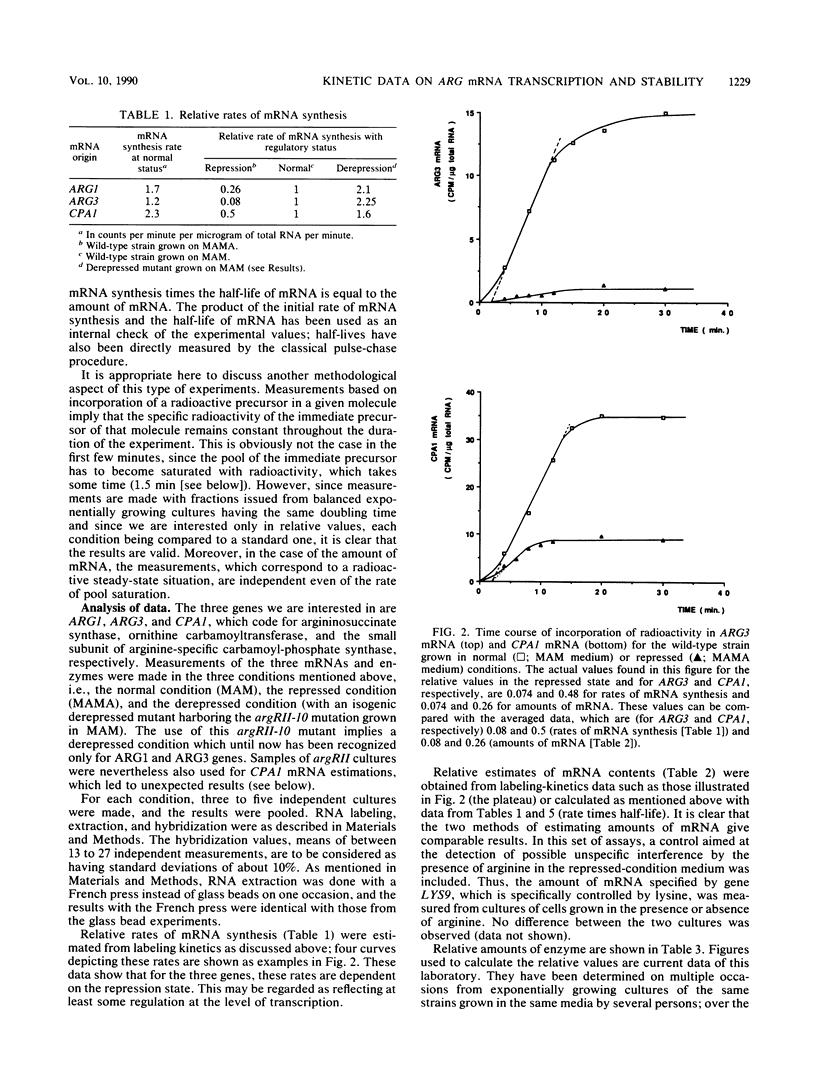
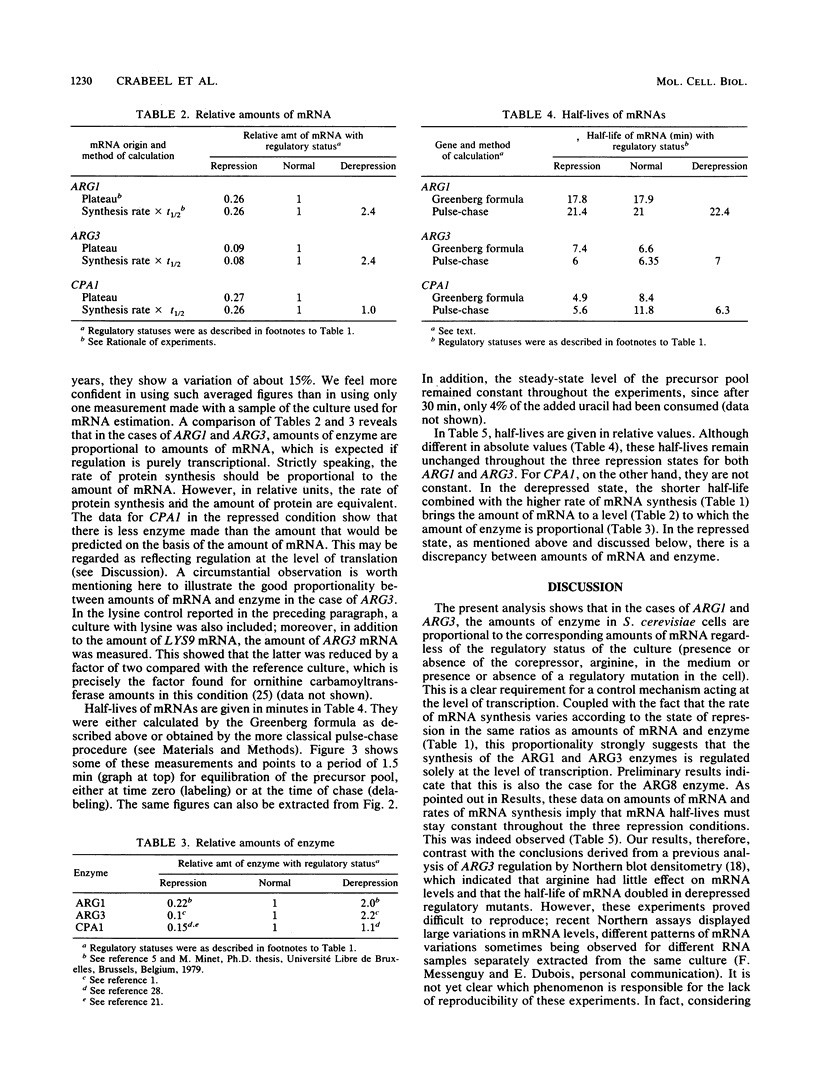
Abstract
A specific repression mechanism regulates arginine biosynthesis in Saccharomyces cerevisiae. The involvement of regulatory proteins displaying DNA-binding features and the location of an operator region between the TATA box and the transcription start of the structural gene ARG3 suggest that this mechanism operates at the level of transcription. A posttranscriptional mechanism has, however, been proposed to account for the conspicuous lack of proportionality between ARG3 mRNA steady-state levels (as determined by Northern [RNA] assays; F. Messenguy and E. Dubois, Mol. Gen. Genet. 189:148-156, 1983) and the cognate enzyme activities. In this work, we have analyzed the time course of the incorporation of radioactive precursors into ARG1 and ARG3 mRNAs and the kinetics of their decay under different regulatory statuses. The results (expressed in terms of relative mRNA levels, relative transcription rates, and mRNA half-lives) give the picture expected from a purely transcriptional control. A similar analysis of expression of the gene CPA1, for which a translational regulation by arginine has been clearly demonstrated (M. Werner, A. Feller, F. Messenguy, and A. Piérard, Cell 49:805-813, 1987), indicates that this gene is also partly regulated at the transcriptional level by the ARGR repressor system. Moreover, the half-life of CPA1 mRNA is reduced twofold in the presence of excess arginine; we suggest that this could be inherent in the mechanism of translational regulation of CPA1.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bechet J., Greenson M., Wiame J. M. Mutations affecting the repressibility of arginine biosynthetic enzymes in Saccharomyces cerevisiae. Eur J Biochem. 1970 Jan;12(1):31–39. doi: 10.1111/j.1432-1033.1970.tb00817.x. [DOI] [PubMed] [Google Scholar]
- Bercy J., Dubois E., Messenguy F. Regulation of arginine metabolism in Saccharomyces cerevisiae: expression of the three ARGR regulatory genes and cellular localization of their products. Gene. 1987;55(2-3):277–285. doi: 10.1016/0378-1119(87)90287-3. [DOI] [PubMed] [Google Scholar]
