Abstract
Here we provide the first report about the rates of muscle evolution derived from Bayesian and parsimony cladistic analyses of primate higher-level phylogeny, and compare these rates with published rates of molecular evolution. It is commonly accepted that there is a ‘general molecular slow-down of hominoids’, but interestingly the rates of muscle evolution in the nodes leading and within the hominoid clade are higher than those in the vast majority of other primate clades. The rate of muscle evolution at the node leading to Homo (1.77) is higher than that at the nodes leading to Pan (0.89) and particularly to Gorilla (0.28). Notably, the rates of muscle evolution at the major euarchontan and primate nodes are different, but within each major primate clade (Strepsirrhini, Platyrrhini, Cercopithecidae and Hominoidea) the rates at the various nodes, and particularly at the nodes leading to the higher groups (i.e. including more than one genera), are strikingly similar. We explore the implications of these new data for the tempo and mode of primate and human evolution.
Keywords: anatomy, evolutionary rates, gradualism, human evolution, molecules, muscles, phylogeny, primates
Introduction
Recent studies suggest that rates of both morphological and molecular evolution vary among taxa (e.g. Chatterjee et al. 2009; Cooper & Purvis, 2009; Perelman et al. 2011; Steiper & Seiffert, 2012). For instance, body mass evolution in carnivores has evolved faster than in primates as a whole (Matilla & Bokma, 2008), while within primates body mass within Strepsirrhines has evolved faster than within Platyrrhini (Purvis et al. 2003). Analyses of 10 markers from putatively neutral, non-coding, non-repetitive regions of the genome show that Old World monkeys evolved significantly faster than hominoids and that, within the cercopithecids, macaques evolved faster than baboons (Peng et al. 2009). With respect to genes related to transcriptional regulation and neuro-development, the human lineage evolved faster than the chimpanzee lineage (Pollard et al. 2006a,b). In contrast, Bakewell et al. (2007) stated that, in general, more genes underwent positive selection in chimpanzee evolution than in human evolution. According to authors such as Cooper & Purvis (2009), population density, metabolic rate, body size, life history, ecological generalization, environmental variables, speciation, geographic range size and the occurrence of competitive interactions are all factors that may explain the different rates of morphological and/or molecular evolution observed at different taxonomic levels. The occurrence of different molecular rates is often used to support the concept of local molecular clocks, which assume significant differences in nucleotide substitution rates in different taxonomic groups. According to Tetushkin (2003: p. 729), primates “provide the most interesting and striking example of such heterogeneity in the tempo of molecular evolution”.
Most recent studies dealing with evolutionary rates within mammals are molecular; in general the few that are non-molecular do not focus on detailed morphology and on phylogenetic characters from explicit cladistic analyses, but mainly on global phenotypes such as body size (see e.g. the recent paper of Venditti et al. 2011). The present study is the first within any vertebrate clade that looks at evolutionary rates using specific phylogenetic morphological characters by focusing specifically on the gross morphology of the muscular system. It is part of a long-term project to investigate the comparative anatomy, homologies and evolution of the striated muscles of all of the major groups of vertebrates (Diogo et al. 2008, 2009a, b, 2010; Diogo & Abdala, 2010 Diogo & Wood, 2011, 2012). The present study focuses on primates, and is based on comprehensive parsimony and Bayesian cladistic analyses of the myology of each of the major primate higher taxa plus a range of outgroups (tree-shrews, dermopterans and rodents; Diogo & Wood, 2011; Fig. 1). A total of 166 characters were extracted from the head, neck (HN) and pectoral and upper limb (PU) musculature, and the most parsimonious tree obtained from the cladistic analysis is congruent with Arnold et al.'s (2010) evolutionary molecular tree of primates. It is also similar to the primate molecular trees obtained by Fabre et al. (2009) and Perelman et al. (2011), except that the two latter studies did not recover the Cebidae as a monophyletic taxon (Fig. 1).
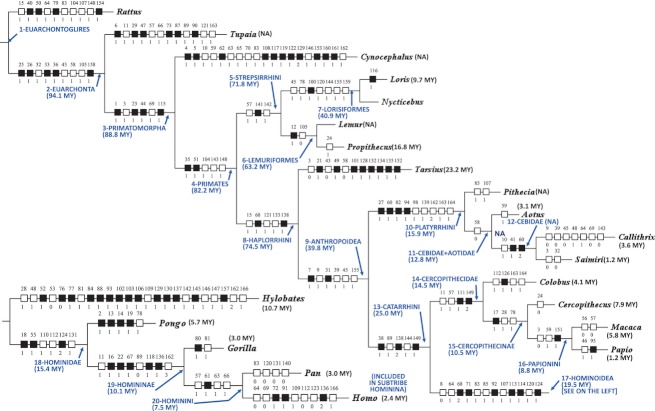
Fig. 1.
Single most parsimonious tree (L 301, CI 58, RI 73) obtained from the analysis of 166 characters of the HN and PU musculature (Diogo & Wood, 2011, 2012). Unambiguous transitions occurring in each branch are shown in white (homoplasic transitions) and black (non-homoplasic transitions) squares (numbers above and below the squares indicate the character and character state, respectively). For each euarchontan clade we show the respective estimated molecular divergence time; the only exception is the genus Homo for which we show the consensus first appearance datum based on the fossil record (for more details, see text).
