Abstract
The mechanosensitive channel of small conductance (MscS) contributes to the survival of bacteria during osmotic downshock by transiently opening large diameter pores for the efflux of cellular contents before the membrane ruptures. Two crystal structures of the Escherichia coli MscS are currently available, the wild type protein in a nonconducting state at 3.7 Å resolution (Bass et al., Science 2002; 298:1582–1587) and the Ala106Val variant in an open state at 3.45 Å resolution (Wang et al., Science 2008; 321:1179–1183). Both structures used protein solubilized in the detergent fos-choline-14. We report here crystal structures of MscS from E. coli and Helicobacter pylori solubilized in the detergent β-dodecylmaltoside at resolutions of 4.4 and 4.2 Å, respectively. While the cytoplasmic domains are unchanged in these structures, distinct conformations of the transmembrane domains are observed. Intriguingly, β-dodecylmaltoside solubilized wild type E. coli MscS adopts the open state structure of A106V E. coli MscS, while H. pylori MscS resembles the nonconducting state structure observed for fos-choline-14 solubilized E. coli MscS. These results highlight the sensitivity of membrane protein conformational equilibria to variations in detergent, crystallization conditions, and protein sequence.
Keywords: mechanosensitive channel, MscS, membrane protein structure, protein structure
Introduction
Bacteria can experience significant environmental stresses that require effective counter measures for survival. Among these stresses are sudden changes in external osmolarity that drives the flow of water across the cell membrane, resulting in either dehydration (osmotic upshock) or swelling (downshock), which can have dire consequences.1,2 Mechanosensitive channels play a major role in the ability of bacteria to survive abrupt osmotic pressure downshock by transiently opening large pores. The accompanying efflux of cellular contents reduces the membrane tension and thereby mitigates the possibility of catastrophic membrane rupture.3 Two families of mechanosensitive channels that confer protection against osmotic downshock have been identified in bacteria,4,5 MscL and MscS, the mechanosensitive channels of large and small conductance, respectively. MscL and MscS both form large, relatively nonselective channels (estimated pore diameter in excess of 10 Å) with conductances several orders of magnitude greater than ion selective channels. While proteins related to MscS and MscL are predicted to exist in all three domains of life,6,7 MscS family members in particular are widely distributed throughout bacterial and archaeal genomes, and multiple family members are often present (e.g. the Escherichia coli genome encodes five MscS-related proteins in addition to MscS itself8,9).
Since the original discovery of MscS by Booth and coworkers,5 an increasingly detailed understanding of the molecular basis for MscS function has been achieved,10 including two crystal structures of E. coli MscS (EcMscS), the wild type channel at 3.7 Å resolution (PDB 2OAU11,12) and the Ala106Val (A106V) variant at 3.45 Å resolution (PDB 2VV513), both solubilized in the detergent fos-choline-14 (FC14). These structures have defined the basic molecular architecture for this protein family, which exists as a heptamer of 286 residue subunits [Fig. 1(a)]. Each subunit contains an N-terminal transmembrane (TM) domain consisting of three membrane spanning helices (denoted TM1, TM2, and TM3), with the remainder of the protein forming a cytoplasmic domain composed of a five-stranded β-sheet domain, two α-helices and a three-stranded β-sheet domain. In the heptamer, the permeation pathway is formed by the packing of TM3s, which may be further described in terms of two helical segments, TM3a and TM3b, separated by a kink or hinge at Gly113. The permeation pathway is flanked by TM1-TM2 helical hairpins, while the cytoplasmic domains form a large fenestrated chamber connecting the permeation pathway to the intracellular environment. Despite the similarities in the cytoplasmic domains of these two structures, significant changes are evident in the TM helices. These include a rearrangement of the TM1-TM2 hairpins that accompany a pivoting of the TM3a helix around Gly113, the hinge position in TM3. As a consequence of these changes, the diameter of the pore increases from ∼5 Å in the wild type structure to ∼13 Å in the A106V variant, and accordingly the wild type 2OAU and A106V 2VV5 structures have been identified with a nonconducting, likely inactivated, state14,15 and open state,13 respectively. Open-state conformations have subsequently been reported16 at lower resolutions for two EcMscS variants with spin label modified cysteines incorporated at positions Asp67 and Leu124 and solubilized in the detergent n-dodecyl-β-D-maltopyranoside (DDM), at resolutions of 4.84 Å (PDB 4AGE) and 4.70 Å (PDB 4AGF), respectively. Quite recently, the structure of a nonconducting conformation of the Thermoanaerobacter tengcongensis MscS (TtMscS; PDB 3T9N) solubilized in DDM has been reported at 3.44 Å resolution.17
Figure 1.

