Abstract
Influenza A(H1N1)pdm virus caused the first human pandemic of the 21st century. Although various probiotic Lactobacillus species have been shown to have anti-microbial effects against pneumonia-inducing pathogens, the prophylactic efficacy and mechanisms behind their protection remain largely unknown. Here, we evaluated the prophylactic efficacy of heat-killed Lactobacillus pentosus b240 against lethal influenza A(H1N1)pdm virus infection in a mouse model. To further define the protective responses induced by b240, we performed virologic, histopathologic, and transcriptomic analyses on the mouse lungs. Although we did not observe an appreciable effect of b240 on virus growth, cytokine production, or histopathology, gene expressional analysis revealed that oral administration of b240 differentially regulates antiviral gene expression in mouse lungs. Our results unveil the possible mechanisms behind the protection mediated by b240 against influenza virus infection and provide new insights into probiotic therapy.
Influenza A viruses are zoonotic agents that cause epizootics and epidemics in domestic animals and humans, respectively. Occasionally, they cause pandemics in humans when new strains emerge with substantial antigenic changes in their hemagglutinin (HA). In the spring of 2009, a swine-origin influenza A(H1N1)pdm virus emerged in Mexico that rapidly spread worldwide, causing the first human pandemic of the 21st century1,2.
To defend against influenza virus infection, various interventions have been undertaken. The current first line of defense is vaccination3. Although vaccination does not provide reliable immunogenic protection unless the antigenicity of the vaccine strains matches that of circulating strains, there is merit in the phylactic effect of inducing virus-specific immune responses. The antiviral drugs oseltamivir and zanamivir are used to treat patients infected with influenza viruses; however, the emergence of drug-resistant viruses4,5 suggests the need for alternative therapeutic approaches.
Probiotics are live microorganisms that benefit humans by maintaining an appropriate balance among the bacteria that live in the gut6. During the last few decades, many clinical trials have evaluated probiotic therapy. Previous studies have demonstrated that various Lactobacillus species, represented by the Lactobacillus casei Shirota, rhamnosus GG, gasseri TMC0356, and plantarum strains, have antiviral effects against lethal doses of influenza viruses7,8,9. Although the effectiveness of probiotics against infectious diseases remains largely unexplored, the strong movement toward preventive medicine has increased the importance of developing probiotic therapy.
Lactobacillus pentosus b240 was originally isolated from fermented tea leaves10. This strain enhances IgA production from Peyer's patch cells in mouse gut11 and accelerates salivary IgA secretion in humans12. Recent studies have shown that oral administration of heat-killed b240 protects mice from bacterial and viral infections, such as those caused by Streptococcus pneumoniae and an influenza H1N1 virus (mouse-adapted laboratory strain, A/PR8/1934), by enhancing the innate immune resopnses13,14. However, the mechanisms underlying this protection are poorly understood.
Here, to examine the antiviral effects of oral administration of heat-killed Lactobacillus pentosus b240 against lethal influenza A(H1N1)pdm virus infection in mice, we investigated the morbidity and mortality of mice orally treated with heat-killed Lactobacillus pentosus b240 for 21 days and then infected with a lethal influenza A(H1N1)pdm virus. Further, to define the host responses mediated by b240 administration, we analyzed virus replication, cytokine expression, histopathology, and gene expression in the lungs of mice orally treated with heat-killed Lactobacillus pentosus b240.
Results
Heat-killed Lactobacillus pentosus b240 partially protects mice against lethal influenza A(H1N1)pdm virus infection
A previous study reported that oral administration of heat-killed Lactobacillus pentosus b240 prolonged survival and decreased virus titers in the lungs of mice infected with A/PR8/1934 (H1N1) virus14. To define the prophylactic effects of b240 administration against influenza A(H1N1)pdm virus infection, we orally administered heat-killed Lactobacillus pentosus b240 to mice daily for 21 days and then infected them with mouse-adapted A/California/04/2009 (CA04) virus (Fig. 1a). Oral administration of b240 was continued for 14 days post-infection. Morbidity and mortality were monitored daily for 14 days post-infection.
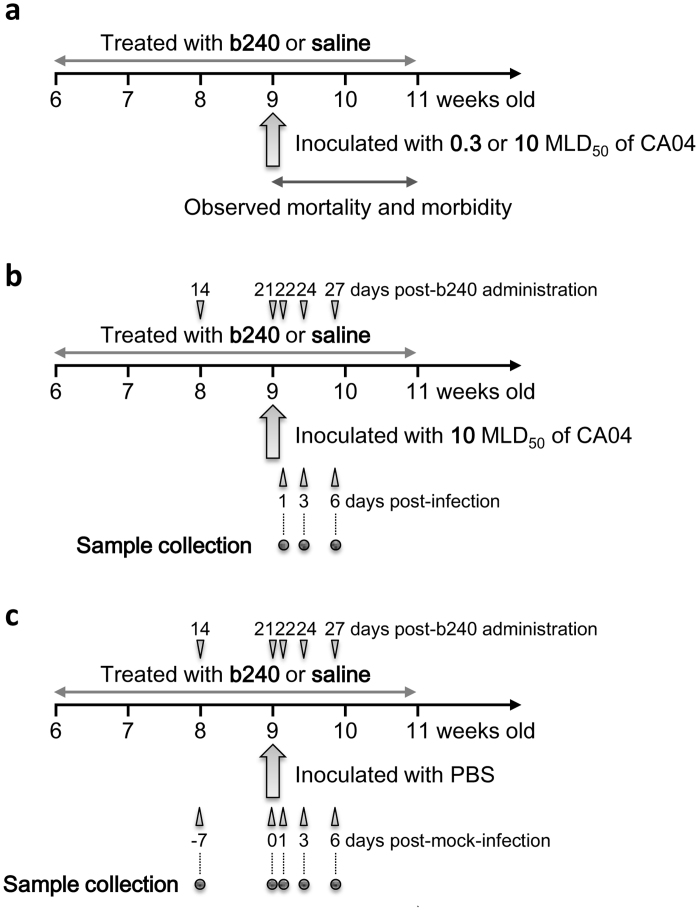
Figure 1. Schedule for the animal experiments.