In the present paper we calculate the rates of muscle evolutionary change within the primate clade resulting from our myology-based cladistic analyses and compare these rates with the molecular rates obtained by other authors, including the rates of molecular nucleotide substitution reported by Perelman et al. (2011). We explore the implications of these new data for the tempo and mode of primate and human evolution. In particular, we address the following specific questions. (a) Are the rates of muscle evolution the same or different across different primate taxa and across different geological time-periods? (b) Are the muscle rates in general similar to the molecular rates provided in recent papers, or is there a mismatch between morphological and molecular rates as predicted by the neutral evolution model of Kimura (1968)?
Materials and methods
The evolutionary changes in striated muscles occurring at each of the nodes shown in Figs 1 and 2 are based on the phylogenetic results of Diogo & Wood (2011, 2012). It should be noted that only ‘unambiguous transitions’ occurred in each branch are shown in Figs 1 and 2 [namely in white (homoplasic transitions) and black (non-homoplasic transitions) squares (numbers above and below the squares indicate the character and character state, respectively)] and used for the calculations of the muscle evolutionary rates (see below). For more details about the phylogenetic methodology employed to search and detect those unambiguous transitions, see Diogo & Wood (2011). The estimated divergence dates for the euarchontan taxa shown in that tree follow Fabre et al. (2009), who used fossil-calibrated rate-autocorrelated Bayesian molecular clock methods to estimate divergence dates while accounting for changes in evolutionary rate over time. The times shown for each clade and for each genus of Fig. 1 refer to minimum divergence times in the sense that they correspond to the divergence times of the most distantly-related species of the respective taxon that were included in Fabre et al.'s (2009) study. There are two reasons for using the estimated times provided by Fabre et al. (2009). First, that study included estimated times for additional euarchontan clades; and secondly, it included data obtained from the analysis of more primate species than are included in other recent molecular studies of primates (e.g. Perelman et al. 2011). By including several species of the same genus in the analysis, it is possible to generate a more reliable estimate of the minimum time of origin of each genus (e.g. if one would only include two highly derived sister-group species A and B of a genus X, the minimum time of origin would obviously be more recent than the minimum time of origin obtained if one would also include the phylogenetically more plesiomorphic species C and D). The rates of muscle evolution given in the tree of Fig. 2 for the nodes leading to the terminal taxa (genera) were measured from the time each genus diverged from its sister-group to the estimated time of origin of that genus. This is because the apomorphies shown in those nodes refer to apomorphies of the ‘genus as a whole’ (i.e. to ‘synapomorphies’ shared by all of the species of that genus). That is, Diogo & Wood (2011) only listed apomorphies that are found in all or at least most of the species of a genus (i.e. if a feature was found in one species of Papio but not in all/most species of that genus, then that feature would not be coded and would be listed as an apomorphy of the genus Papio). Thus, the character state changes were not accumulated during the evolutionary history of a single extant species of that genus, but were changes accumulated at the node leading to the last common ancestor (LCA) of the extant species of this genus. The taxa for which Fabre et al. (2009) only examined a single species (e.g. the genera Lemur and Pithecia) as well as the family Cebidae (which was not recovered as a monophyletic group in Fabre et al.'s study) are indicated as ‘NA’ (non-applicable) in Fig. 2. As H. sapiens is the only extant species of this genus, we cannot tell whether the character apomorphies listed for Homo were already present in the LCA of the various Homo species ca. 2.4 Ma, or whether they accumulated up until the origin of H. sapiens ca. 200 000 years ago (Wood & Baker, 2012). So in this case we provide two rates of evolution for the node leading to modern humans, one (1.77) resulting from the division of the nine character state changes accumulated in that node by 5.1 (i.e. 7.5–2.4 Ma), the other (1.24) resulting from the division of the nine character states by 7.3 (i.e. 7.5–0.2 Ma). It is important to stress that a strength of the Diogo & Wood (2011, 2012) cladistic analysis is that it explicitly avoided using an arbitrary selection of characters or characters. The only bias in our character selection was the intentional one that we used as our evidence the gross morphology of ‘all’ of the striated muscles in the HN and PU regions; we were careful not to cherry-pick that evidence for characters whose distribution was consistent with a preferred a priori phylogenetic or functional hypothesis, and included features that are autapomorphic and features that are non-autapomorphic, without restrictions. Therefore, the type of random, non-selected characters used by Diogo & Wood (2011, 2012) is particularly useful for the analyses provided in the present paper. The molecular substitution rates shown in the blue circles of Fig. 2 follow Perelman et al.'s (2011) study. Although each author would probably argue that the results of his molecular clock studies are more accurate than those of other authors, we decided to use Perelman et al. (2011) because this is the most recent and detailed report specifically focused on nucleotide substitution rates in primates, calculating rates from 54 nuclear gene regions (total of 34 927 sequence sites) of 186 primate species representing about 90% of the described genera. In this respect, it is important to note that Fabre et al. (2009) did not publish specific rates for the specific nodes examined by them because the methods used by them were not adequate to infer these type of data, i.e. that was not the purpose of that paper as recognized by the authors themselves (F. Pierre-Henri, perssional communication). However, it should also be noted that in the Discussion we will also compare the myological evolutionary rates obtained by us with the molecular substitution rates obtained by various authors other than Perelman et al. (2011). Although the absolute rates will not affect the comparisons of the patterns of morphological and genetic changes specifically shown for the different nodes in Fig. 2, it should be noted that, as reviewed by Gibbons (2012), estimates of mutation rates in primate and human evolution remain the subject of much controversy. Also, although our sampling of primate taxa is particularly detailed for a myological study (Diogo & Wood, 2011), it is less limited that that used by genetic analyses such as Perelman et al. (2011), e.g. for strepsirrhines and platyrrhines. It is also worthy to note that consideration of different anatomical regions might yield different phylogenetic results, a subject that was discussed in detail by Diogo & Wood (2011, 2012), and that is addressed in this paper and in Fig. 2 where we show the different evolutionary rates for the HN vs. the PU muscles.