(a) Superposition of EcMscS DDM (green) and HpMscS DDM (blue), with one subunit highlighted from each heptamer, including the representation of the transmembrane (TM) helices as cylinders. The principal differences between these structures occur in the positioning of the transmembrane helices TM1, TM2, and TM3a, while TM3b and the cytoplasmic domain are similar in both structures. This figure was created using Chimera.35 (b) Protein sequence alignment of EcMscS and HpMscS emphasizing the transmembrane organization, with TM1 highlighted in blue, TM2 highlighted in green, and the two sections of TM3 (TM3a and TM3b) highlighted in yellow. The red stars and the red circle denote the pore blocking residues (Leucines 105 and 109 in EcMscS) and the hinge position (Gly113) in TM3, respectively. The sequence alignment was performed using TCoffee36 and the sequence display output using multiple align show.37 [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
To identify additional conformational states and/or improved diffraction quality crystals of MscS, we have screened multiple constructs and detergents for expression and purification. After an extensive search and optimization, we were able to obtain diffraction data to ∼4.5 Å resolution for both Helicobacter pylori MscS (HpMscS) and wild type EcMscS solubilized in DDM. While this resolution is inadequate for detailed modeling, it is sufficient to position the TM helices and establish the overall conformational state. Intriguingly, DDM-solubilized wild type EcMscS resembles the open state structure of the A106V EcMscS, while the DDM-solubilized HpMscS resembles the FC14 solubilized structure of wild type EcMscS in a nonconducting state.
Results and Discussion
The expression, purification, and crystallization of DDM-solubilized EcMscS and HpMscS are described in the Materials and Methods section. The sequence identity between EcMscS and HpMscS (286 and 274 residues, respectively) is 33% [Fig. 1(b)]. DDM-solubilized EcMscS crystallized in space group P212121 with cell dimensions a = 128.2 Å, b = 150.0 Å, and c = 175.8 Å (isomorphous to those described in Ref.16), while HpMscS crystallized in space group P212121 with cell dimensions a = 113.9 Å, b = 143.1 Å, and c = 178.4 Å. These crystal forms are distinct from those found in the earlier studies of EcMscS (2OAU space group P43212, a = 184.3 Å, c = 260.5 Å, and 2VV5 space group C2, a = 230.9 Å, b = 126.6 Å, c = 123.2 Å, and β = 90.4°) as well as TtMscS (3T9N space group P212121, a = 95.0 Å, b = 138.6 Å, and c = 214.6 Å); the packing arrangements in these crystals also differ from each other. Data collection statistics for EcMscS and HpMscS solubilized in DDM are summarized in Table I. The structures were solved by molecular replacement using the conserved cytoplasmic domain of the EcMscS structure, which prevents any bias from the TM helices. This truncated model was used to phase electron density maps to identify the conformational state of the TM domains. The crystallographic refinements were then performed using complete models generated from either the 2OAU or 2VV5 structures. The crystal structure of wild type EcMscS solubilized in DDM was determined at 4.4 Å resolution (PDB 4HWA) and revealed that the TM helix arrangement resembles that of the open state structure (2VV5). Figure 2a illustrates the electron density map for DDM-solubilized EcMscS with the TM domains of the 2OAU and 2VV5 structures superimposed, clearly indicating that the 2VV5 model provides a superior fit. Conversely, the structure of DDM solubilized HpMscS determined at 4.2 Å resolution (PDB 4HW9) corresponds to the nonconducting state of the 2OAU structure [Fig. 2(b)].
Table I.