We conducted three types of experiments to determine the prophylactic effects of b240 administration on pathogenesis and host responses in mice. In all experiments, mice were orally mock-administered with saline or administered with heat-killed Lactobacillus pentosus b240 at a dose of 10 mg/mouse daily for 21 days prior to infection and for 14 days after infection or mock infection. (a) To define the prophylactic effect of b240 administration on mouse survival, mice were infected with 0.3 or 10 MLD50 of mouse-adapted CA04 virus on day 21 post-b240 administration and mortality and morbidity were observed for 14 days post-infection. (b) To define the effects of b240 administration on virus replication, histopathology, cytokine expression, and gene expression, mice were infected with 10 MLD50 of CA04 virus on day 21 post-b240 administration and euthanized on days 1, 3, and 6 post-infection. Their lungs were subjected to virus titration, histopathological examination, cytokine measurement, and microarray analysis. (c) To investigate the immune responses induced by b240 administration in the lungs of mice, mice were mock-infected with PBS on day 21 post-b240 administration and euthanized on days −7, 0, 1, 3, and 6 post-infection (14, 21, 22, 24, and 27 post-b240 administration). Their lungs were subjected to cytokine measurement and microarray analysis.
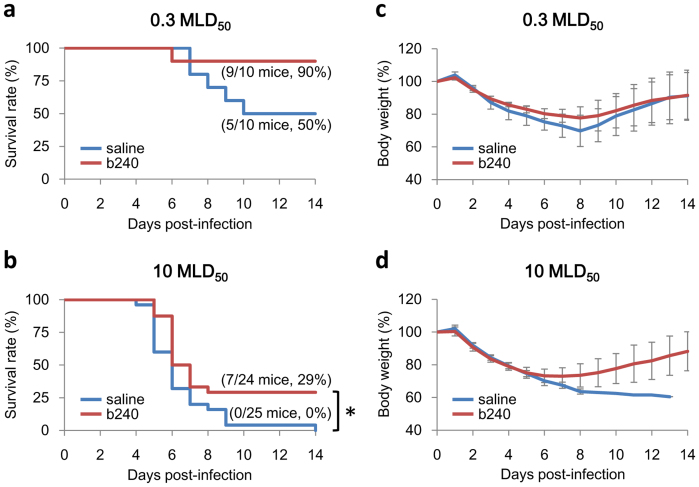
Mice infected with 0.3 mouse LD50 (MLD50) of CA04 virus exhibited a 40% higher survival rate relative that of the control group (P value = 0.076), although there were no substantial differences in body weight between the two groups (Fig. 2a, c). In mice infected with 10 MLD50, there were statistically significant differences in survival (P value = 0.0079) between the two groups (Fig. 2b, d). Based on our finding that oral administration of b240 statistically significantly prolonged mouse survival, we choose the 10 MLD50 dose for downstream analyses.
Figure 2. Efficacy of oral b240 administration in CA04-infected mice.
Mice were administered heat-killed b240 at a dose of 10 mg/mouse daily for 21 days prior to infection and for 14 days after infection. Mice in the control group received saline. They were then intranasally infected with 0.3 or 10 MLD50 of mouse-adapted CA04 virus on day 21 post-b240 administration. Mortality and morbidity were monitored daily for 14 days post-infection. Percent survival (a) and body weight (c) are shown for each group of ten mice infected with 0.3 MLD50 of virus. Percent survival (b) and body weight (d) are shown for b240- or saline-treated groups of 24 or 25 mice infected with 10 MLD50 of virus, respectively. The body weight values are means ± SD for the mice that were alive at each time point. Asterisk: P value < 0.05, significant difference compared to the virus-infected group not treated with b240 (log-rank test).
Heat-killed Lactobacillus pentosus b240 does not affect virus growth or histopathology in the lungs of mice infected with lethal influenza A(H1N1)pdm virus infection
To understand the mechanism by which oral administration of heat-killed b240 protects mice from lethal influenza A(H1N1)pdm virus infection (Fig. 2b, d), we first examined virus growth and the histopathology in the lungs of mice infected with CA04 virus after b240 treatment on days 1, 3, and 6 post-infection (Fig. 1b). The virus growth assay revealed that virus titers in the lungs of b240-treated mice were not statistically significantly different from those in the lungs of control mice at any time point tested (Table 1), indicating that b240 administration had no effect on virus growth in mouse lungs. Furthermore, we found that there were no apparent differences in the extent of pneumonia or viral antigen expression between the lungs of b240-treated mice and those of control mice at all time points tested (Table 2). These results indicate that oral administration of heat-killed b240 augments protection against a lethal dose of influenza A(H1N1)pdm virus by mechanisms that do not substantially affect virus replication or histopathology.
Table 1. Effect of oral administration of heat-killed b240 on virus titers in the lungs of mice infected with CA04 virus.
| Treatment | Virus titer (mean log10 p.f.u. ± SD/g) | ||
|---|---|---|---|
| b240 | 8.9 ± 0.15 | 7.8 ± 0.17 | 7.4 ± 0.32 |
| saline | 9.0 ± 0.13 | 7.9 ± 0.12 | 7.4 ± 0.20 |
Nine mice per group were infected with 10 MLD50 of CA04 virus and three mice in each group were euthanized on days 1, 3, and 6 post-infection. Virus in the lungs was titrated by using plaque assay in MDCK cells. The values are means ± SD (n = 3). Statistical significance was determined with a two-tailed Student's t-test comparing b240- and saline-treated mice at each time point; values were considered to be significantly different when the P value was less than 0.05.
Table 2. Histological examination of the lungs of mice.
| Days | 1 | 3 | 6 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| #* | 1 | 2 | 3 | 1 | 2 | 3 | 1 | 2 | 3 |
| b240 | ++/++ | +++/+++ | ++/++ | +++/+++ | ++/++ | +++/+++ | +++/+++ | +++/+++ | +++/+++ |
| saline | ++/++ | +++/+++ | +++/+++ | +++/+++ | +++/+++ | +++/+++ | +++/+++ | +++/+++ | ND† |
Nine mice per group were infected with 10 MLD50 of CA04 virus and three mice per group were euthanized on days 1, 3, and 6 post-infection. The extent of pneumonia and viral antigen expression was evaluated by using a distribution scoring system. 0%, -; 0% < 40%, +; 40% < 60%, ++; and 60% < 100%, +++.
*The number of the individual animals.
†Not determined.