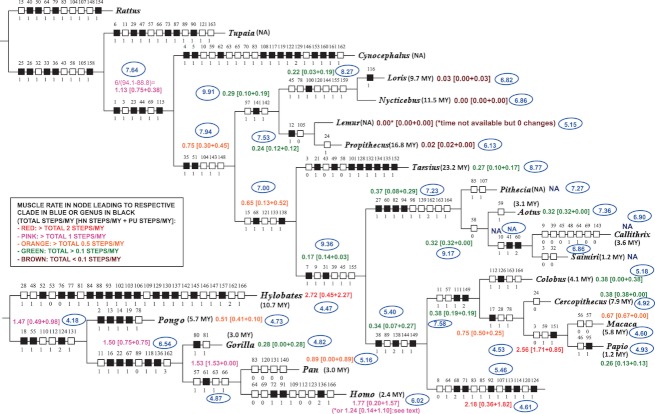
Fig. 2.
Single most parsimonious tree shown in Fig. 1, the muscle evolutionary rates [number of total unambiguous character state changes as well as of unambiguous HN (chars. 1–67) and PU (chars. 68–166) character state changes per 1 million years (1 MY)] for each node within the primate clade and for the node leading to the Primatomorpha being given next to these nodes. For comparison, the nucleotide substitution rates (unit = number of substitutions/site/MY × 104; see text for more details) calculated by Perelman et al. (2011) is shown inside a blue oval next to each node (for more details, see text).
Results
The most parsimonious tree obtained from the cladistic analysis of the 166 muscle-based characters from the HN and PU examined by us (CI 58, RI 73) has a total length of 301 steps, of which 220 are unambiguously optimized in the tree (the squares in Figs 1 and 2). Detailed descriptions and photographs of the muscles for each primate taxon included in the cladogram of Fig. 1 are given in Diogo & Wood (2012). Detailed descriptions of each of the phylogenetic characters used are given in Diogo & Wood (2011). A detailed list of the synapomorphies/apomorphies obtained in our phylogenetic analyses is given in Appendix S1. The results of the rate of muscle evolutionary changes per million of years (Ma) for each primate node as well as for the nodes leading to the Primatomorpha (using the molecular divergence times provided by Fabre et al. 2009; see above) calculated in the present study are shown in Fig. 2 and are briefly summarized below.
The rate of evolution of HN and PU muscles at the node leading to the Primatomorpha (1.13) is substantially higher than that at the node leading to primates (0.75), suggesting that muscle evolution was already high in other euarchontan clades and thus in at least some mammalian groups well before the extinction of dinosaurs ca.65 Ma. The rates of muscle evolution at the nodes leading to the Strepsirrhini (0.29), Lemuriformes (0.24) and Lorisiformes (0.22), and particularly to each of the strepsirrhine terminal taxa (0.03, 0.00, 0.00 and 0.02) are all low. The rate at the node leading to Tarsius (0.27) is not as low as that at the nodes leading to the terminal strepsirrhine taxa. That is, several muscles of Tarsius are markedly plesiomorphic, but this taxon had a long evolutionary history independent from that of the other extant primate taxa and therefore did accumulate a series of peculiar, and often unique, apomorphies (e.g. two sets of contrahentes; Diogo & Wood, 2011, 2012). The node leading to anthropoids (0.17) has a lower rate of muscle evolution than that leading to the haplorrhines (0.65). As might be expected from the distinctive morphology of the New World monkeys, the node leading to the Platyrrhini has a higher rate of muscle evolution than does the node leading to anthropoids. The rate of muscle evolution at the node leading to the terminal taxon Aotus (0.32) is greater than that at any of the nodes leading to any of the strepsirrhine terminal taxa included in the analysis. Remarkably, the rate of muscle evolution at the node leading to the clade including the platyrrhine families Cebidae plus Aotidae (0.32) is the same as that at the node leading to a single aotid genus, Aotus (0.32), and is very similar to that at the node leading to the whole Platyrrhini clade (0.37). Moreover, the rate at the node leading to the Cercopithecidae (0.38) is the same as that at the node leading to the cercopithecid Colobus (0.38) and at the node leading to Cercopithecus (0.38). The rate of muscle evolution at the node leading to the Papionini (2.56) is much higher than that at the nodes leading to Macaca (0.67) and to Papio (0.26).