- Crabeel M., Huygen R., Cunin R., Glansdorff N. The promoter region of the arg3 gene in Saccharomyces cerevisiae: nucleotide sequence and regulation in an arg3-lacZ gene fusion. EMBO J. 1983;2(2):205–212. doi: 10.1002/j.1460-2075.1983.tb01406.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabeel M., Huygen R., Verschueren K., Messenguy F., Tinel K., Cunin R., Glansdorff N. General amino acid control and specific arginine repression in Saccharomyces cerevisiae: physical study of the bifunctional regulatory region of the ARG3 gene. Mol Cell Biol. 1985 Nov;5(11):3139–3148. doi: 10.1128/mcb.5.11.3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabeel M., Seneca S., Devos K., Glansdorff N. Arginine repression of the Saccharomyces cerevisiae ARG1 gene. Comparison of the ARG1 and ARG3 control regions. Curr Genet. 1988 Feb;13(2):113–124. doi: 10.1007/BF00365645. [DOI] [PubMed] [Google Scholar]
- Davis R. H. Compartmental and regulatory mechanisms in the arginine pathways of Neurospora crassa and Saccharomyces cerevisiae. Microbiol Rev. 1986 Sep;50(3):280–313. doi: 10.1128/mr.50.3.280-313.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delforge J., Messenguy F., Wiame J. M. The regulation of arginine biosynthesis in Saccharomyces cerevisiae. The specificity of argR- mutations and the general control of amino-acid biosynthesis. Eur J Biochem. 1975 Sep 1;57(1):231–239. doi: 10.1111/j.1432-1033.1975.tb02295.x. [DOI] [PubMed] [Google Scholar]
- Donahue T. F., Daves R. S., Lucchini G., Fink G. R. A short nucleotide sequence required for regulation of HIS4 by the general control system of yeast. Cell. 1983 Jan;32(1):89–98. doi: 10.1016/0092-8674(83)90499-3. [DOI] [PubMed] [Google Scholar]
- Furter R., Braus G., Paravicini G., Mösch H. U., Niederberger P., Hütter R. Regulation of the TRP4 gene of Saccharomyces cerevisiae at the transcriptional level and functional analysis of its promotor. Mol Gen Genet. 1988 Jan;211(1):168–175. doi: 10.1007/BF00338409. [DOI] [PubMed] [Google Scholar]
- Gillespie D., Spiegelman S. A quantitative assay for DNA-RNA hybrids with DNA immobilized on a membrane. J Mol Biol. 1965 Jul;12(3):829–842. doi: 10.1016/s0022-2836(65)80331-x. [DOI] [PubMed] [Google Scholar]
