culture media, sodium bicarbonate, sodium pyruvate, nonessential amino acids and HEPES buffer used in this study were purchased from Cellgro Mediatech (Manassas, VA). Neurobasal media, N2, B27, penicillin–streptomycin, Oligofectamine, were purchased from Invitrogen (Carlsbad, CA). Human recombinant bFGF and EGF were purchased from R&D systems (Minneapolis, MN). Fetal bovine serum (FBS) was purchased from Gemini Bio-Products (West Sacramento, CA). Human type IV collagen was purchase from Sigma-Aldrich (St Louis, MO). Crystal Violet was purchased from Promega Corp (Madison, WI). All microRNAs were obtained from Ambion-Biosystems (Huston, TX). The miRNeasy kit, miScript reverse transcription kit and human U6B probe, miR-10b and universal primers were purchased from Qiagen (Valencia, CA). The different antibodies for immunoblotting were purchased from Cell Signaling (Carlsbad, CA), Santa-Cruz Biotechnology (Santa Cruz, CA) and Abcam (Cambridge, MA).
Real-time PCR
GBM tissues (n = 20), normal brain tissues (n = 5), GBM cell lines and GSCs were lysed using Qiazol, transferred to QIAshredder columns, centrifuged at 13,000 × g for 3 min, and then RNA was isolated using the miRNeasy kit according to the manufacturer's instructions (Qiagen). RT–PCR on 500 ng of RNA using the miScript reverse transcription kit was used to generate cDNA. From 16 ng of cDNA template, quantitative real-time PCR analyses for miR-10b and U6B were performed using miR-10b and U6B-specific forward primers and a universal reverse primer according to the manufacture's protocol (Qiagen). U6B was used as a control to normalize the levels of miR-10b. Applied Biosystems (StepOnePlus) real-time PCR system was used to carry out the quantitative PCR, using hot start, with annealing at 55 °C (30 s), extension 70 °C (30 s) for 40 cycles, followed by a melt curve analysis. Data analysis was carried out using StepOne software V2.1 (Applied Biosystems).
miR-10b inhibition
An antisense (anti-miR-10b) was used to inhibit miR-10b expression in astrocytes, A172, T98G, SF767 cells and GSCs 0380, 0822 and 1228. For GSC transfection, the plates were coated with polyornithine for 24 h and washed with PBS prior to transfection procedure. Cells were plated at 50–70 % confluence and transfected with anti-miR-10b using oligofectamine as a transfection reagent. Anti-miR-control (Anti-miR-Cont) was used as a control. Inhibition of miR-10b was verified by quantitative RT-PCR.
Immunoblotting
Immunoblotting was performed using antibodies specific for TIAM1, SDC-1, STAT3, NF-1, PTEN and β-Actin as a loading control. To assess the effect of anti-miR-10b on the expression of different proteins promoting migration and/or invasion in GBM, different GBM cell lines expressing high levels of miR-10b were treated as follows: 50–60 % confluent cells were treated with anti-miR-10b and anti-miR-Con, or miR-10b and miR-Cont for 48 h and subsequently immunoblotted for different predicted targets/proteins of miR-10b that were selected based on target prediction software (TargetScan). The cells were subsequently lysed with RIPA buffer (1 % Igepal, 0.5 % sodium deoxycholate and 0.1 % SDS in PBS), and equal amounts of protein were electrophoretically separated in a polyacrylamide gradient gel (Invitrogen, Carlsbad, CA), trans-blotted to a nitrocellulose membrane, and then incubated overnight with primary antibodies at 4 °C. The primary antibody-bound membranes were washed three times with tween-PBS before incubation with the corresponding horseradish-conjugated secondary antibodies. After the final wash, the immunoreactive signals were detected by enhanced chemiluminescence.
Proliferation assay
To assess the effect of miR-10b inhibition on cell growth, 30,000 GBM and GSC cells were seeded in triplicates in medium containing 10 % FBS, and transfected either with anti-miR-10b or anti-miR-Cont (20 nM). 48 h later, the cells were trypsinized and harvested every day for 4 days and counted with a hemocytometer and growth curves were established.
Migration assay
A172, SF767, T98G GBM cells, and 0308, 0822, 1228 GSCs were seeded at 60 % confluence and transfected with anti-miR-10b or anti-miR-Cont (20 nM) for 24 h, or with miR-10b or miR-Cont (20 nM) before a scratch was made in each well using a sterile pipette tip. Cells that migrated into the scratch 48 h post-treatment were photographed at 40× magnification.