The rates of muscle evolution at the nodes leading to the Hominidae (1.47), Homininae (1.50) and Hominini (1.53) are similar, but the rate at the node leading to Homo (1.77 or 1.24: see Materials and methods) is higher than at the node leading to Pan (0.89) and much higher than at the node leading to Gorilla (0.28). Among the seven primate nodes with faster rates (Fig. 2; rates > 1.00), seven concern nodes that lead to, or are nodes within the, hominoid clade, the rates at the nodes leading to the hominoids (2.18) and Hylobates (2.72) being particularly high; the only non-hominoid rate included in the seven faster rates concerns the node leading to the Papionini. Thus, in general with respect to the muscles in the anatomical regions we investigated, the rates at the origin of the hominoids, and then within the hominoid clade, are faster than the rates elsewhere in the primate clade. At the other extreme, all the four primate nodes having the slowest rates in the tree (Fig. 2; rates < 0.10) are inside the clades Strepsirrhini.
There are some striking differences in rates of change at the same node when the data are divided and analyzed by anatomical region. For example, the rate of the PU state changes at the node leading to Hylobates (2.27), the highest in the tree, is five times greater than the rate of the HN changes at this same node (0.45). In contrast, the rate of the HN changes at the node leading to Pongo (0.41) is four times greater than that of the PU changes at the same node (0.10). These new data about partial rates of muscle evolution support and expand the idea of primate mosaic muscle evolution proposed by Diogo & Wood (2011, 2012).
Discussion
Muscle vs. molecular rates
The neutral theory predicts that the vast majority of molecular (e.g. DNA) evolution is hidden from selection, i.e. that molecular evolution occurs at a constant evolutionary rate and is mainly decoupled from morphological change (Kimura, 1968). However, as explained above, contrary to the predictions of neutral theory, molecular rates are known to differ substantially across different taxa, and this has lead to the development of methods that accept and model molecular rate variation across lineages (e.g. the ‘relaxed clocks’ methods). But the neutral theory prediction that there should be no significant correlation between genetic and morphological rates of change was supported, i.e. most investigators have been unable to find evidence of significant correlation between molecular (e.g. DNA sequences) and morphological (of global phenotypes, e.g. body size: see above) rates in the several vertebrate datasets analyzed by them (e.g. Bromham et al. 2002; Davies & Savolainen, 2006, and references therein).
With respect to the striated muscles we examined in this study, we found significant differences between muscle and molecular evolutionary rates at the same nodes. For example, whereas the genome-wide analysis of 14 000 genes undertaken by Bakewell et al. (2007, p. 7492) suggested that ‘there were more adaptive genetic changes during chimp evolution than during human evolution’, the results of our parsimony and Bayesian analyses suggest that with respect to the gross morphology of the HN and PU muscles, the human lineage evolved at a faster rate than the chimpanzee lineage. In this case concerning chimpanzees and humans, the muscle evolutionary rates are therefore more in agreement with the molecular substitution rates obtained by Perelman et al. (2011), where the human and chimpanzee lineages have substitution rates of 6.02 and 5.16 substitutions/site/MY × 104, respectively. However, whereas Perelman et al. (2011) found high nucleotide substitution rates at non-catarrhine nodes, in our dataset eight of the 11 primate nodes with the lowest rates (i.e. rates < 0.30) are those leading to, or within, the Strepsirrhini and Tarsioidea, and most of the highest rates were at nodes leading to, or within, the hominoid clade (Fig. 2; see above). These data thus support the hypothesis (e.g. Bonner, 1988) that large-sized taxa (e.g. hominoids and particularly hominids) evolve faster than small-sized taxa (e.g. Tarsius and various strepsirrhines, particularly lorisiforms). Some authors such as Stanley (1979) and Simpson (1953) argued that large-sized species evolve more quickly because their low population size and low fecundity restrict gene flow, although they also noted that small-sized species tend to have faster life histories, which in theory could increase the rate of evolution. Further studies are needed to test if the differences observed in the evolutionary rates of the primate lineages shown in Fig. 2 are, or are not, strongly and directly related to differences in overall body size. In this respect it is interesting to note, for instance, that Conroy (2003) pointed out that in primates there is, in general, no inverse relationship between species diversity and body mass.
The data presented above could also be used to support the hypothesis that evolution is faster in the direction of greater anatomical complexity in the higher primates including modern humans, following a ‘scalae naturae’ conception of the natural order. That is, to support the idea that the faster myological evolutionary rates seen in the hominoids are leading to a greater anatomical complexity of these primates. However, as explained by Diogo & Wood (2011), faster evolutionary rates do not necessarily correspond to more muscles, or more complex muscles. In fact, hominoids such as modern humans and chimpanzees have fewer muscles than most primates and particularly ‘plesiomorphic’ primates such as those in the strepsirrhine and tarsioid clades. For example, modern humans usually have 123 HN and PU striated muscles (not including the extrinsic facial muscles of the ear and the muscles of the eye), whereas Tarsius and Nycticebus may have up to 138 or 139 muscles, respectively. This is because many of the character state changes that are inferred to have occurred in the hominoid clade, including the human clade, concern the loss, and not the gain, of muscles.