Data Collection and Refinement Statistics
| Data processing statistics | EcMscS | HpMscS |
|---|---|---|
| Unit cell dimensions | 128.2 | 113.9 |
| (a, b, and c) (Å) | 150 | 143.1 |
| Space group P212121 | 175.8 | 178.4 |
| Wavelength (Å) | 0.9795 | 1 |
| Resolution (Å)a | 46.6–4.37 | 46.1–4.14 |
| (4.48–4.37) | (4.25–4.14) | |
| Unique reflections | 22,521 | 22,539 |
| Redundancy | 6.6 | 3.6 |
| Completeness (%) | 99.6(99.8) | 98.8(98.4) |
| I/σ | 18.1(3.9) | 13.6(2.2) |
| Rmergea,b | 0.053(0.552) | 0.048(0.755) |
| Refinement statistics | ||
| Resolution(Å) a | 37.9–4.37 | 44.5–4.14 |
| (4.57–4.37) | (4.36–4.14) | |
| R-worka,c | 0.267(0.295) | 0.283(0.393) |
| R-freea,c | 0.288(0.331) | 0.311(0.444) |
| Average B factor (Å b) | 192 | 172 |
| rmsd bond length (Å) | 0.006 | 0.015 |
| rmds bond angle (°) | 1.11 | 2.22 |
| Ramachandran plot (favored, allowed, outliers, %) d | 91, 8, 1 | 94, 6, 0 |
Numbers in parentheses represent data in the highest resolution shell.
Rmerge(I) = ∑hkl((∑i|Ihkl,i − <Ihkl>|)/∑iIhkl,i).
Rcryst = ∑hkl (|Fobs| − |Fcalc|) /∑hkl|Fobs|. Rfree was computed identically, except that ∼5% of the reflections were omitted as a test set.
Assigned with PHENIX.28
Figure 2.
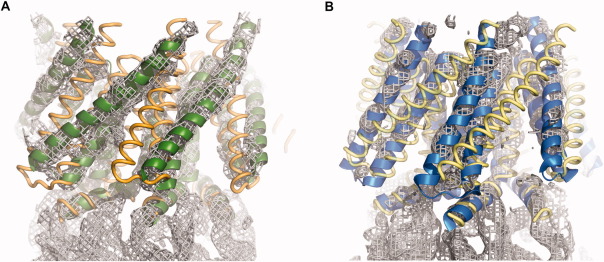
Electron density maps in the transmembrane domain regions of DDM solubilized EcMscS (a) and DDM solubilized HpMscS (b). The electron density maps were calculated following molecular replacement with PHENIX28 using only the cytoplasmic domain to minimize model bias in the TM region, followed by averaging of the density with MAIN.38 The 2OAU and 2VV5 structures are superimposed to illustrate the correspondence between the electron density maps and the nonconducting and open state structures. For DDM solubilized EcMscS (a), the 2VV5 and 2OAU structures are illustrated as green ribbons and gold coils, respectively. For DDM solubilized HpMscS (b), the 2OAU and 2VV5 structures are depicted as blue ribbons and yellow coils, respectively. This figure was created with PyMol.39
In comparing these structures, a seemingly reasonable expectation would be that larger the detergent micelle, the larger should be the nonprotein volume of the unit cell. Further assuming that the micelle size of the pure detergent is representative of that observed in the protein-detergent complex, the detergent with the larger micelle size would then be anticipated to have a larger Matthews' coefficient, VM, which gives the ratio of the unit cell volume to total protein molecular weight.18 This is not the case, however; while the molecular weight of a DDM micelle is ∼50% greater than for FC14 (72 kD vs. 47 kD, respectively19), the FC14 solubilized EcMscS structures have larger Matthews' coefficients (VM = 4.8 and 4.2 Å3 Da−1 for 2OAU and 2VV5, respectively) than the DDM solubilized EcMscS structures (3.8 Å3 Da−1). Furthermore, while one might expect that the open state would have a larger volume and consequently a larger VM than a nonconducting state, there is no correlation between conformational state and VM. Specifically, the nonconducting state structures (2OAU, HpMscS and TtMscS) have VM = 4.8, 3.2 and 3.2 Å3 Da−1, respectively, while the open state structures (2VV5 and DDM solublized EcMscS) are 4.2 and 3.8 Å3 Da−1, respectively.