Invasion assay
GBM cells and GSCs were treated with 20 nM anti-miR-10b or anti-miR-control for 24 h. The following day, the cells (2 × 105) were seeded in serum-free or growth factor-free media in collagen IV-coated inserts. 600 μL of complete medium was placed in the lower chamber as a chemoattractant. After 12 h of incubation at 37 °C in 5 % CO2, invaded cells were stained with 0.1 % crystal violet solution and photographed at 40×. The cells were then counted under the microscope in five randomly chosen fields and the number of invaded cells was used for subsequent comparative analyses by one-way ANOVA with the least significant different correction. Each experiment was performed three times.
In vivo experiments
The effect of miR-10b inhibition on in vivo tumor growth was tested in an intracranial GBM xenograft model. 1228 GSCs were transfected with anti-miR-10b or anti-miR-Control, at the concentration of 20 nM, for 24 h. Transfected cells (3 × 105) were stereotactically implanted into the striata of immunodeficient mice (n = 8). Tumor growth was monitored by MRI every week. The animals were sacrificed 4 weeks post-tumor implantation. The brains were removed, sectioned, and stained with hematoxylin and eosin (H&E). Maximal tumor cross-sectional areas were measured by computer-assisted image analysis.
Statistics
All experiments were performed at least in triplicates. Numerical data were expressed as mean ± standard deviation. Two group comparisons were analyzed by two-sided Student's t test. Multiple group comparisons were analyzed with Bonferroni/Dunn multiple comparisons tests. p values were determined for all analyses and p < 0.05 was considered significant.
Results
TCGA data analysis reveals a correlation between miR-10b expression and poor patient prognosis
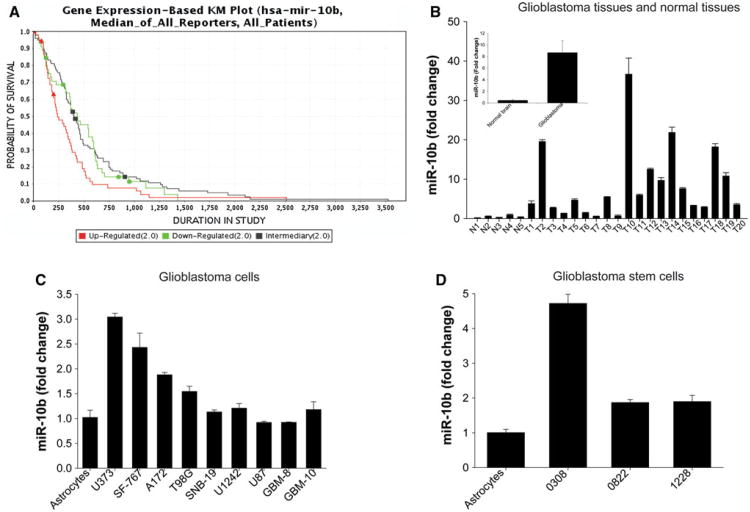
We used the Cancer Molecular Analysis Portal of the NCI (https://cma.nci.nih.gov/cma-tcga/geneView) to analyze The Cancer Genome Atlas (TCGA) data for potential correlations between miR-10b expression levels and patient survival [25]. The analysis and Kaplan–Meier survival curve showed that high levels of miR-10b are associated with poor survival in GBM patients (Fig. 1a). These data suggest that miR-10b is an important regulator of GBM malignancy. By the time this manuscript was submitted for publication, the Cancer Molecular Analysis Portal was discontinued by the NCI. Therefore, the above data are presented with the limitation and disclaimer that they cannot be reproduced using the same analysis portal.
Fig. 1.
miR-10b is upregulated in GBM specimens, tumor cell lines and cancer stem cell lines (GSC), and expression levels correlate with patient survival. a Gene expression-based Kaplan–Meier plot shows an inverse correlation between miR-10b levels and patient survival. b miR-10b levels were measured by quantitative RT-PCR in GBM tissues (T), normal brain (N), normal human astrocytes, GBM cell lines (A172, U87, U373, SNB19, T98G, SF767, U1242), primary GBM cells (GBM8 and GBM10) and GSCs (0308, 0822,1228). The results show that average levels of miR-10b in GBM tissues were significantly higher than normal brain tissues (p < 0.01). c miR-10b levels in U373, SF767, A172, and T98G GBM cell lines were significantly higher than in astrocytes (p < 0.05). d miR-10b levels in GSCs were significantly higher than in astrocytes (p < 0.05)
miR-10b is overexpressed in GBM tissues, GBM cell lines and GBM stem cell lines
We measured the expression of miR-10b in human GBM specimens and GBM cell and stem cell lines using quantitative RT-PCR. miR-10b expression was significantly higher (up to 36-fold) in human GBM specimens as compared to normal brain, where miR-10b levels were very low or barely detectable in some samples. miR-10b levels were on average ∼ninefold higher in GBM tissues than in normal brain (Fig. 1b). We also found that miR-10b expression was higher (up to threefold) in some (U373, A172, SF767 and T98G) but not all GBM cell lines in comparison with human astrocytes (Fig. 1c). Importantly, miR-10b level in GSCs 0308, 0822 and 1228 were significantly higher (up to 4.5-fold increase) than in established GBM cells and in normal human astrocytes (Fig. 1d).