Effect of Lactobacillus pentosus b240 administration on cytokine/chemokine expression
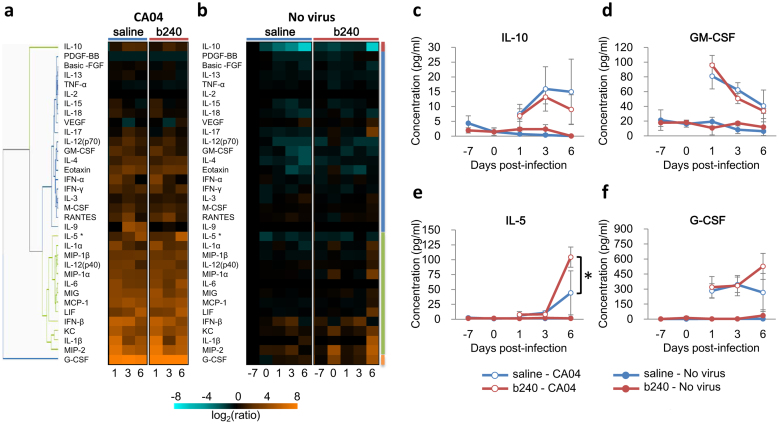
Cytokines and chemokines are important mediators of the host defense against bacterial and viral infection and play a proinflammatory role in pulmonary inflammation during the influenza virus infection15,16,17. We therefore examined the effects of oral administration of b240 on the expression levels of 34 different cytokines/chemokines in mice infected with CA04 virus (Figs. 1b and 3a). In addition, to investigate the effect of oral b240 administration on the immune system in the lungs of uninfected mice, we also analyzed cytokine expression levels in the lungs of mice treated with b240 at 14, 21, 22, 24, and 27 days post-b240 administration (−7, 0, 1, 3, and 6 days post-mock-infection) (Figs. 1c and 3b).
Figure 3. Cytokine/chemokine expression profiles in the lungs of mice treated with b240.
(a, b) The expression of cytokines/chemokines in mouse lungs is shown separately for the experiments described in Fig. 1a and b. Expression was visualized by using a heatmap and by using hierarchal clustering with the UPGMA method in TIBCO Silver Spotfire ver. 3.2. Expression of the representative cytokines in distinct clusters is shown: (c) IL-10, (d) GM-CSF, (e) IL-5, and (f) G-CSF. All values were normalized to the mean value of the saline-treated, PBS-inoculated mice at 14 days post-b240 administration (−7 days post-infection). The values are means ± SD (n = 3). Asterisk: P value < 0.05, significant difference compared with the control group (two-way ANOVA).
Our cytokine expression assay revealed that virus infection substantially changed the expression levels of most of cytokines tested regardless of whether the mice were administered b240 (Fig. 3a, b). In virus-infected animals (Fig. 3a), there were no statistically significant differences between the expression levels of cytokines measured in the lungs of animals treated with b240 and those treated with saline, with the exception of IL-5, the expression of which was significantly higher in the lungs of animals treated with b240 than in those treated with saline (Fig. 3c–f). Furthermore, in the lungs of uninfected mice (Fig. 3b), no cytokines were significantly differentially expressed following b240 administration, indicating no apparent effects of b240 on the immune system as determined by cytokine and chemokine secretion.
Changes in gene expression mediated by oral administration of Lactobacillus pentosus b240 in the lungs of mice
To gain further insights into the possible mechanism by which heat-killed b240 mediates the recovery of mice infected with influenza A(H1N1)pdm virus, we performed gene transcriptional analyses on the lungs of mice by using oligonucleotide-based DNA microarrays. To identify the genes whose expression was statistically significantly differentially regulated in the lungs of mice, we applied two types of statistical tests: a T-test at each time point, and a two-way ANOVA test (factors of day and treatment) with the Benjamini-Hochberg correction for false discovery rate, to the entire data set. We first asked whether oral administration of b240 induced differential host responses in the lungs of mice on days 1, 3, and 6 post-infection with CA04 (Fig. 1b). Among the 55821 probes tested, only one gene was statistically significantly differentially regulated between the lungs of b240-treated mice and those of control mice at 1 day post-infection (P < 0.05) and no genes were universally differentially regulated as a result of oral administration of b240 throughout the infection time (Table S1; microarray data is available at the GEO database under the accession number GSE43764).
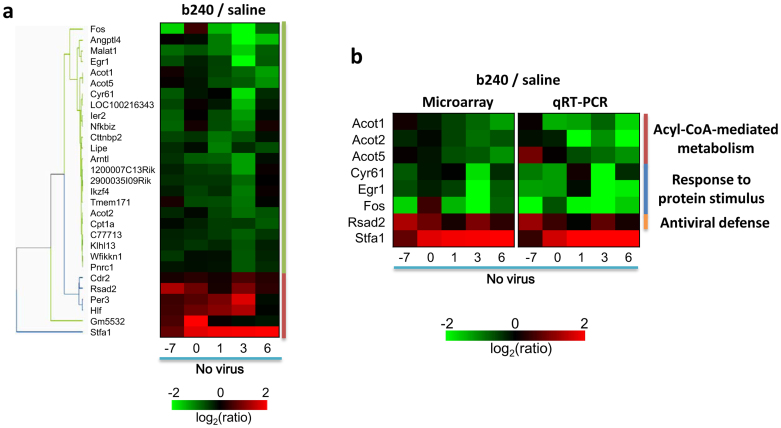
We then attempted to identify genes that were differentially regulated in the lungs of uninfected mice after oral administration of b240 by investigating b240-induced host responses in the lungs on days 14, 21, 22, 24, and 27 post-b240 administration (i.e., −7, 0, 1, 3, and 6 days post-mock-infection) (Figs. 1c and 4). We found, by using the two-way ANOVA test, that 85 probes (76 distinct genes) were significantly differentially regulated in the lungs of uninfected mice following oral administration of b240 (P value < 0.05) (Tables S1 and S2). These genes were further filtered to include genes whose expression changed 1.5-fold relative to the level in the saline-treated group at at least one time point and to exclude genes that could not be functionally annotated. Application of these criteria led us to identify 29 genes that were significantly differentially regulated following oral administration of b240 (P value < 0.05, Fold change > 1.5) (Fig. 4a). Of these 29 differentially expressed genes, most were down-regulated, although six were up-regulated in the lungs of uninfected mice after oral administration of b240. Of these six genes, the Stefin A1 gene (Stfa1) was most up-regulated (1.71 – 14.36-fold increase) (Table S2 and Fig. 4a).
Figure 4. Expression profiles of genes differentially regulated in the lungs of mice treated with b240.