- Greenberg J. R. High stability of messenger RNA in growing cultured cells. Nature. 1972 Nov 10;240(5376):102–104. doi: 10.1038/240102a0. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. G. A hierarchy of trans-acting factors modulates translation of an activator of amino acid biosynthetic genes in Saccharomyces cerevisiae. Mol Cell Biol. 1985 Sep;5(9):2349–2360. doi: 10.1128/mcb.5.9.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. GCN4 protein, synthesized in vitro, binds HIS3 regulatory sequences: implications for general control of amino acid biosynthetic genes in yeast. Cell. 1985 Nov;43(1):177–188. doi: 10.1016/0092-8674(85)90022-4. [DOI] [PubMed] [Google Scholar]
- Lacroute F., Piérard A., Grenson M., Wiame J. M. The biosynthesis of carbamoyl phosphate in Saccharomyces cerevisiae. J Gen Microbiol. 1965 Jul;40(1):127–142. doi: 10.1099/00221287-40-1-127. [DOI] [PubMed] [Google Scholar]
- Losson R., Lacroute F. Interference of nonsense mutations with eukaryotic messenger RNA stability. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5134–5137. doi: 10.1073/pnas.76.10.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messenguy F. Concerted repression of the synthesis of the arginine biosynthetic enzymes by aminoacids: a comparison between the regulatory mechanisms controlling aminoacid biosyntheses in bacteria and in yeast. Mol Gen Genet. 1979 Jan 16;169(1):85–95. doi: 10.1007/BF00267549. [DOI] [PubMed] [Google Scholar]
- Messenguy F., Dubois E., Descamps F. Nucleotide sequence of the ARGRII regulatory gene and amino acid sequence homologies between ARGRII PPRI and GAL4 regulatory proteins. Eur J Biochem. 1986 May 15;157(1):77–81. doi: 10.1111/j.1432-1033.1986.tb09640.x. [DOI] [PubMed] [Google Scholar]
- Messenguy F., Dubois E. Participation of transcriptional and post-transcriptional regulatory mechanisms in the control of arginine metabolism in yeast. Mol Gen Genet. 1983;189(1):148–156. doi: 10.1007/BF00326068. [DOI] [PubMed] [Google Scholar]
- Messenguy F., Dubois E. The yeast ARGRII regulatory protein has homology with various RNases and DNA binding proteins. Mol Gen Genet. 1988 Jan;211(1):102–105. doi: 10.1007/BF00338399. [DOI] [PubMed] [Google Scholar]