miR-10b regulates GBM cell and stem cell proliferation
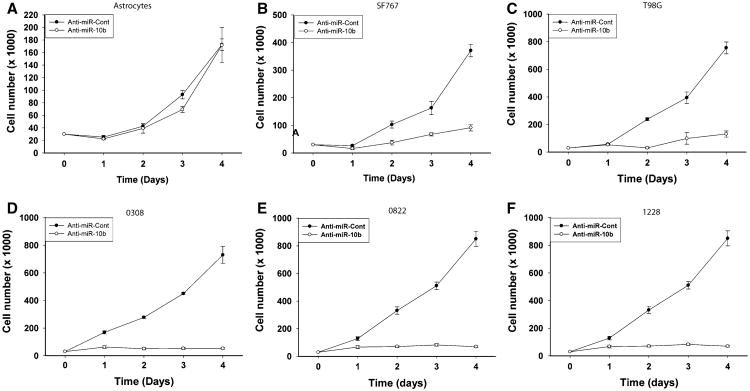
The effects of miR-10b on GSC proliferation have not been investigated to date. We assessed the effects of miR-10b inhibition on GBM cell and stem cell proliferation by cell counting. Astrocytes, T98G, A172, SF767 GBM cells, and GSCs 0308, 0822, and 1228 were transfected with anti-miR-10b or anti-miR-control and the cells were counted for 4 days. miR-10b inhibition did not have any effect on the growth on human Astrocytes (Fig. 2a). However, miR-10b inhibition significantly inhibited the proliferation of SF767, T98G GBM cells and 0308, 0822, 1228 GSCs (n = 3; p < 0.05). The effect of miR-10b inhibition on cell growth was significantly stronger in GSCs when compared to established GBM cell lines. In SF767 cells, inhibition of miR-10b reduced cell proliferation from 370 ± 22.76 in control to 91.67 ± 11.67 (p < 0.05) (Fig. 2b). Similarly, in T98G cells, the rate of cell proliferation significantly decreased from 755 ± 42.72 in control cells to 131.25 ± 22.42 in anti-miR-10b treated cells (Fig. 2c). Inhibition of miR-10b in 0308 and 1228 GSCs revealed a strong inhibitory effect on cell proliferation. Indeed, miR-10b inhibition decreased cell numbers from 730 ± 61.67 in control to 52.50 ± 6.29 in 0308 treated cells (Fig. 2d) and from 1130 ± 55 to 28.33 ± 4.17 in 1228 cells treated with anti-miR-10b (Fig. 2e). Similar results were observed when miR-10b was silenced in 0822 GSCs, with a decrease of cell growth from 850 ± 54.78 in control cells to 70 ± 6.43 in anti-miR-10b treated cells (Fig. 2f).
Fig. 2.
miR-10b regulates GBM cell and stem cell proliferation. miR-10b expression was inhibited in astrocytes, T98G, SF-767 GBM cells and 0308, 0822, 1288 GSCs by transfection with anti-miR-10b (20 nM). Control cells were transfected with anti-miR-Control (anti-miR-Cont) (20 nM) for 48 h and then cells were counted every day for 4 days. a The results show that inhibition of miR-10b did not affect cell growth in astrocytes, b but significantly decreased cell growth in SF767 cells and c T98G cells. A significantly stronger anti-proliferative effect was obtained upon anti-miR-10b treatment in d 0308 GSCs, e 0822 GSCs and f 1228 GSCs
miR-10b significantly decreases migration and invasion in GBM cell lines and GBM stem cells
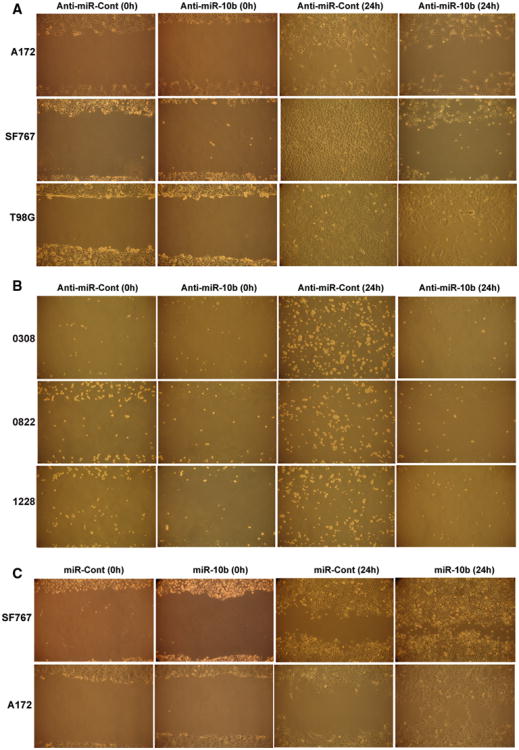
The effects of miR-10b on the invasion and migration of GSCs has to our best knowledge not been reported to date. We tested the effect of miR-10b inhibition on GBM cell and GSC migration and invasion using the scratch assay and transwell assay, respectively. Inhibition of miR-10b decreased the migration of A172, SF767 and T98G (Fig. 3a). A similar inhibitory effect on cell migration upon miR-10b inhibition was found in the three tested GSCs 0308, 0822 and 1228 (Fig. 3b). Conversely, overexpression of miR-10b in SF767 and A172 significantly increased cell migration compared to control, confirming the role of miR-10b in inducing the migration of GBM cells (Fig. 3c).