Animal experiments were conducted as described in Fig. 1c. Genes that were differentially regulated following b240 administration were identified by comparing the gene expression in the lungs of mice treated with b240 and those given saline. (a) Twenty-nine genes were selected by using two-way ANOVA (treatment effect, P < 0.05) and by filtering the genes whose expression changed by at least 1.5-fold relative to the level in the saline-treated group at at least one time point. Gene expression was visualized by means of a heatmap and by using hierarchal clustering with the UPGMA method in TIBCO Silver Spotfire ver. 3.2. The heatmap was generated by using the mean expression values for the b240-treated animals, which were normalized to the values of the time-matched, saline-treated controls in the PBS-inoculated group (n = 3). (b) Validation of microarray findings by using qRT-PCR. The expression of the following eight genes was measured by using qRT-PCR: Acot1, Acot2, Acot5, Cyr61, Egr1, Fos, Rsad2, and Stfa1. The gene expression levels of three biological replicates from three animals per group are represented as mean values. The microarray data presented are those from Fig. 4a.
Next, to determine the biological relevance of the differential regulation of these genes, we performed functional enrichment analyses according to Gene Ontology (GO) specifications and the Ingenuity Pathways Analysis (IPA) application. GO analysis is widely used to classify genes based on their known functions, whereas IPA analysis is used to identify the molecular pathways enriched by the genes based on known their interactions18,19, thereby enabling the functional classification of and pathway enrichment analysis for genes that are differentially regulated.
GO analysis identified 7 GO terms that statistically significantly enriched the gene set (Table 3). Among the 7 GO terms, the top 6 were commonly associated with acyl-CoA (palmitoyl-CoA)-mediated metabolism. Three acyl-CoA thioesterase (Acots) genes (Acot1, Acot2, and Acot5) were significantly down-regulated in mouse lungs following oral administration of b240. GO analysis also revealed that the gene set was enriched by the FBJ osteosarcoma oncogene (Fos), early growth response 1 (Egr1), and cysteine-rich, angiogenic inducer, 61 (Cyr61) genes, which are involved in the "response to protein stimulus" (Table 3). The expression of these genes was down-regulated by b240 administration.
Table 3. Top GO terms enriched by genes that were significantly differentially expressed following oral administration of b240 in the lungs of uninfected mice.
| GO term | P value | Molecules |
|---|---|---|
| Palmitoyl-CoA Hydrolase Activity | 0.001 | Acot1↓, Acot2↓, Acot5↓ |
| Thioester Metabolic Process | 0.001 | Acot1↓, Acot2↓, Acot5↓ |
| Acyl-CoA Thioesterase Activity | 0.001 | Acot1↓, Acot2↓, Acot5↓ |
| Acyl-CoA Metabolic Process | 0.001 | Acot1↓, Acot2↓, Acot5↓ |
| CoA Hydrolase Activity | 0.001 | Acot1↓, Acot2↓, Acot5↓ |
| Carboxylesterase Activity | 0.002 | Acot1↓, Acot2↓, Acot5↓, Lipe↓ |
| Response to Protein Stimulus | 0.037 | Cyr61↓, Egr1↓, Fos↓ |
Genes statistically significantly regulated in the lungs of uninfected mice treated with b240 compared with saline-treated mice were determined by using two-way ANOVA (Fold change > 1.5, P < 0.05). The set of genes differentially regulated by b240 administration was functionally annotated by means of GO grouping. Statistical significance was determined by using Fisher's exact test (P < 0.05). Arrows next to each gene name indicate the up- or down-regulation induced by b240 administration.
IPA analysis identified 14 canonical pathways that statistically significantly enriched the gene set (Table 4). Remarkably, the Rsad2 gene, which is involved in the "role of lipids/lipid rafts in the pathogenesis of influenza" canonical pathway20, was up-regulated in the lungs of uninfected mice after oral administration of b240 (Table 4). Thus, we found that oral administration of heat-killed b240 induced differential regulation of several genes with shared functions and a specific gene that has known antiviral activity against influenza virus infection.
Table 4. Top ingenuity canonical pathways enriched by genes that were significantly differentially expressed following oral administration of b240 in the lungs of uninfected mice.
| Ingenuity Canonical Pathways | P value | Ratio | Molecules |
|---|---|---|---|
| Circadian Rhythm Signaling | 0.001 | 0.057 (2/35) | Per3↑, Arntl↓ |
| IL-17A Signaling in Fibroblasts | 0.001 | 0.050 (2/40) | Fos↓, Nfkbiz↓ |
| IGF-1 Signaling | 0.006 | 0.019 (2/107) | Fos↓, Cyr61↓ |
| Role of Tissue Factor in Cancer | 0.008 | 0.018 (2/114) | Egr1↓, Cyr61↓ |
| GNRH Signaling | 0.010 | 0.014 (2/145) | Fos↓, Egr1↓ |
| AMPK Signaling | 0.011 | 0.012 (2/166) | Cpt1a↓, Lipe↓ |
| CXCR4 Signaling | 0.014 | 0.012 (2/169) | Fos↓, Egr1↓ |
| RAR Activation | 0.018 | 0.011 (2/183) | Fos↓, Pnrc1↓ |
| Role of Lipids/Lipid Rafts in the Pathogenesis of Influenza | 0.029 | 0.036 (1/28) | Rsad2↑ |
| IL-17A Signaling in Gastric Cells | 0.030 | 0.040 (1/25) | Fos↓ |
| TNFR2 Signaling | 0.035 | 0.029 (1/34) | Fos↓ |
| April Mediated Signaling | 0.045 | 0.023 (1/43) | Fos↓ |
| B Cell Activating Factor Signaling | 0.047 | 0.022 (1/45) | Fos↓ |
| MIF Regulation of Innate Immunity | 0.048 | 0.020 (1/50) | Fos↓ |
Genes statistically significantly regulated in the lungs of uninfected mice treated with b240 compared with saline-treated mice were determined by using two-way ANOVA (Fold change > 1.5, P < 0.05). Functional enrichment analysis for the set of genes differentially regulated by b240 administration was determined by using IPA software. Statistical significance was determined by using Fisher's exact test (P < 0.05). Arrows next to each gene name indicate the up- or down-regulation induced by b240 administration.
To validate the transcriptional findings from our microarray analysis, we conducted a quantitative real-time PCR (qRT-PCR) assay for the following eight genes: Stfa1 as the most up-regulated gene; three genes for "Acyl-CoA-mediated metabolism", represented by Acot1, Acot2, and Acot5; three genes for "response to protein stimulus", represented by Cyr61, Egr1, and Fos; and Rsad2 for the antiviral response. The expression levels determined by qRT-PCR were consistent with those obtained by microarray analysis (Fig. 4b), confirming the down-regulation of "Acyl-CoA-mediated metabolism" and "response to protein stimulus", and the up-regulation of Stfa1 and Rsad2 gene expression. Thus, we identified specific genes that are differentially regulated in the lungs of mice orally treated with heat-killed Lactobacillus pentosus b240.