Surprisingly, despite differences in conformational states and sequences, the same basic crystal packing is observed for both DDM solubilized EcMscS and DDM solubilized HpMscS [Figs. 3(a,b)]. In both structures, MscS heptamers are packed in a herringbone pattern in the crystallographic ac plane with the sevenfold axis roughly perpendicular to the b axis; adjacent planes are related by the crystallographic 21 screw axes along b [Fig. 3(c)]. Interactions between planes are primarily stabilized by lattice contacts between the cytoplasmic domains, while interactions within a plane involve contacts between the cytoplasmic domain and the periplasmic surface of the translocation pathway. While the lattice contacts cannot be identified in detail at this resolution, they do not appear to involve highly conserved regions. That distinct conformations can be observed within the same basic crystal packing suggests that it might be possible to drive the transformation between conformational states in the crystal by suitable manipulations, perhaps through judicious choice of amphiphilic additives to the crystal that could partition into the micelle and alter their size or stability.
Figure 3.
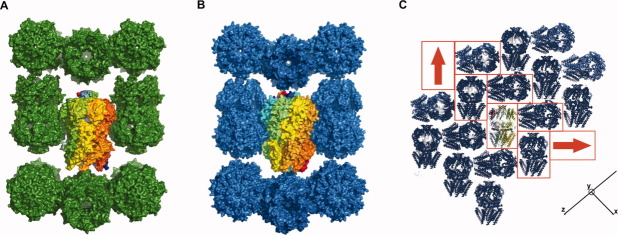
Similarities in the crystal packings of DDM solubilized EcMscS (a) and HpMscS (b). The surface representation with a highlighted central channel indicates possible lattice contacts between the cytoplasmic domains, with more limited contact between the N-terminal and C-terminal domains. The crystallographic b axis is horizontal, with the sevenfold axis of the central MscS heptamer (along diagonal of the ac plane) in the vertical direction. (c) Crystal packing in the ac plane of HpMscS, with a single channel at the center highlighted with multicolored chains. The red boxes identify the herringbone pattern from each stacking subunit. The red arrow denotes the orientation of each channel, with the head corresponding to the C-terminal, cytoplasmic domain while the tail end denotes the N-terminal, transmembrane domains. The x, y, and z axes indicate the orientation of the structure along the crystallographic a, b, and c axes, respectively. A similar packing arrangement is also present in EcMscS. This figure was created with PyMol.39
Detergents play key roles in the structural biology of membrane proteins, both for solubilization from the membrane and for maintaining the structural stability of membrane proteins outside of the natural lipid bilayer. Unfortunately, detergents are not passive by-standers in this process, and as a consequence, they can destabilize–or more generally perturb–the structural state of membrane proteins. Detergents have been classified as either “mild” or “harsh” according to their tendency to destabilize membrane proteins. This classification of detergents can be anecdotal but leads to some general trends (see Ref.20 for a clear discussion). Mild detergents are generally characterized by large, neutral head groups and long alkyl tails, while harsher detergents have small, charged head groups and short tails. DDM is a mild detergent, while fos--choline detergents are typically considered harsher. FCs are often found to solubilize overexpressed membrane proteins, particularly eukaryotic ones,21,22 which gives rise to some concern that they may be partially denaturing.23 Nevertheless, this family of detergents has been used in the successful structure determinations of the 2OAU and 2VV5 forms of EcMscS, as well as an OmpF structure.24 While the displacement of the TM1-TM2 helices from the permeation pathway in 2OAU for wild type EcMscS could potentially reflect partial destabilization of those helices by FC14, the observation that HpMscS could be solublized in the mild detergent DDM reinforces that this is likely a functionally relevant conformational state.
The similarities of the DDM-solubilized structures of EcMscS and HpMscS to previously determined FC14-solubilized structures underscore the mechanistic significance of these two conformational states. The demonstration that these particular conformations can exist under diverse experimental conditions indicates that they are not artifacts of a single set of solution conditions (as also discussed in Ref.16). Since the open and nonconducting state structures have been observed under a variety of conditions and with distinct protein sequences, this suggests that MscS behaves as a bistable system where the conformational equilibrium between the two states is sensitive to the precise experimental conditions. These observations consequently provide both a cautionary tale for establishing the functional relevance of a particular structure and highlight an approach for sampling conformational space by screening different detergents, constructs, homologs, and crystallization conditions.