Fig. 3.
miR-10b regulates GBM cell and stem cell migration. GBM cells A172, SF767 and T98G and GSCs 0308, 0822 and 1228 were transfected with anti-miR-10b or anti-miR-Cont (20 nM) for 24 h. GBM cells SF767 and A172 were treated with miR-10b or miR-Cont (20 nM) for 24 h. The cells were assessed for migration using the scratch/wound assay. a The results show that miR-10b inhibition reduces cell migration in A172, AF767 and T98G cell lines, b as well as in 0308, 0822 and 1228 GSCs. c The results also show that miR-10b overexpression significantly increases cell migration
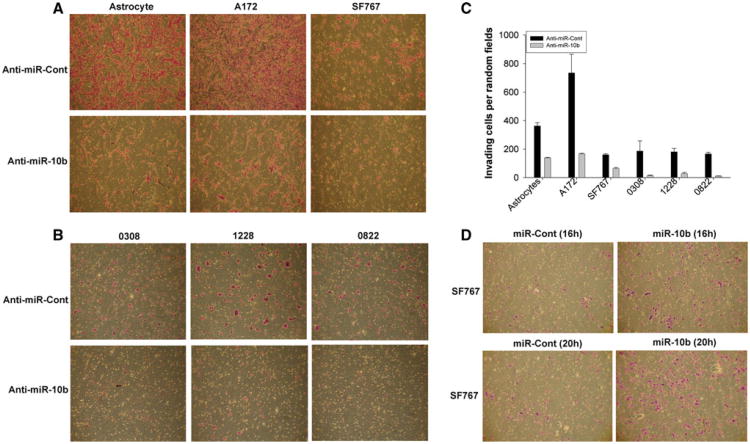
Inhibition of miR-10b reduced A172, SF767, 0308, 0822 and 1228 cell invasion (Fig. 4a, b). Indeed, the number of invading cells per random field was quantified for each cell line, and was significantly decreased from 362 ± 23 to 138.5 ± 3.5 in astrocytes, from 734.5 ± 130.5 to 165.5 ± 4.5 in A172 cells, and 159.5 ± 7.5 to 63.5 ± 6.5 in SF767 cells. Upon miR-10b inhibition, cell invasiveness was also significantly reduced in 0308, 0822 and 1228 GSCs, where the number of invading cells was reduced from 186 ± 71 to 13.5 ± 3.5 in 0308 cells, from 166.5 ± 10.5 to 10.5 ± 1.5 in 0822 cells and 180.5 ± 23.5 to 27.5 ± 8.5 in 1228 cells (Fig. 4c). Comparing the effect of miR-10b inhibition on invasion between different cells also showed that the invasion inhibitory effect of miR-10b inhibition was higher in GSCs than in established cell lines and astrocytes (Fig. 4c). Conversely, miR-10b overexpression in SF767 led to a significant increase of cell invasion (Fig. 4d). The efficiency of miR-10b inhibition or overexpression was verified by qRT-PCR. Altogether, these data show that miR-10b might enhance GBM malignancy by inducing GBM cell and stem cell infiltrative behavior.