Discussion
Here, we demonstrated that oral administration of b240 slightly augments protection against a lethal influenza A(H1N1)pdm virus in a mouse model. We also unveiled the host responses that are induced in mouse lungs after oral administration of b240.
Although mice orally administered with b240 and infected with a lethal dose of A(H1N1)pdm virus exhibited prolonged survival, we did not detect a significant difference between virus titers in the lungs of animals treated with b240 and those treated with saline. Kobayashi et al.14 previously reported that the virus titers were significantly reduced in the lungs of mice orally administered with b240 prior to virus challenge. However, they investigated virus titers in the lungs of mice infected with a non-lethal dose of A/PR8/1934, which suggests that this discrepancy may be attributable to the different virus dosages used in the two studies.
Oral administration of b240 did not appreciably affect histopathology, cytokine/chemokine expression, or gene expression in the lungs of mice infected with lethal A(H1N1)pdm virus. However, gene transcriptional analysis revealed that oral administration of b240 did differentially regulate specific genes, represented by Acots (Acot1, Acot2, and Acot5), Cyr61, Egr1, Fos, Stfa1, and Rsad2, in the lungs of uninfected mice.
Acots, which were functionally classified into "Acyl-CoA-mediated metabolism" by GO grouping, were down-regulated in mouse lungs following oral administration of b240. Acots are involved in two-major metabolic pathways21: the catabolism of fatty acid beta-oxidation and the anabolic pathway that converts fatty acids into cellular lipids. The expression of Acot genes is regulated by peroxisome proliferator-activated receptors (PPARs), which function in the metabolism of lipids, carbohydrates, bile acids, and amino acids, as well as in inflammation22,23,24. Moreover, Acots play an important role in the generation of arachidonic acid, the precursor to eicosanoids, which are responsible for the physiological manifestations of inflammation25,26, suggesting a mechanism by which the differential regulation of Acots by b240 might contribute to the recovery of mice from CA04 infection.
Furthermore, Cyr61, Egr1, and Fos, which were functionally classified into "response to protein stimulus" by GO grouping, were down-regulated after b240 administration. Cyr61 and Egr1 are potential molecular biomarkers for severity of lung injury27,28. Cyr61 encodes a multifunctional protein that can activate IL-6 production, resulting in inflammation and disease progression29. In addition, the transcription factor Egr1 and Fos are early responders during influenza infection30. In particular, Egr1 is a critical regulator of host inflammatory chemokines and is associated with CD8+ T cell-mediated lung injury in influenza infection31. Given that blocking Cyr61 with a specific mAb ameliorated the inflammatory reaction in mice29, and that genetic deficiency of Egr1 significantly abrogates both chemokine expression and the immunopathological injury associated with T cell recognition31, the down-regulation of these genes induced by oral administration of b240 may play a role in alleviating pulmonary injury caused by the inflammatory response.
Stfa1 was the most up-regulated gene induced by oral administration of b240, although we did not functionally characterize it here. Stfa1 inhibits cysteine endo- and exopeptidases, such as cathepsin L and S, which are involved in antigen processing32. Given that increased expression of specific cathepsins has been shown to be associated with autoimmune diseases and bacterial infection33,34, the up-regulation of the Stfa1 gene may contribute to protection from self-inflicted damage.
Importantly, Rsad2 was up-regulated after b240 administration. Rsad2 is radical S-adenosyl methionine domain-containing protein 2, also known as viperin35,36. Rsad2 (viperin) is an interferon-stimulated gene (ISG), induced by type I, II, and III interferons after infection with a broad range of DNA and RNA viruses36,37,38,39,40,41,42. Therefore, the viperin activation is involved in defenses against a broad range of viruses, including HIV-1, HCV, human cytomegalovirus, alpha-virus, West Nile virus, dengue and influenza virus20,43. Viperin is also induced by non-viral microbial products, such as lipopolysaccharide, and by a wide range of bacteria, suggesting a broad role in innate antimicrobial defenses44,45. A previous study demonstrated that mucosal stimulation with reovirus primes lymphocytes in the gut potentially protecting against subsequent respiratory challenge46, which indicates that immune priming could be triggered by mucosal administration47. Our finding that the Rsad2 gene is up-regulated in mouse lungs could also be the result of immune priming induced by the oral administration of b240 in the gut.
The immune priming induced by oral administration of heat-killed Lactobacillus pentosus b240 may have several important implications. First, oral administration of b240 augments protection against not only different types of influenza viruses but also Streptococcus pneumoniae infection13,14. During infections with gram-positive bacteria, such as Streptococcus agalactiae and pneumoniae, interferon production is believed to play a critical role in the clearance of the infection by the host48. Therefore, the differential regulation of the interferon-related gene Rsad2, induced by oral administration of b240, likely plays a role in the alleviation of S. pneumoniae-mediated pneumonia. Second, immune priming was caused by heat-killed "unvital" Lactobacillus pentosus b240. This indicates that immune priming was caused in response to the stimulus of proteins, nucleic acids, and lipids that are components of Lactobacillus pentosus b240. Therefore, one could argue that immune priming might be a common feature, to various degrees, of broad ranges of Lactobacilli that have anti-microbial activity.
In general, live Lactobacillus strains induce the same or stronger immune responses than do heat-killed strains14,49,50,51. Since the efficacy of live bacteria may be influenced by the number of bacteria that reach the intestine alive and this number may vary from one experiment to the next, we determined that we would have greater control and/or consistency in our experiments if we used killed bacteria rather than live bacteria. We, therefore, used heat-killed Lactobacillus pentosus b240; however, live Lactobacillus pentosus b240 may induce stronger immune responses than the heat-killed bacterium and may be a more efficient probiotic.
Our study raises the following questions: (i) is heat-killed b240 efficacious against infections by other influenza viruses with different virulence or subtypes, or against other pneumonia-causing viruses, (ii) are the genes that are up-regulated or down-regulated upon b240 treatment of mice indeed responsible for the protective efficacy, and (iii) are other Lactobacillus species also efficacious? Further studies are needed to answer these questions.