Materials and Methods
Cloning of MscS homologs
MscS homologs were identified by a BLAST search using the NCBI genome database. Twenty-five homologs were selected, the genes amplified by PCR from genomic DNA, and then cloned into pET expression vectors (pET19b+, pET26b+, and pET28b+; see Table S1). Constructs in pET28b+ have an N-terminal 6×-histidine tag, MscS, and a C-terminal FLAG tag. Constructs in pET26b+ have a pelB signal sequence, MscS, and a C-terminal 6×-histidine tag. The last construct, pET19b+ has an N-terminal 6×-histidine tag and MscS. The constructs were transformed into BL21 DE3 Gold cells (Stratagene) for expression screening.
Expression and monodispersity screening
Initial screenings of MscS expression were conducted using Western blotting analysis to determine which constructs had the highest expression. Constructs for 14 of the 25 MscS homologs exhibited reasonable expression levels in our E. coli expression system (as assessed in comparison to EcMscS.) These constructs were then screened to assess extraction efficiency from a panel of 25 detergents (Table S2). Detergent selection was further refined by performing small-scale expression in two liters of Terrific Broth media with 1% glycerol. Whole cells were solublized using a subset of the 25 detergents and purified using 1 mL gravity flow Ni-NTA columns. Three buffers were used (column equilibration, wash, and elution buffers) consisting of 20 mM Tris pH 7.5, 150 mM NaCl, 10% glycerol, and imidazole at concentrations of 30 mM, 60 mM, and 300 mM, respectively. Size exclusion chromatography (Superdex S200 column) was then used to determine whether constructs were aggregated or monodisperse. The sizing buffer used for EcMscS consisted of 50 mM Tris pH 7.5, 150 mM NaCl, 10% glycerol, 0.05% DDM, and 5 mM dithiothreitol. The same buffer was used for HpMscS, omitting the dithiothreitol. FC14 and DDM provided the most monodisperse samples, while Cymal 5, LDAO, and C12E8 exhibited monodisperse samples with certain MscS homologs but generally showed poor extraction properties. Of the 25 MscS homologs, only seven (Table S1) were sufficiently monodisperse under at least some conditions to proceed to large scale expression and purification.
Large scale expression and purification of EcMscS and HpMscS
Subsequent large-scale growth and expression for crystallization experiments were performed using a 60-L fermenter. Only three of the 25 MscS homologs were successfully scaled up (in addition to EcMscS); these were the MscS from Archaeoglobus fulgidus, H. pylori, and Thermoplasma volcanium. Of these, only the H. pylori and E. coli MscS gave any crystallization leads. Expression was induced with 1 mM IPTG for 1 h, resulting in ∼700–800 g of wet cells. For each HpMscS prep, ∼150–200 g of cells were homogenized using a microfluidizer with buffer containing 20 mM Tris, pH 7.5, 20 mM NaCl, 4 mM MgCl2 and extracted with 1% DDM. Subsequent purification steps were performed in the same buffer with 0.05% DDM. Detergent extracted protein was bound to a large 100-mL Ni-NTA column, washed with five column volumes of 100 mM imidazole, and eluted with 0.5 column volumes of 300 mM imidazole. The eluent was then run on a FLAG affinity column using 20 mM Tris, pH 7.5, 150 mM NaCl, and 0.05% DDM and eluted with 0.1 M glycine, pH 3.5, 150 mM NaCl, and 0.05% DDM. The final eluent was injected onto a Sephacryl S300 26/10 size exclusion column. Concentration of the protein was performed using the Amicon Ultra-15 Centrifugal Filter units (Millipore) with a 100-kDa molecular weight cut off. The N-terminal 6×-histidine tag was removed by thrombin digestion for 2 days at 4°C in thrombin buffer (2 mM Tris pH 7.5, HCl, pH 8.4, 150 mM NaCl, and 2.5 mM CaCl2). For EcMscS, ∼50 g of cells were used for each purification. The purification protocol was modified slightly from above, with an anion exchange column (Source 30Q) replacing the FLAG column before separation on the Sephacryl S300 26/10 column. The eluent from the Ni affinity column was diluted 10 times before loading on a Source 30Q column. The loading was done overnight, and the next morning, the protein was concentrated using the same method as HpMscS. For EcMscS, the N-terminal 6×-histidine tag was left uncleaved for crystallization trials.