Fig. 4.
miR-10b regulates GBM cell and stem cell invasion. GBM cells (SF767 and T98G), GSCs (0308, 0822 and 1228), and astrocytes were transfected with anti-miR-10b or anti-miR-Contol (20 nM) for 24 h. GBM SF767 cells were transfected with miR-10b or miR-cont (20 nM) for 24 h. Cell invasion was assessed using a transwell assay. a The results show that mir-10b inhibition inhibits the invasion of astrocytes, SF767 and T98G cells, b as well as 0308, 0822 and 1228 GSCs. c The invasion inhibition is significantly higher in GSCs than GBM cells. d The results also show that miR-10b overexpression significantly increases the invasion in GBM cells
Verification of miR-10b predicted targets
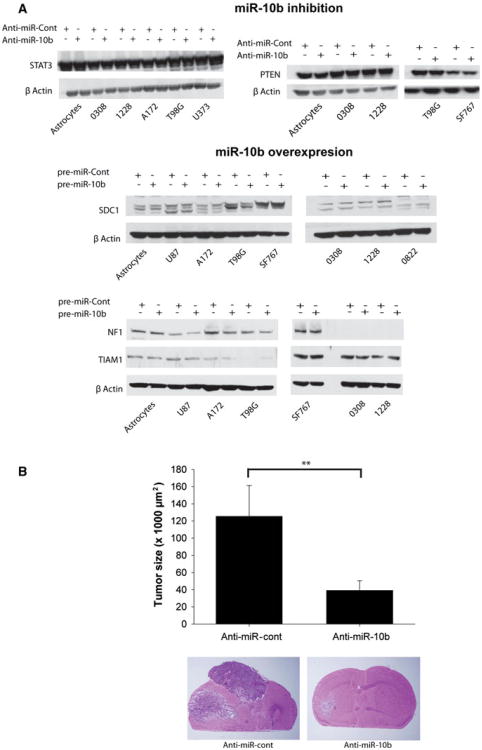
Analysis of miR-10b predicted targets using target prediction software (Targetscan) revealed potential target proteins that are known to regulate proliferation, invasion and migration in GBM. Predicted miR-10b targets included PTEN, STAT3, SDC1, TIAM1, NF1. However we were unable to experimentally verify the regulation of any of the above potential targets upon miR-10b inhibition or miR-10b overexpression despite repeated attempts (Fig. 5a).
Fig. 5.
miR-10b does not affect the levels of predicted target proteins and miR-10b inhibition significantly inhibits the in vivo growth of human glioblastoma stem cell line-derived xenografts. a Immunoblots showing the absence of changes in miR-10b predicted target proteins' expression upon miR-10b inhibition or miR-10b overexpression in glioblastoma cells and GSCs as well as astrocytes used as a control. The blots were stripped and hybridized for β-Actin as a loading control. b GSCs (1228) were transfected in vitro with 20 nM anti-miR-10b or anti-miR-control for 24 h. 3 × 105 of transfected cells were intracranially implanted into the brains of SCID/BALBc immunodeficient mice (n = 8). Tumor growth was monitored by MRI for 4 weeks. Animals were sacrificed and tumor size was assessed by measuring tumor cross-sectional area on H&E stained slides, with computer-assisted image analysis. The results show that anti-miR-10b treatment significantly inhibits in vivo GSC xenograft growth (** p < 0.01)
miR-10b inhibition inhibits the in vivo growth of human GSC-derived xenografts
We tested the effects of targeting miR-10b on the growth of GSC-derived xenografts. 1228 GSCs were transfected with anti-miR-10b or anti-miR-Control (20 nM) for 24 h and the cells were then intracranially implanted in SCID/BALBc mice. Tumor growth was monitored by MRI and the animals were euthanized after 4 weeks. Tumor size quantification was carried out on H&E stained brain frozen sections. While control animals developed very large tumors averaging 125.52 ± 35.82 mm2 cross-sectional area, the inhibition of miR-10b led to inhibition of tumor growth and reduced tumor size to an average of 39.16 ± 11.27 mm2 (n = 8, p < 0.01) (Fig. 5b). These data show for the first time that inhibition of an oncogenic microRNA by its specific antagomiR in GSCs leads to inhibition of in vivo GBM growth. These data suggest that the targeting of the oncogenic miRNA miR-10b is a promising approach targeting glioblastoma stem cells and for future GBM therapy.
Discussion
Our study suggests an important role of the oncogenic miR-10b in GBM stem cell malignancy. We show that miR-10b is highly expressed in GBM tissues, established cell lines and stem cell lines. We find that inhibiting the expression of miR-10b reduces GBM cell growth and significantly decreases GSC proliferation, migration and invasion. We also show for the first time that miR-10b inhibition in GSCs significantly decreases in vivo tumor growth, thus supporting the potential usefulness of targeting oncogenic miR-10b for GBM therapy.
One main mechanism of GBM resistance to therapy is the ability of tumor cells to migrate and invade the surrounding brain and the spinal cord, rendering total surgical removal of the tumor practically impossible [1]. Furthermore, GSCs have been described by numerous groups as strong contributors to therapeutic resistance, responsible for GBM tumor initiation and recurrence [26, 27]. Tumor-initiating cells contribute to tumor progression and invasion in human hepatocellular carcinoma and pancreatic cancer [28, 29]. Stem cell markers have been shown to be predictors of oral cancer invasion as well as retinoblastoma invasion [30, 31]. Similarly, cancer stem cells were proven to mediate invasion/metastasis and poor clinical outcome in inflammatory breast cancer [32]. Therefore, considering the infiltrative phenotype of GBMs, targeting GSCs might be a promising approach to more successful cancer therapies.