In summary, we examined the effect of oral administration of b240 on influenza A(H1N1)pdm virus infection and found that b240 augments protection against a lethal dose of CA04 virus. Although oral administration of b240 induced no substantial differences in virus replication, histopathology, cytokine/chemokine expression, or gene expression in the lungs of mice infected with CA04 virus, it did cause the down-regulation of genes involved in "Acyl-CoA-mediated metabolism" and "response to protein stimulus", and the up-regulation of the antiviral gene Rsad2 in the lungs of uninfected mice. Importantly, genes known to have antiviral activities against influenza virus, such as Egr1 and Rsad2, were differentially regulated by b240, suggesting that the activation of innate immunity may be the mechanism behind the protection of influenza virus-infected mice afforded by b240. Our results provide a clue toward a better understanding of the antiviral activities induced by oral administration of heat-killed Lactobacillus pentosus b240.
Methods
Ethics statement
Our research protocol for the use of mice followed the University of Tokyo's Regulations for Animal Care and Use, which was approved by the Animal Experiment Committee of the Institute of Medical Science, the University of Tokyo (approval number; 19–28).
Cells and viruses
Madin-Darby canine kidney (MDCK) cells were maintained in Eagle's minimal essential medium (MEM) containing 5% newborn calf serum (NCS) and incubated at 37°C with 5% CO2. A mouse-adapted influenza A(H1N1)pdm virus strain, A/California/04/2009 (CA04), was generated by serial passage in the lungs of mice as previously described52 and propagated in MDCK cells for use in this study.
Administration of Lactobacillus pentosus b240 in mice
Six-week-old female BALB/c mice (Japan SLC Inc., Shizuoka, Japan) were used in the study. Oral administration of b240 was initiated in mice at six weeks of age. Mice were orally administered heat-killed Lactobacillus pentosus b240 every day at a dose of 10 mg/mouse, which corresponds to 1010 cell counts of heat-killed microbe, in 200 μl of buffered saline for 5 weeks. The control group received saline. The b240 dose was determined based on a previous study14.
Experimental infection of mice
The experimental schedule is briefly described in Fig. 1. On day 21 post-b240 administration, mice were anesthetized with sevoflurane and intranasally infected with PBS, or with 0.3 or 10 MLD50 of CA04 virus [MLD50 = 3.8 × 104 p.f.u.]. To determine the effects of oral administration of b240 on mouse survival, ten mice per group were infected with 0.3 MLD50 of CA04 virus and were monitored daily for morbidity and mortality for up to 14 days post-infection (Fig. 1a). For the infection with 10 MLD50 of virus, b240- or saline-treated groups of 24 or 25 mice were similarity used. To investigate the effects of oral administration of b240 on viral replication and host immune responses to CA04 virus infection, 18 mice per group were infected with 10 MLD50 of CA04 virus (Fig. 1b) on day 21 post-b240 administration. Three mice per group were euthanized on days 1, 3, and 6 post-infection and their lungs were collected. These lung tissues were then cut into several pieces for virus titration, cytokine measurement, and microarray analysis; the lung samples were immediately frozen at −80°C until processing. For the histopathological analysis, three mice per group were similarly euthanized on days 1, 3, and 6 post-infection and their lungs were collected. These lung tissues were fixed with 10% neutral buffered formalin and subjected to histopathological analysis as described below. To investigate the immune responses induced by oral administration of b240 in the lungs of uninfected mice, 15 mice per group were mock-infected with PBS on day 21 post-b240 administration (Fig. 1c). Three mice per group were euthanized on days 14, 21, 22, 24, and 27 post-b240 administration (−7, 0, 1, 3, and 6 days post-mock infection) and their lungs were collected. These lung tissues were then cut into several pieces for cytokine measurement and microarray analysis.
Histopathological analyses
Fixed mouse lungs were serially cut into 1.5 mm-thick pieces and embedded into paraffin for microscopic examination. Five μm-thick paraffin sections were stained with hematoxylin and eosin; additional sections were cut for immunohistological staining with rabbit polyclonal antiserum against an H1N1 virus (A/WSN/1933). Specific antigen-antibody reactions were visualized by using the Dako EnVision system, with 3,3' diaminobenzidine tetrahydrochloride (Dako Japan Inc., Tokyo). The extent of pneumonia and viral antigen expression was evaluated by means of a distribution scoring system (0%, −; 0% < 40%, +; 40% < 60%, ++; and 60% < 100%, +++).
Cytokine analysis
Mouse lung samples were treated with the Bio-Plex Cell Lysis Kit (Bio-Rad Laboratories, Richmond, CA) according to the manufacturer's instructions. The concentrations of IFN-α and -β in mouse lung homogenates were determined by using the Mouse IFN Alpha or beta ELISA kit (Invitrogen, Carlsbad, CA). The concentrations of other cytokines/chemokines were determined by using the Bio-Plex Mouse Cytokine 23-Plex and 9-Plex panel (Bio-Rad Laboratories) and by using an array analysis with the Bio-Plex protein multi-array system (Bio-Rad Laboratories) as previously described53. Statistically significant differences in cytokine expression between the saline- and b240-treated groups in the PBS- and CA04-infected groups were determined by using two-way analysis of variance (ANOVA); values were considered to be significantly different when the P value was less than 0.05.
RNA isolation and integrity
The lungs of mice were homogenized with TRIzol Reagent (Invitrogen) at a final concentration of 5% w/v. The homogenized materials were transferred to a fresh 1.5 ml tube to which chloroform-isoamyl alcohol (5:1) was added. The tube was vortexed and then centrifuged for 15 min at 12,000 rpm at 4°C. The resulting aqueous layer was subjected to RNA extraction by using RNeasy Mini kit columns (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Isolated total RNA quality was assessed by determining UV 260/280 absorbance ratios and examining 28 S/18 S ribosomal RNA bands with an Agilent 2100 bioanalyzer (Agilent Technologies).