Crystallization and x-ray crystallography of EcMscS and HpMscS
HpMscS crystals were prepared by hanging drop vapor diffusion at 20°C using a final protein concentration of 10–15 mg/mL. Over 2000 conditions were screened, with multiple hits from a variety of conditions. The final condition contained 100 mM citrate pH 5.0, 200 mM lithium chloride, 28% PEG 400, and 5% glycerol. The crystals grew in size between ∼200 and 300 μm in about 1 week. EcMscS crystals were grown by hanging drop vapor diffusion at 4°C also using a protein concentration of 10–15 mg/mL. After optimization, the final crystallization condition contained 100 mM Tris HCl pH 8.5, 30% PEG 400, and 200 mM MgCl2. Crystals grew to ∼200 μm in size. The original conditions for crystallizing EcMscS solubilized in FC14 did not produce any crystals, and attempts to crystallize HpMscS in FC14 were also unsuccessful.
Screening and diffraction data collection of EcMscS and HpMscS were conducted at the Stanford Synchrotron Radiation Lightsource on beamline 12-2 using the automated sample mounting system using remote access controlled by Blu-Ice.25–27 Data process and crystallographic calculations were performed using a combination of PHENIX, XIA2, XDS, and CCP4.28–31
Molecular replacement for EcMscS was performed with PHENIX, using the 2VV5 cytosolic domain as the search model with residues 128–280 on each chain. The TM domains from 2VV5 aligned well against the subsequently generated density map, as inspected with COOT.32 The mutated residue at 106 was then changed back from Val to Ala, to use as the input model for refinement. For HpMscS, SCWRL (an algorithm to predict side chain conformation) was used to generate a homology model for HpMscS from a single subunit of the EcMscS model.33,34 For the molecular replacement calculation, the cytosolic domain of 2OAU was utilized with the same residues as for the EcMscS molecular replacement. The full length HpMscS structure generated from SCRWL was superimposed onto the molecular replacement model. Refinements of both EcMscS and HpMscS were performed with PHENIX, incorporating noncrystallographic symmetry restraints. Individual B factors were refined with the weighting scheme determined by PHENIX. For HpMscS, the Ramachandran plot was manually optimized in COOT and then subsequently used as a reference model for further refinement in PHENIX.
The root mean square deviations (rmsd's) between Cα positions of the EcMscS-DDM structure to equivalent residues in the 2VV5 and 2OAU heptameric structures are 0.5 and 7.9 Å, respectively; the corresponding rmsd's for the HpMscS structures are 7.6 Å and 1.8 Å, respectively, indicative of the close similarities of the DDM solubilized EcMscS and HpMscS structures to 2VV5 and 2OAU, respectively. For comparison, the equivalent values for the TtMscS structure are 6.5 Å and 2.9Å, respectively. With the exception of the 2OAU, the other EcMscS structures and HpMscS exhibit nearly sevenfold molecular symmetry, with rmsd's < 0.5 Å when comparing the rotated versions of a structure to the unrotated form (i.e., subunits ABCDEFG superimposed onto BCDEFGA, etc.). For 2OAU, the corresponding values are ∼2.0 Å, which reflects the breakdown from sevenfold symmetry in the TM region.12
Electronic supplementary material
The supplementary material contains Tables S1 and S2 detailing the MscS homologs cloned in this study and the detergents screened for solubilization, respectively. Supplemental Figure S1 illustrates the electron density for DDM solubilized EcMscS and HpMscS calculated from the final refined models. All supplementary material is contained in a single file (supplemental.pdf).
Acknowledgments
We thank the Gordon and Betty Moore Foundation for their generous support of the Molecular Observatory at Caltech. Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource (SSRL), a Directorate of SLAC National Accelerator Laboratory, and an Office of Science User Facility operated for the U.S. Department of Energy Office of Science by Stanford University. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program (P41RR001209), and the National Institute of General Medical Sciences. Coordinates and structure factors have been deposited in the Protein Data Bank of the Research Collaboratory for Structural Bioinformatics, with IDs 4HWA and 4HW9 for the EcMscS and HpMscS structures, respectively.
Supplementary material
Additional Supporting Information may be found in the online version of this article.