Our study shows a significant up-regulation of miR-10b expression in primary GBM tissues compared to normal brain specimens where this miRNA was almost undetectable. These data confirm previous reports showing that miR-10b is highly expressed in different grades and types of gliomas [19, 20]. Importantly, in this study we show for the first time that miR-10b is strongly up-regulated in three different and well-characterized GSCs (0308, 0822, 1228), thus suggesting the involvement of miR-10b in GSC biology. The above GSC lines have been previously characterized and shown to have marked phenotypic and genotypic differences with established glioma cell lines. Unlike the established lines, these GSCs harbored extensive similarities to normal neural stem cells and recapitulated the genotype, gene expression patterns, and in vivo biology of human glioblastomas [5]. We found that miR-10b inhibition did not affect the growth of human astrocytes used as a control, nor A172 GBM cells (data not shown). This effect was minimal on standard GBM cell growth, confirming a recently reported finding in oral cancer [33]. However, the three GSC lines 1228, 0822 and 0308, which express the highest miR-10b levels among all cell lines screened, consistently responded to miR-10b inhibition with a significant inhibition of cell proliferation. miR-10b inhibition also reduced established GBM cell invasion and strongly inhibited GSC invasion. These data support previous findings that miR-10b is associated with cell migration, motility and invasiveness in human esophageal cancer cells [34, 35], plays a critical role in breast cancer invasion [36] and metastasis [37] and promotes cell invasion in gastric cancer [38]. However, our data showing a strong inhibition of invasion in A172 and SF767 GMB cells upon miR-10b inhibition differ from Gabriely's reports showing that miR-10b expression modulation in A172 and U251 GBM cell lines did not affect cell invasion in vitro, a discrepancy that might be attributed to technical differences in performing the inhibition experiments [19]. Altogether, our data confirm previous reports on the oncogenic role of miR-10b in GBM cells and further show for the first time a potential role for miR-10b in GBM stem cell malignancy.
miR-10b has been previously shown to target molecules that modulate several cellular functions, including migration, invasion, and proliferation. Furthermore, studies have reported that miR-10b targeting could also impact tumor growth and metastasis by targeting several metastasis genes such as HOXD10 [16, 38], TIAM1 [39], uPAR and RhoC/Akt [20, 38], KLF4 [35] and NF-1 [34] in different cancers. However, a recent study reported that miR-10b operates not by repressing HOXD10 which controls cell migration and invasion, but by controlling cell cycle and apoptosis [19, 40]. Despite repeated attempts, we could not confirm any of these targets in GBM stem cells. We also searched and investigated other predicted miR-10b targets known to be associated with migration and/or invasion such as TIAM and Syndecan1 (SDC1), but could not experimentally verify any of these targets.
Conclusion
Our study showed for the first time that inhibition of miR-10b can lead to the inhibition of glioblastoma stem cell-derived xenografts. Considering the potential importance of cancer stem cells in tumor initiation, recurrence and resistance to therapy, our data suggest that targeting miR-10b is a promising approach for a more efficient future GBM therapy. In conclusion, our study provides evidence for a role of miR-10b in GBM malignancy in general, and cancer stem cells in particular, and suggests miR-10b as a potential target for GBM therapy.
Acknowledgments
Supported by NIH RO1 NS045209 (R. Abounader) and NIH R01 CA134843 (R. Abounader).
Footnotes
Conflict of interest: None of the authors has any conflict of interest associated with the present work.
Contributor Information
Fadila Guessous, Departments of Microbiology, Immunology and Cancer Biology, University of Virginia, PO Box 800168, Charlottesville, VA 22908, USA.
Melissa Alvarado-Velez, Departments of Microbiology, Immunology and Cancer Biology, University of Virginia, PO Box 800168, Charlottesville, VA 22908, USA.
Lukasz Marcinkiewicz, Departments of Microbiology, Immunology and Cancer Biology, University of Virginia, PO Box 800168, Charlottesville, VA 22908, USA.
Ying Zhang, Departments of Microbiology, Immunology and Cancer Biology, University of Virginia, PO Box 800168, Charlottesville, VA 22908, USA.
Jungeun Kim, Departments of Microbiology, Immunology and Cancer Biology, University of Virginia, PO Box 800168, Charlottesville, VA 22908, USA.
Simon Heister, Departments of Microbiology, Immunology and Cancer Biology, University of Virginia, PO Box 800168, Charlottesville, VA 22908, USA.