Microarray analyses
Cy3-labeled cRNA probe synthesis was initiated with 100 ng of total RNAs by using the Agilent Low Input Quick Amp Labeling kit, one color (Agilent Technologies, Santa Clara, CA). The Agilent SurePrint G3 Gene Mouse GE 8 × 60 K microarrays (4852A) were used according to the manufacturer's instructions. Slides were scanned with an Agilent's High-Resolution Microarray Scanner, and image data were processed by using Agilent Feature Extraction software ver. 10.7.3.1. All data were subsequently uploaded into GeneSpring GX ver. 11.5 (Agilent Technologies) for data analysis. In accordance with proposed MIAME (minimum information about a microarray experiment) standards, microarray data obtained in this study are publically available at the GEO database under the accession number GSE43764. For the microarray data analysis, each gene expression array data set was normalized to the in silico pool for time-matched, saline-treated PBS-inoculated animals (n = 3). Statistically significant differences in gene expression between the saline- and b240-treated groups in the PBS-inoculated and CA04-infected groups were determined by using the T-test at each time point or two-way ANOVA (P < 0.05) with the Benjamini-Hochberg false discovery rate (FDR) post-test. Differentially expressed genes were further filtered to include genes whose expression changed 1.5-fold relative to the level in the saline-treated group at at least one time point. Genes that passed the statistical tests were further assigned to a GO grouping or uploaded into IPA (Ingenuity Systems; http://www.ingenuity.com/) to identify the functions and pathways that enrich the gene set. GO analysis was carried out using GeneSpring Gx ver. 11.5, where Benjamini-Hochberg corrected P values were used to determine the GO terms that were significant (P < 0.05). IPA analysis was carried out using the default setting, where P values were calculated by using Fisher's exact test to identify IPA canonical pathways that were significant (P < 0.05). In the data import from GeneSpring to the IPA software, gene annotation was carried out based on the Agilent probe ID extracted from GeneSpring Gx ver. 11.5. Genes that were significantly differentially regulated and enriched GO groups or IPA canonical pathways were selected for qRT-PCR analyses.
qRT-PCR analyses
qRT-PCR was performed to validate transcriptional findings determined by using microarray analysis. RNA was reverse transcribed with oligonucleotide dT and SuperScriptTM III reverse transcriptase (Invitrogen). qRT-PCR was conducted with the SYBR Green PCR master mix (Invitrogen) and carried out on the ABI 7900HT Fast Real-time PCR System platform. Target gene mRNA levels were normalized to 18 s ribosomal RNA according to the 2−ΔΔCt calculation as described previously54. The qRT-PCR primers for each target were designed by using IDT SciTools RealTime PCR at http://www.idtdna.com/scitools/Applications/RealTimePCR/. Sequences are available upon request.
Author Contributions
M.K., N.K. and Y.K. conceived and designed the research. M.K., R.T., S.S., H.K., K.S., R.U. and S.W. performed the experiments. M.K., R.T., S.S., H.K., K.S., H.S. and M.T. analyzed the data. H.S., M.T. and N.K. contributed reagents. R.T. prepared the manuscript and figures. All authors reviewed the manuscript.
Supplementary Material
Supplementary Information
Acknowledgments
We thank Prof. Sanae Okada, Faculty of Applied Bioscience, Tokyo University of Agriculture for providing us with Lactobacillus pentosus b240 and Susan Watson for editing the manuscript. This work was supported by a grant-in-aid for Specially Promoted Research and by a contract research fund for the Program for Funding Research Centers for Emerging and Reemerging Infectious Diseases from the Ministries of Education, Culture, Sports, Science, and Technology, and by grants-in-aid of Health, Labor, and Welfare of Japan, by ERATO (Japan Science and Technology Agency), and by National Institute of Allergy and Infectious Diseases Public Health Service Research grants. RT was supported by Research Fellowships from the Japan Society for the Promotion of Science for Young Scientists.
References
- Perez-Padilla R. et al. Pneumonia and respiratory failure from swine-origin influenza A(H1N1) in Mexico. N. Engl. J. Med. 361, 680–689 (2009). [DOI] [PubMed] [Google Scholar]
- WHO. Statements 2009. World now at the start of 2009 influenza pandemic. 11 June 2009. Accessed at http://www.who.int/mediacentre/news/statements/2009/en/.
- Van der Wouden J. C., Bueving H. J. & Poole P. Preventing influenza: an overview of systematic reviews. Respir. Med. 99, 1341–1349 (2005). [DOI] [PubMed] [Google Scholar]
- Guabareva L. V., Kaiser L. & Hayden F. G. Influenza virus neuraminidase inhibitors. Lancet 355, 827–835 (2000). [DOI] [PubMed] [Google Scholar]
- Maeda N. & Uede T. Swine-origin influenza-virus-induced acute lung injury: Novel or classical pathogenesis? World J. Biol. Chem. 1, 85–94 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid G. et al. Microbiota restoration: natural and supplemented recovery of human microbial communities. Nat. Rev. Microbiol. 9, 27–38 (2011). [DOI] [PubMed] [Google Scholar]
- Hori T., Kiyoshima J., Shida K. & Yasui H. Effect of intranasal administration of Lactobacillus casei Shirota on influenza virus infection of upper respiratory tract in mice. Clin. Diagn. Lab. Immunol. 8, 593–597 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawase M., He F., Kubota A., Harata G. & Hiramatsu M. Oral administration of lactobacilli from human intestinal tract protects mice against influenza virus infection. Lett. Appl. Microbiol. 51, 6–10 (2010). [DOI] [PubMed] [Google Scholar]
- Youn H. N. et al. Intranasal administration of live Lactobacillus species facilitates protection against influenza virus infection in mice. Antiviral Res. 93, 138–143 (2012). [DOI] [PubMed] [Google Scholar]
- Okada S., Daengsubha W., Uchimura T., Ohara N. & Kozaki M. Flora of lactic acid bacteria in Miang produced in northern Thailand. J. Gen. Appl. Microbiol. 32, 57–65 (1986). [Google Scholar]
- Yamahira S., Toba M., Kishi K. & Okamatsu H. Stimulation of mucosal immune system by lactic acid bacteria originating in tea. Jpn. J. Lactic Acid Bact. 17, 57–60 (2006). [Google Scholar]
- Kotani Y. et al. Oral intake of Lactobacillus pentosus strain b240 accelerates salivary immunoglobulin A secretion in the elderly: A radomized, placebo-controlled, double-blind trial. Immun. Aging 7, 11 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A. et al. Lactobacillus pentosus strain b240 suppresses pneumonia induced by Streptococcus pneumoniae in mice. Lett. Appl. Microbiol. 53, 35–43 (2011). [DOI] [PubMed] [Google Scholar]
- Kobayashi N. et al. Oral administration of heat-killed Lactobacillus pentosus strain b240 augments protection against influenza virus infection in mice. Int. Immunopharmacol. 11, 199–203 (2011). [DOI] [PubMed] [Google Scholar]