There are several possible reasons why there are differences between the rates of molecular and morphological evolution at the same node. One concerns the neutral model of evolution proposed by Kimura (1968), as explained above. It is possible that a majority of variation is due to mutations in housekeeping and general regulatory genes; this may appear as variability in individual molecular characters, but is hardly good material for building morphological adaptations (e.g. Hansen & Houle, 2004). Galis and colleagues suggest an intriguing example, where apparent genetic variability in the number of mammalian neck vertebrae is rendered useless by pleiotropic effects that greatly elevate cancer risk (e.g. Galis et al. 2006; Galis & Metz, 2007; see above). Other possible reasons include the occurrence of epistatic constraints and other phenomena listed by authors such as Bromham et al. (2002), Hansen & Houle (2004) and Davies & Savolainen (2006).
However, it is important to note that there are various cases where the muscle evolutionary rates obtained in our study are similar to the molecular evolutionary rates published in the literature. For instance, the regions of the genome associated with genes related to transcriptional regulation and neuro-development (e.g. HAR1) show an accelerated rate of substitutions in the human lineage compared with the chimpanzee lineage, as the divergence from the common ancestor of modern humans and chimpanzees (Pollard et al. 2006a,b). As explained above, the substitution rates obtained by Perelman et al. (2011) are also higher in the human lineage than in the chimpanzee lineage. Accordingly, in our study the muscle evolutionary rates are also higher in the former lineage (Fig. 2). The muscle evolutionary rate in the Lemur lineage is extremely low (0.00: Fig. 2), and molecular studies also suggest that rates of mtDNA evolution in the Lemur lineage are remarkably low (e.g. Hasegawa et al. 1990). Poux & Douzery (2004) also reported a low rate of evolution of the nuclear gene encoding IRBP in lemuriforms but, contrary to our study of muscles, in their molecular study the lorisiforms displayed a significantly higher evolutionary rate than lemuriforms.
In a study of a subset of 10 markers from putatively neutral, non-coding, non-repetitive regions of the genome, Peng et al. (2009) reported that the lineage including Macaca and Papio has an evolutionary rate 1.36–1.44 times higher than the lineage that includes hominoids. Although in general the muscle evolutionary rates of hominoids obtained in our study are higher than in other primate clades (Fig. 2; see above), the node leading to the Papionini has a higher muscle evolutionary rate than at any hominoid node (Fig. 2). The study of Peng et al. (2009) also showed that in their 10 markers the macaque lineage has evolved almost twice as fast as the baboon lineage, and in our study the muscle evolutionary rate leading to Macaca is about 2.5 times higher than that leading to Papio. Analyses of ENCODE data have also shown that the branch leading to macaque is significantly longer than that leading to baboon (Margulies et al. 2007). Another study using over 8 million base pairs of aligned genomic sequences among several Old World monkeys also indicates significant rate differences between macaques and baboons (Elango et al. 2009). However, within the 10 markers examined by Peng et al. (2009), New World monkeys evolved at a faster rate overall than the hominoids, while within our muscle data all of the hominoid nodes, except the one leading to Gorilla, evolved at rates that are faster than those at the nodes leading to the Plaryrrhini, the Cebidae + Aotidae and to Aotus (Fig. 2). As stressed by Peng et al. (2009), these and other findings indicate that differences in molecular rates among lineages are a common feature of primate genome evolution. In particular, Peng et al.'s (2009) results show that evolutionary molecular rate variation is a common phenomenon even in putatively neutral genomic regions. The nucleotide substitution rates obtained by Perelman et al. (2011) are also similar to the muscle rates obtained by us, in the sense that the rates at the nodes leading to each strepsirrhine genus shown in the figure are lower than the rates at the nodes leading to the Lemuriformes, Lorisiformes and Strepsirrhini. However, the substitution rates obtained by them at the nodes leading to the Lorisiformes (8.27 substitutions/site/MY × 104) and Strepsirrhni (9.91) are higher than the rate at the node leading to the Primates (7.94), while the muscle rate at the node leading to the latter clade (0.75) is substantially higher than that at any of the nodes leading to the strepsirrhine clades.