References
- 1.Wood JM. Osmosensing by bacteria: signals and membrane-based sensors. Microbiol Mol Biol Rev. 1999;63:230–262. doi: 10.1128/mmbr.63.1.230-262.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Booth IR, Louis P. Managing hypoosmotic stress: aquaporins and mechanosensitive channels in Escherichia coli. Curr Opin Microbiol. 1999;2:166–169. doi: 10.1016/s1369-5274(99)80029-0. [DOI] [PubMed] [Google Scholar]
- 3.Kung C, Martinac B, Sukharev S. Mechanosensitive channels in microbes. Annu Rev Microbiol. 2010;64:313–329. doi: 10.1146/annurev.micro.112408.134106. [DOI] [PubMed] [Google Scholar]
- 4.Sukharev SI, Blount P, Martinac B, Blattner FR, Kung C. A large-conductance mechanosensitive channel in E. coli encoded by mscL alone. Nature. 1994;368:265–268. doi: 10.1038/368265a0. [DOI] [PubMed] [Google Scholar]
- 5.Levina N, Totemeyer S, Stokes NR, Louis P, Jones MA, Booth IR. Protection of Escherichia coli cells against extreme turgor by activation of MscS and MscL mechanosensitive channels: identification of genes required for MscS activity. EMBO J. 1999;18:1730–1737. doi: 10.1093/emboj/18.7.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pivetti CD, Yen MR, Miller S, Busch W, Tseng YH, Booth IR, Saier MH., Jr Two families of mechanosensitive channel proteins. Microbiol Mol Biol Rev. 2003;67:66–85. doi: 10.1128/MMBR.67.1.66-85.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balleza D, Gomez-Lagunas F. Conserved motifs in mechanosensitive channels MscL and MscS. Eur Biophys J. 2009;38:1013–1027. doi: 10.1007/s00249-009-0460-y. [DOI] [PubMed] [Google Scholar]
- 8.Schumann U, Edwards MD, Rasmussen T, Bartlett W, van West P, Booth IR. YbdG in Escherichia coli is a threshold-setting mechanosensitive channel with MscM activity. Proc Natl Acad Sci U S A. 2010;107:12664–12669. doi: 10.1073/pnas.1001405107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Booth IR, Edwards MD, Black S, Schumann U, Miller S. Mechanosensitive channels in bacteria: signs of closure? Nat Rev Microbiol. 2007;5:431–440. doi: 10.1038/nrmicro1659. [DOI] [PubMed] [Google Scholar]
- 10.Naismith JH, Booth IR. Bacterial mechanosensitive channels-MscS: evolution's solution to creating sensitivity in function. Annu Rev Biophys. 2012;41:157–177. doi: 10.1146/annurev-biophys-101211-113227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bass RB, Strop P, Barclay M, Rees DC. Crystal structure of Escherichia coli MscS, a voltage-modulated and mechanosensitive channel. Science. 2002;298:1582–1587. doi: 10.1126/science.1077945. [DOI] [PubMed] [Google Scholar]
- 12.Steinbacher S, Bass RB, Strop P, Rees DC. Structures of the prokaryotic mechanosensitive channels MscL and MscS. In: Hamill OP, editor. Mechanosensitive ion channels, part A. San Diego: Academic Press; 2007. pp. 1–24. [Google Scholar]
- 13.Wang WJ, Black SS, Edwards MD, Miller S, Morrison EL, Bartlett W, Dong CJ, Naismith JH, Booth IR. The structure of an open form of an E. coli mechanosensitive channel at 3.45 Å resolution. Science. 2008;321:1179–1183. doi: 10.1126/science.1159262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anishkin A, Sukharev S. Water dynamics and dewetting transitions in the small mechanosensitive channel MscS. Biophys J. 2004;86:2883–2895. doi: 10.1016/S0006-3495(04)74340-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belyy V, Anishkin A, Kamaraju K, Liu NL, Sukharev S. The tension-transmitting ‘clutch’ in the mechanosensitive channel MscS. Nat Struct Mol Biol. 2010;17:451–458. doi: 10.1038/nsmb.1775. [DOI] [PubMed] [Google Scholar]
- 16.Pliotas C, Ward R, Branigan E, Rasmussen A, Hagelueken G, Huang H, Black S, Booth IR, Schiemann O, Naismith JH. Conformation state of the MscS mechanosensitive channel in solution revealed by pulsed electron-electron double resonance (PELDOR) spectroscopy. Proc Natl Acad Sci USA. 2012;109:E2675–E2682. doi: 10.1073/pnas.1202286109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang X, Wang J, Feng Y, Ge J, Li W, Sun W, Iscla I, Blount P, Li Y, Yang M. Structure and molecular mechanism of an anion-selective MscS-like ion channel. Proc Natl Acad Sci USA. 2012;44:18180–18185. doi: 10.1073/pnas.1207977109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matthews BW. Solvent content of protein crystals. J Mol Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 19.Strop P, Brunger AT. Refractive index-based determination of detergent concentration and its application to the study of membrane proteins. Protein Sci. 2005;14:2207–2211. doi: 10.1110/ps.051543805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prive GG. Detergents for the stabilization and crystallization of membrane proteins. Methods. 