- Messenguy F., Feller A., Crabeel M., Piérard A. Control-mechanisms acting at the transcriptional and post-transcriptional levels are involved in the synthesis of the arginine pathway carbamoylphosphate synthase of yeast. EMBO J. 1983;2(8):1249–1254. doi: 10.1002/j.1460-2075.1983.tb01577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messenguy F. Regulation of arginine biosynthesis in Saccharomyces cerevisiae: isolation of a cis-dominant, constitutive mutant for ornithine carbamoyltransferase synthesis. J Bacteriol. 1976 Oct;128(1):49–55. doi: 10.1128/jb.128.1.49-55.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Passmore S., Maine G. T., Elble R., Christ C., Tye B. K. Saccharomyces cerevisiae protein involved in plasmid maintenance is necessary for mating of MAT alpha cells. J Mol Biol. 1988 Dec 5;204(3):593–606. doi: 10.1016/0022-2836(88)90358-0. [DOI] [PubMed] [Google Scholar]
- Ramos F., Dubois E., Piérard A. Control of enzyme synthesis in the lysine biosynthetic pathway of Saccharomyces cerevisiae. Evidence for a regulatory role of gene LYS14. Eur J Biochem. 1988 Jan 15;171(1-2):171–176. doi: 10.1111/j.1432-1033.1988.tb13773.x. [DOI] [PubMed] [Google Scholar]
- Ramos F., Thuriaux P., Wiame J. M., Bechet J. The participation of ornithine and citrulline in the regulation of arginine metabolism in Saccharomyces cerevisiae. Eur J Biochem. 1970 Jan;12(1):40–47. doi: 10.1111/j.1432-1033.1970.tb00818.x. [DOI] [PubMed] [Google Scholar]
- Thuriaux P., Ramos F., Piérard A., Grenson M., Wiame J. M. Regulation of the carbamoylphosphate synthetase belonging to the arginine biosynthetic pathway of Saccharomyces cerevisiae. J Mol Biol. 1972 Jun 20;67(2):277–287. doi: 10.1016/0022-2836(72)90241-0. [DOI] [PubMed] [Google Scholar]
- Waldron C., Lacroute F. Effect of growth rate on the amounts of ribosomal and transfer ribonucleic acids in yeast. J Bacteriol. 1975 Jun;122(3):855–865. doi: 10.1128/jb.122.3.855-865.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner M., Feller A., Messenguy F., Piérard A. The leader peptide of yeast gene CPA1 is essential for the translational repression of its expression. Cell. 1987 Jun 19;49(6):805–813. doi: 10.1016/0092-8674(87)90618-0. [DOI] [PubMed] [Google Scholar]
- Wolfner M., Yep D., Messenguy F., Fink G. R. Integration of amino acid biosynthesis into the cell cycle of Saccharomyces cerevisiae. J Mol Biol. 1975 Aug 5;96(2):273–290. doi: 10.1016/0022-2836(75)90348-4. [DOI] [PubMed] [Google Scholar]