Benjamin Kefas, Department of Neurology, University of Virginia, PO Box 800168, Charlottesville, VA 22908, USA.
Jakub Godlewski, Laboratory for Neuro-oncology and Neurosciences, The Ohio State University, Columbus, OH, USA.
David Schiff, Department of Neurology, University of Virginia, PO Box 800168, Charlottesville, VA 22908, USA.
Benjamin Purow, Department of Neurology, University of Virginia, PO Box 800168, Charlottesville, VA 22908, USA.
Roger Abounader, Email: ra6u@virginia.edu, Departments of Microbiology, Immunology and Cancer Biology, University of Virginia, PO Box 800168, Charlottesville, VA 22908, USA; Department of Neurology, University of Virginia, PO Box 800168, Charlottesville, VA 22908, USA.
References
- 1.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, Chin L, DePinho RA, Cavenee WK. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 3.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 4.Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, Fiocco R, Foroni C, Dimeco F, Vescovi A. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64:7011–7021. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- 5.Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W, Park JK, Fine HA. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 6.Hadjipanayis CG, Van Meir EG. Brain cancer propagating cells: biology, genetics and targeted therapies. Trends Mol Med. 2009;15:519–530. doi: 10.1016/j.molmed.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang Z, Cheng L, Guryanova OA, Wu Q, Bao S. Cancer stem cells in glioblastoma–molecular signaling and therapeutic targeting. Protein Cell. 2010;1:638–655. doi: 10.1007/s13238-010-0078-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 9.Nicoloso MS, Spizzo R, Shimizu M, Rossi S, Calin GA. MicroRNAs—the micro steering wheel of tumour metastases. Nat Rev Cancer. 2009;9:293–302. doi: 10.1038/nrc2619. [DOI] [PubMed] [Google Scholar]
- 10.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 11.Godlewski J, Newton HB, Chiocca EA, Lawler SE. MicroRNAs and glioblastoma; the stem cell connection. Cell Death Differ. 2010;17:221–228. doi: 10.1038/cdd.2009.71. [DOI] [PubMed] [Google Scholar]
- 12.Li Y, Guessous F, Zhang Y, Dipierro C, Kefas B, Johnson E, Marcinkiewicz L, Jiang J, Yang Y, Schmittgen TD, Lopes B, Schiff D, Purow B, Abounader R. MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res. 2009;69:7569–7576. doi: 10.1158/0008-5472.CAN-09-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guessous F, Zhang Y, Kofman A, Catania A, Li Y, Schiff D, Purow B, Abounader R. MicroRNA-34a is tumor suppressive in brain tumors and glioma stem cells. Cell Cycle. 2010;9:1031–1036. doi: 10.4161/cc.9.6.10987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Dutta A, Abounader R. The role of microRNAs in glioma initiation and progression. Front Biosci. 2012;17:700–712. doi: 10.2741/3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baffa R, Fassan M, Volinia S, O'Hara B, Liu CG, Palazzo JP, Gardiman M, Rugge M, Gomella LG, Croce CM, Rosenberg A. MicroRNA expression profiling of human metastatic cancers identifies cancer gene targets. J Pathol. 2009;219:214–221. doi: 10.1002/path.2586. [DOI] [PubMed] [Google Scholar]
- 16.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 17.Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, Hagan JP, Liu CG, Bhatt D, Taccioli C, Croce CM. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–1908. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- 18.Ciafre SA, Galardi S, Mangiola A, Ferracin M, Liu CG, Sabatino G, Negrini M, Maira G, Croce CM, Farace MG. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334:1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 19.Gabriely G, Yi M, Narayan RS, Niers JM, Wurdinger T, Imitola J, Ligon KL, Kesari S, Esau C, Stephens RM, Tannous BA, Krichevsky AM. Human glioma growth is controlled by microRNA-10b. Cancer Res. 2011;71:3563–3572. doi: 10.1158/0008-5472.CAN-10-3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sasayama T, Nishihara M, Kondoh T, Hosoda K, Kohmura E. MicroRNA-10b is overexpressed in malignant glioma and associated with tumor invasive factors, uPAR and RhoC. Int J Cancer. 2009;125:1407–1413. doi: 10.1002/ijc.24522. [DOI] [PubMed] [Google Scholar]