2007;41:388–397. doi: 10.1016/j.ymeth.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 21.Chaudhary S, Pak JE, Pedersen BP, Bang LJ, Zhang LB, Ngaw SMM, Green RG, Sharma V, Stroud RM. Efficient expression screening of human membrane proteins in transiently transfected human embryonic kidney 293S cells. Methods. 2011;55:273–280. doi: 10.1016/j.ymeth.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lebon G, Bennett K, Jazayeri A, Tate CG. Thermostabilisation of an agonist-bound conformation of the human adenosine A2A receptor. J Mol Biol. 2011;409:298–310. doi: 10.1016/j.jmb.2011.03.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewinson O, Lee AT, Rees DC. The funnel approach to the precrystallization production of membrane proteins. J Mol Biol. 2008;377:62–73. doi: 10.1016/j.jmb.2007.12.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kefala G, Ahn C, Krupa M, Esquivies L, Maslennikov I, Kwiatkowski W, Choe S. Structures of the OmpF porin crystallized in the presence of foscholine-12. Protein Sci. 2010;19:1117–1125. doi: 10.1002/pro.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soltis SM, Cohen AE, Deacon A, Eriksson T, Gonzalez A, McPhillips S, Chui H, Dunten P, Hollenbeck M, Mathews I, Miller M, Moorhead P, Phizackerley RP, Smith C, Song J, van dem Bedem H, Ellis P, Kuhn P, McPhillips T, Sauter N, Sharp K, Tsyba I, Wolf G. New paradigm for macromolecular crystallography experiments at SSRL: automated crystal screening and remote data collection. Acta Crystallogr D Biol Crystallogr. 2008;64:1210–1221. doi: 10.1107/S0907444908030564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cohen AE, Ellis PJ, Miller MD, Deacon AM, Phizackerley RP. An automated system to mount cryo-cooled protein crystals on a synchrotron beamline, using compact sample cassettes and a small-scale robot. J Appl Crystallogr. 2002;35:720–726. doi: 10.1107/s0021889802016709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McPhillips TM, McPhillips SE, Chiu HJ, Cohen AE, Deacon AM, Ellis PJ, Garman E, Gonzalez A, Sauter NK, Phizackerley RP, Soltis SM, Kuhn P. Blu-Ice and the Distributed Control System: software for data acquisition and instrument control at macromolecular crystallography beamlines. J Synchrotron Radiat. 2002;9:401–406. doi: 10.1107/s0909049502015170. [DOI] [PubMed] [Google Scholar]
- 28.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winter G. Xia2: an expert system for macromolecular crystallography data reduction. J Appl Crystallogr. 2010;43:186–190. [Google Scholar]
- 30.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Q, Canutescu AA, Dunbrack RL., Jr SCWRL and MolIDE: computer programs for side-chain conformation prediction and homology modeling. Nat Protoc. 2008;3:1832–1847. doi: 10.1038/nprot.2008.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Canutescu AA, Shelenkov AA, Dunbrack RL., Jr A graph-theory algorithm for rapid protein side-chain prediction. Protein Sci. 2003;12:2001–2014. doi: 10.1110/ps.03154503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 36.Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 37.Stothard P. The sequence manipulation suite: javascript programs for analyzing and formatting protein and DNA sequences. BioTechniques. 2000;28:1102–1104. doi: 10.2144/00286ir01. [DOI] [PubMed] [Google Scholar]
- 38.Turk D. Density modification in MAIN. In: Read RJ, Sussman J, editors. Proceedings of the NATO Advanced Study Institute on Evolving Methods for Macrmolecular Crystallography. Vol. 254. Berlin: Springer-Verlag; 2007. pp. 111–122. [Google Scholar]
- 39.Schrödinger L. The PyMOL molecular graphics system. http://www.pymol.org/citing. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