Evolutionary rates and internal constraints
As explained above, a particularly striking and intriguing result of our study is that the muscle rate at the platyrrhine node leading to the Cebidae plus Aotidae is the same as that at the subsequent node leading to the genus Aotus (and is very similar to that at the node leading to the whole Platyrrhini clade), and that the rate at the node leading to the Cercopithecidae is the same as that at the subsequent nodes leading to Colobus and to Cercopithecus. As these are the only rates that are exactly similar to each other across the whole tree set out in Fig. 2, and as this happens in two different clades, it seems that for some reason the number of morphological evolutionary changes accumulated per period of time in at least some nodes in the same clade is essentially constant. This would be the expectation for molecular evolutionary changes according to the neutral model of evolution (see below), but to our knowledge this has not been reported for any type of morphological evolutionary changes, at least within the order Primates. In fact, the muscle rate at the node leading to strepsirrhines (0.29) and then at the subsequent nodes leading to the lorisiforms (0.22) and to the lemuriforms (0.24) are also very similar to each other, particularly when one compares the differences with the range of different rates within all the primate nodes shown in Fig. 2 (0.00–2.72). The same can be said about the rates at the nodes leading to the Hominidae (1.47), Homininae (1.50) and Hominini (1.53). These results could be used to support the proposal by authors such as Gould (2002) that ‘internal’ (e.g. ontogenetic) constraints play an important role in evolution. That is, in the last 25 Ma there have been for instance major climate and environmental changes in Africa and Asia, yet the rate of muscle changes accumulated during that period at the nodes leading to the Cercopithecidae and then to Colobus, and also to Cercopithecus, is exactly the same. One could argue that this could be explained by the occurrence of functional constraints that would lead to a scenario where muscles of a same functional group/anatomical region are evolving in a correlated fashion. However, what is particularly striking is the fact that these similarities in overall rates do not necessarily correspond to similarities in partial rates for the HN and PU regions, because at the node leading to the Cercopithecidae the rate for the HN changes is 0.19, at the node leading to Colobus it is 0.00, and at the node leading to Cercopithecus it is 0.38: the respective rates for the PU changes are 0.19, 0.38 and 0.00.
For those authors that support the paradigm of ‘internal constraints’ the analysis of these partial rates would be viewed as evidence that ontogenetic constraints are so strong and interconnected that the potential for overall change accumulated in the different regions of the body is limited. This is in line with the results of recent studies showing that in early organogenesis, and particularly during the so-called ‘phylotypic stage’, there is substantial interactivity among different body modules and thus there is low effective modularity (e.g. Galis & Metz, 2007). It has also been argued that from a developmental perspective, if extensive somatic investment is made in one structure of one body module, this could limit investment dedicated to the formation of another structure from not only that module but also from other body modules; it is also possible that constructional trade-offs constrain investment in whole phenotypes because the structural space in organisms is limiting (e.g. Hulsey & Hollingsworth, 2011).
According to Galis & Metz (2007), examples of stable (gradualism) and static (stasis) evolution are mainly associated with strong pleiotropic constraints. But if these pleiotropic constraints are broken the rate of evolution can increase, due to the following major reasons: (i) heterochronic events (e.g. the weaker constraint on variation in the number of cervical vertebrae in birds, compared with mammals, may in part be due to the later stage at which the cervico-thoracic boundary is determined); (ii) the incidence of disease (e.g. one of the negative pleiotropic effects associated with cervical ribs in modern humans is childhood cancer: see above); and (iii) the relaxation of stabilizing selection, particularly in combination with strong directional selection. Within our study, the constant muscle rate of 0.38 at the nodes leading to cercopithecids and then to Colobus and to Cercopithecus can be given as an example of slow, gradual evolution, whereas the fast rate of change at the node leading to the Papionini (2.56) and then the subsequent slowing down of muscle evolution at the nodes leading to Macaca (0.67) and to Papio (0.26) could be used as an example of the type of evolution proposed in Eldredge & Gould's (1972) theory of punctuated equilibrium (see also Gould, 1977, 2002). The fact that in the node leading to the Papionini the muscle rate of total changes (2.56) is almost seven times faster than the rates of 0.38 at the other cercopithecid nodes mentioned above can be due to a breaking of pleiotropic constraints followed by a strong increase of the evolutionary rate, as proposed by Galis & Metz (2007). We plan to undertake future work to specifically test if there are any events, within our phylogenetic results, that do represent true instances of punctuacted equilibrium or of evolutionary stasis.
Conclusions
With respect to the two main questions we addressed in this study, the most remarkable and surprising outcome of our analysis of muscle evolutionary rates is that there are several cases in which the rates of various lineages of each of the major primate clades are strikingly similar. However, there are also examples of a shift towards a faster rate of muscle evolution that is then followed by a slow down in the rate of muscle evolution. With respect to the second question, there are several examples of substantial differences between muscle and molecular evolutionary rates at the same primate nodes, as would be predicted by the neutral model of evolution. But at other places on the tree the muscle evolutionary rates obtained in our study are similar to the published rates of molecular evolution. Our study therefore suggests that the tempo and mode of primate and human evolution is complex, and provides examples of different models of evolution occurring in the very same clade, the Primates. It suggests that at the level of such major mammalian and probably vertebrate clades, simplistic dichotomies such as ‘gradual vs. punctuated’ and ‘neutral vs. non-neutral’ are not useful.