- 21.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 22.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, Subramaniam A, Propp S, Lollo BA, Freier S, Bennett CF, Bhanot S, Monia BP. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 23.Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen HF, Berger U, Gullans S, Kearney P, Sarnow P, Straarup EM, Kauppinen S. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 24.Jin H, Yu Y, Chrisler WB, Xiong Y, Hu D, Lei C. Delivery of microRNA-10b with polylysine nanoparticles for inhibition of breast cancer cell wound healing. Breast Cancer (Auckl) 2012;6:9–19. doi: 10.4137/BCBCR.S8513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Network CGAR. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 27.Rich JN, Bao S. Chemotherapy and cancer stem cells. Cell Stem Cell. 2007;1:353–355. doi: 10.1016/j.stem.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 28.Yang W, Wang C, Lin Y, Liu Q, Yu LX, Tang L, Yan HX, Fu J, Chen Y, Zhang HL, Tang L, Zheng LY, He YQ, Li YQ, Wu FQ, Zou SS, Li Z, Wu MC, Feng GS, Wang HY. OV6(+) tumor-initiating cells contribute to tumor progression and invasion in human hepatocellular carcinoma. J Hepatol. 2012;57(3):613–620. doi: 10.1016/j.jhep.2012.04.024. [DOI] [PubMed] [Google Scholar]
- 29.Lenz J, Karasek P, Jarkovsky J, Muckova K, Dite P, Kala Z, Veselska R, Hermanova M. Clinicopathological correlations of nestin expression in surgically resectable pancreatic cancer including an analysis of perineural invasion. J Gastrointest Liver Dis. 2011;20:389–396. [PubMed] [Google Scholar]
- 30.Siu A, Lee C, Dang D, Lee C, Ramos DM. Stem cell markers as predictors of oral cancer invasion. Anticancer Res. 2012;32:1163–1166. [PubMed] [Google Scholar]
- 31.Mohan A, Kandalam M, Ramkumar HL, Gopal L, Krishnakumar S. Stem cell markers: ABCG2 and MCM2 expression in retinoblastoma. Br J Ophthalmol. 2006;90:889–893. doi: 10.1136/bjo.2005.089219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Charafe-Jauffret E, Ginestier C, Iovino F, Tarpin C, Diebel M, Esterni B, Houvenaeghel G, Extra JM, Bertucci F, Jacquemier J, Xerri L, Dontu G, Stassi G, Xiao Y, Barsky SH, Birnbaum D, Viens P, Wicha MS. Aldehyde dehydrogenase 1-positive cancer stem cells mediate metastasis and poor clinical outcome in inflammatory breast cancer. Clin Cancer Res. 2010;16:45–55. doi: 10.1158/1078-0432.CCR-09-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu YC, Chen YJ, Wang HM, Tsai CY, Chen WH, Huang YC, Fan KH, Tsai CN, Huang SF, Kang CJ, Chang JT, Cheng AJ. Oncogenic function and early detection potential of miRNA-10b in oral cancer as identified by microRNA profiling. Cancer Prev Res (Phila) 2012;5(4):665–674. doi: 10.1158/1940-6207.CAPR-11-0358. [DOI] [PubMed] [Google Scholar]
- 34.Chai G, Liu N, Ma J, Li H, Oblinger JL, Prahalad AK, Gong M, Chang LS, Wallace M, Muir D, Guha A, Phipps RJ, Hock JM, Yu X. MicroRNA-10b regulates tumorigenesis in neurofibromatosis type 1. Cancer Sci. 2010;101:1997–2004. doi: 10.1111/j.1349-7006.2010.01616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tian Y, Luo A, Cai Y, Su Q, Ding F, Chen H, Liu Z. MicroRNA-10b promotes migration and invasion through KLF4 in human esophageal cancer cell lines. J Biol Chem. 2010;285:7986–7994. doi: 10.1074/jbc.M109.062877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bourguignon LY, Wong G, Earle C, Krueger K, Spevak CC. Hyaluronan-CD44 interaction promotes c-Src-mediated twist signaling, microRNA-10b expression, and RhoA/RhoC upregulation, leading to Rho-kinase-associated cytoskeleton activation and breast tumor cell invasion. J Biol Chem. 2010;285:36721–36735. doi: 10.1074/jbc.M110.162305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma L. Role of miR-10b in breast cancer metastasis. Breast Cancer Res. 2010;12:210. doi: 10.1186/bcr2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Z, Zhu J, Cao H, Ren H, Fang X. miR-10b promotes cell invasion through RhoC-AKT signaling pathway by targeting HOXD10 in gastric cancer. Int J Oncol. 2012;40(5):1553–1560. doi: 10.3892/ijo.2012.1342. [DOI] [PubMed] [Google Scholar]
- 39.Moriarty CH, Pursell B, Mercurio AM. miR-10b targets Tiam1: implications for Rac activation and carcinoma migration. J Biol Chem. 2010;285:20541–20546. doi: 10.1074/jbc.M110.121012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gabriely G, Teplyuk NM, Krichevsky AM. Context effect: microRNA-10b in cancer cell proliferation, spread and death. Autophagy. 2011;7:1384–1386. doi: 10.4161/auto.7.11.17371. [DOI] [PubMed] [Google Scholar]