In this sense, our study is in agreement with recent molecular studies showing that a same clade might have very different evolutionary rates for different parts of the genome. For instance, authors often refer to a ‘molecular hominid slowdown’, but a study of MT-CYB (better known as cytochrome b) revealed that humans (together with the African Elephant) have the highest rate among the 10 sequences (representing eight orders) examined (Bininda-Emonds, 2007). Also, the overall study of 44 genes undertook by Bininda-Emonds (2007) did support the idea that most hominids have in general slow rates of evolution, but revealed that orangutans have a much faster rate than other hominids. A still different scenario was pointed out by a large-scale analysis of lineage-specific rates of single-nucleotide substitutions among hominoids (Elango et al. 2006), which, in contrast to the results obtained in various other molecular studies as well as to the results obtained from our myological study (Fig. 2), revealed that orangutans, but also gorillas, evolved faster than chimpanzees and humans. In this respect, an important aspect of our study is that it revealed that, contrary to the idea of a ‘general molecular slow-down of hominoids’, the muscle evolutionary rates at the nodes leading to, or within, the hominoids are among the highest within the whole Primate clade. In order to expand our study of evolutionary rates and of the complexity of primate evolution, we intend to include the muscles of other regions (e.g. the thorax, back, pelvic region and lower limbs) plus the hard tissues of all of the major regions of the body.
Acknowledgments
The authors thank the numerous institutions and individuals that have allowed us to obtain and dissect the primate specimens studied by us, as well as our colleagues and the Editor and two anonymous reviewers for insightful comments. The contributions of RD were supported by a George Washington University (GW) Presidential Merit Fellowship and by a Howard University College of Medicine start-up package, those of ZP by a grant from the National Natural Science Foundation of China (31071903), and those of BW by the GW University Professorship in Human Origins and support from the GW Provost via the GW Signature Program.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Appendix S1. Synapomorphies of clades and apomorphies of terminal taxa.
References
- Arnold C, Matthews LJ, Nunn CL. The 10kTrees Website: a new online resource for primate phylogeny. Evol Anthropol. 2010;19:114–118. [Google Scholar]
- Bakewell MA, Shi P, Zhang J. More genes underwent positive selection in chimpanzee evolution than in human evolution. Proc Natl Acad Sci USA. 2007;104:7489–7494. doi: 10.1073/pnas.0701705104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bininda-Emonds ORP. Fast genes and slow clades: comparative rates of molecular evolution in mammals. Evol Bioinformatics. 2007;3:59–85. [PMC free article] [PubMed] [Google Scholar]
- Bonner T. The Evolution of Complexity. Princeton: Princeton University Press; 1988. [Google Scholar]
- Bromham L, Woolfit M, Lee MSY, et al. Testing the relationship between morphological and molecular rates of change along phylogenies. Evolution. 2002;56:1921–1930. doi: 10.1111/j.0014-3820.2002.tb00118.x. [DOI] [PubMed] [Google Scholar]
- Chatterjee HJ, Ho SYM, Barnes I, et al. Estimating the phylogeny and divergence times of primates using a supermatrix approach. BMC Evol Biol. 2009;9:259. doi: 10.1186/1471-2148-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conroy GC. The inverse relationship between species diversity and body mass: do primates play by the “rules”? J Hum Evol. 2003;45:43–55. doi: 10.1016/s0047-2484(03)00082-4. [DOI] [PubMed] [Google Scholar]
- Cooper N, Purvis A. What factors shape rates of phenotypic evolution? A comparative study of cranial morphology of four mammalian clades. J Evol Biol. 2009;22:1024–1035. doi: 10.1111/j.1420-9101.2009.01714.x. [DOI] [PubMed] [Google Scholar]
- Davies TJ, Savolainen V. Neutral theory, phylogenies, and the relationship between phenotypic change and evolutionary rates. Evolution. 2006;60:476–483. [PubMed] [Google Scholar]
- Diogo R, Abdala V. Muscles of Vertebrates – Comparative Anatomy, Evolution, Homologies and Development. Oxford: Taylor & Francis; 2010. [Google Scholar]
- Diogo R, Wood BA. Soft-tissue anatomy of the primates: phylogenetic analyses based on the muscles of the head, neck, pectoral region and upper limb, with notes on the evolution of these muscles. J Anat. 2011;219:273–359. doi: 10.1111/j.1469-7580.2011.01403.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diogo R, Wood BA. Comparative Anatomy and Phylogeny of Primate Muscles and Human Evolution. Oxford: Taylor & Francis; 2012. [Google Scholar]
- Diogo R, Abdala V, Lonergan NL, et al. From fish to modern humans – comparative anatomy, homologies and evolution of the head and neck musculature. J Anat. 2008;213:391–424. doi: 10.1111/j.1469-7580.2008.00953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diogo R, Abdala V, Aziz MA, et al. From fish to modern humans – comparative anatomy, homologies and evolution of the pectoral and forelimb musculature. J Anat. 2009a;214:694–716. doi: 10.1111/j.1469-7580.2009.01067.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diogo R, Wood BA, Aziz MA, et al. On the origin, homologies and evolution of primate facial muscles, with a particular focus on hominoids and a suggested unifying nomenclature for the facial muscles of the Mammalia. J Anat. 2009b;215:300–319. doi: 10.1111/j.1469-7580.2009.01111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diogo R, Potau JM, Pastor JF, et al. Photographic and Descriptive Musculoskeletal Atlas of Gorilla. Oxford: Taylor & Francis; 2010. [Google Scholar]
- Elango N, Thomas JW, Program NCS, et al. Variable molecular clocks in hominoids. Proc Natl Acad Sci USA. 2006;103:1370–1375. doi: 10.1073/pnas.0510716103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elango N, Lee J, Peng Z, et al. Evolutionary rate variation in Old World monkeys. Biol Letters. 2009;5:405–408. doi: 10.1098/rsbl.2008.0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldredge N, Gould SJ. Punctuated equilibria: an alternative to phyletic gradualism. In: Schopf TJM, editor. Models in Paleobiology. San Francisco: Freeman Cooper; 1972. pp. 82–115. [Google Scholar]
- Fabre P-H, Rodrigues A, Douzery EJP. Patterns of macroevolution among Primates inferred from a supermatrix of mitochondrial and nuclear DNA. Mol Phylogenet Evol. 2009;53:808–825. doi: 10.1016/j.ympev.2009.08.004. [DOI] [PubMed] [Google Scholar]
- Galis F, Metz JAJ. Evolutionary novelties: the making and breaking of pleiotropic constraints. Int Comp Biol. 2007;47:409–419. doi: 10.1093/icb/icm081. [DOI] [PubMed] [Google Scholar]
- Galis F, Van Dooren TJ, Feuth JD, et al. Extreme selection in humans against homeotic transformations of cervical vertebrae. Evolution. 2006;60:2643–2654. [PubMed] [Google Scholar]
- Gibbons A. Turning back the clock: slowing the pace of prehistory. Science. 2012;338:189–191. doi: 10.1126/science.338.6104.189. [DOI] [PubMed] [Google Scholar]
- Gould SJ. Ontogeny and Phylogeny. Cambridge: Harvard University Press; 1977. [Google Scholar]
- Gould SJ. The Structure of Evolutionary Theory. Belknap: Harvard; 2002. [Google Scholar]
- Hansen TF, Houle D. Evolvability, stabilizing selection, and the problem of stasis. In: Pigliucci M, Preston K, editors. The Evolutionary Biology of Complex Phenotypes. Oxford: Oxford University Press; 2004. pp. 130–150. [Google Scholar]
- Hasegawa M, Kishino H, Hayasaka K, et al. Mitochondrial DNA evolution in primates: transition rate has been extremely low in lemur. J Mol Evol. 1990;31:113–121. doi: 10.1007/BF02109480. [DOI] [PubMed] [Google Scholar]
- Hulsey CD, Hollingsworth PR. Do constructional constraints influence cyprinid (Cyprinidae: Leuciscinae) craniofacial evolution? Biol J Linn Soc. 2011;103:136–146. [Google Scholar]
- Kimura M. Evolutionary rate at the molecular level. Nature. 1968;5129:624–626. doi: 10.1038/217624a0. [DOI] [PubMed] [Google Scholar]
- Margulies EH, Cooper GM, Asimenos G, et al. Analyses of deep mammalian sequence alignments and constraint predictions for 1% of the human genome. Genome Res. 2007;17:760–774. doi: 10.1101/gr.6034307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matilla TM, Bokma F. Extant mammal body masses suggest punctuated equilibrium. Proc R Soc B. 2008;275:2195–2199. doi: 10.1098/rspb.2008.0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Z, Elango N, Wildman DE, et al. Primate phylogenomics: developing numerous non-coding, non-repetitive markers for ecological and phylogenetic applications and analysis of evolutionary rate variation. BMC Genomics. 2009;10:247. doi: 10.1186/1471-2164-10-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelman P, Johnson WE, Roos C, et al. A molecular phylogeny of living primates. PLoS Genet. 2011;7:e1001342. doi: 10.1371/journal.pgen.1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, et al. Forces shaping the fastest evolving regions in the human genome. PLoS Genet. 2006a;2:e168. doi: 10.1371/journal.pgen.0020168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006b;443:167–172. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- Poux C, Douzery EJ. Primate phylogeny, evolutionary rate variations, and divergence times: a contribution from the nuclear gene IRBP. Am J Phys Anthropol. 2004;124:1–16. doi: 10.1002/ajpa.10322. [DOI] [PubMed] [Google Scholar]
- Purvis A, Webster A, Agapow P-M. Primate life histories and phylogeny. In: Kappeler PM, Pereira M, et al., editors. Primate Life Histories and Socioecology. Chicago: University of Chicago Press; 2003. pp. 25–40. [Google Scholar]
- Simpson GG. The Major Features of Evolution. New York: Columbian University Press; 1953. [Google Scholar]
- Stanley SM. Macroevolution: Patterns and Process. San Francisco: WH Freeman; 1979. [Google Scholar]
- Steiper ME, Seiffert ER. Evidence for a convergent slowdown in primate molecular rates and its implications for the timing of early primate evolution. Proc Natl Acad Sci USA. 2012;109:6006–6011. doi: 10.1073/pnas.1119506109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetushkin EY. Rates of molecular evolution of primates. Russ J Gen. 2003;39:721–736. [PubMed] [Google Scholar]
- Venditti C, Meade A, Pagel M. Multiple routes to mammalian diversity. Nature. 2011;479:393–396. doi: 10.1038/nature10516. [DOI] [PubMed] [Google Scholar]
- Wood BA, Baker J. Evolution in the genus Homo. Ann Rev Ecol Evol Syst. 2012;42:47–69. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.