- Strieter R. M., Belperio J. A. & Keane M. P. Cytokines in innate host defense in the lung. J. Clin. Invest. 109, 699–705 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. A. Neutrophils, host defense, and inflammation: a double-edged sword. J. Leukoc. Biol. 56, 672–686 (1994). [DOI] [PubMed] [Google Scholar]
- Goodman R. B., Pugin J., Lee L. S. & Matthay M. A. Cytokine-mediated inflammation in acute lung injury. Cytokine Growth Factor Rev. 14, 523–535 (2003). [DOI] [PubMed] [Google Scholar]
- Bradel-Trethway B. G. et al. Comprehensive proteomic analysis of influenza virus polymerase complex reveals a novel association with mitochondrial proteins and RNA polymerase accessory factors. J. Virol. 85, 8569–81 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belisle S. E. et al. Genomic profiling of tumor necrosis factor alpha (TNF-alpha) receptor and interleukin-1 receptor knockout mice reveals a link between TNF-alpha signaling and increased severity of 1918 pandemic influenza virus infection. J. Virol. 84, 12576–12588 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Hinson E. R. & Cresswell P. The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe 2, 96–105 (2007). [DOI] [PubMed] [Google Scholar]
- Hardwick J. P. Cytochrome P450 omega hydroxylase (CYP4) function in fatty acid metabolism and metabolic diseases. Biochm. Pharma. 75, 2263–2275 (2008). [DOI] [PubMed] [Google Scholar]
- Kirkby B., Roman N., Kobe B., Kellie S. & Forwood J. K. Functional and structural properties of mammalian acyl-coenzyme A thioesterases. Prog. Lipid Res. 49, 366–377 (2010). [DOI] [PubMed] [Google Scholar]
- Di Nunzio M., Danesi F. & Bordoni A. n-3 PUFA as regulators of cardiac gene transcription: a new link between PPAR activation and fatty acid composition. Lipids 44, 1073–1079 (2009). [DOI] [PubMed] [Google Scholar]
- Martin G., Schoonjans K., Lefebvre A. M., Staels B. & Auwerx J. Coordinate regulation of the expression of the fatty acid transport protein and acyl-CoA synthetase genes by PPARalpha and PPARgamma activators. J. Biol. Chem. 272, 28210–28217 (1997). [DOI] [PubMed] [Google Scholar]
- Jiang Y. J. et al. Modulation of cytosolic phospholipase A(2) by PPAR activators in human preadipocytes. J. Lipid Res. 42, 716–724 (2001). [PubMed] [Google Scholar]
- Rouzer C. A. et al. Lipid profiling reveals arachidonate deficiency in RAW264.7 cells: Structural and functional implications. Biochemistry 45, 14795–14808 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace M. J. et al. Early biomarkers and potential mediators of ventilation-induced lung injury in very preterm lambs. Respir. Res. 10, 19 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matute-Bello G. et al. Essential role of MMP-12 in Fas-induced lung fibrosis. Am. J. Respir. Cell. Mol. Biol. 37, 210–221 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J. et al. Cyr61 induces IL-6 production by fibroblast-like synoviocytes promoting Th17 differentiation in rheumatoid arthritis. J. Immunol. 188, 5776–5784 (2012). [DOI] [PubMed] [Google Scholar]
- Tatebe K. et al. Response network analysis of differential gene expression in human epithelial lung cells during avian influenza infections. BMC Bioinformatics. 11, 170 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramana C. V., Cheng G. S., Kumar A., Kwon H. J. & Enelow R. I. Role of alveolar epithelial early growth response-1 (Egr-1) in CD8+ cell-mediated lung injury. Mol. Immunol. 47, 623–631 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode W. & Huber R. Structural basis of endoproteinase-protein inhibitor interaction. Biochim. Biophys. Acta. 1477, 241–252 (2000). [DOI] [PubMed] [Google Scholar]
- Menzel K. et al. Cathepsins B, L and D in inflammatory bowel disease macrophages and potential therapeutic effects of cathepsin inhibition in vivo. Clin. Exp. Immunol. 146, 169–180 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C. C. et al. Experimental malaria infection triggers early expansion of natural killer cells. Infect. Immun. 76, 5873–5882 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaveta G., Shi J., Chow V. T. & Song J. Structural characterization reveals that viperin is a radical S-adenosyl-L-methionine (SAM) enzyme. Biochem. Biophys. Res. Commun. 391, 1390–1395 (2010). [DOI] [PubMed] [Google Scholar]
- Fitzgerald K. A. The interferon inducible gene: Viperin. J. Interferon Cytokine Res. 31, 131–135 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin K. C. & Cresswell P. Viperin (cig5), an IFN-inducible antiviral protein directly induced by human cytomegalovirus. Proc. Natl. Acad. Sci. U. S. A. 98, 15125–15130 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feld J. J. et al. S-adenosyl methionine improves early viral responses and interferon-stimulated gene induction in hepatitis C nonresponders. Gastroenterology 140, 830–839 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo J. Y., Yaneva R., Hinson E. R. & Cresswell P. Human cytomegalovirus directly induces the antiviral protein Viperin to enhance infectivity. Science 332, 1093–1097 (2011). [DOI] [PubMed] [Google Scholar]
- Jiang D. et al. Identification of five interferon-induced cellular proteins that inhibit West Nile virus and dengue virus infections. J. Virol. 84, 8332–8341 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y. L., Chang T. H., Liao C. L. & Lin Y. L. The cellular antiviral protein Viperin is attenuated by proteasome-mediated protein degradation in Japanese encephalitis virus-infected cells. J. Virol. 82, 10455–10464 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinson E. R. et al. Viperin is highly induced in neutrophils and macrophages during acute and chronic lymphocytic choriomeningitis virus infection. J. Immunol. 184, 5723–5731 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T. et al. Antiviral protein Viperin promotes Toll-like receptor 7- and Toll-like receptor 9-mediated type I interferon production in plasmacytoid dendritic cells. Immunity 34, 352–363 (2011). [DOI] [PubMed] [Google Scholar]
- Hinson E. R. & Cresswell P. The N-terminal amphipathic alpha-helix of Viperin mediates localization to the cytosolic face of the endoplasmic reticulum and inhibits protein secretion. J. Biol. Chem. 284, 4705–4712 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivieccio M. A. et al. TLR3 ligation activates an antiviral response in human fetal astrocytes: a role for Viperin/cig5. J. Immunol. 177, 4735–4741 (2006). [DOI] [PubMed] [Google Scholar]
- Zuercher A. W., Jiang H. Q., Thurnheer M. C., Cuff C. F. & Cebra J. J. Distinct mechanisms for cross-protection of the upper versus lower respiratory tract through intestinal priming. J. Immunol. 169, 3920–3925 (2002). [DOI] [PubMed] [Google Scholar]
- Kau A. L., Ahern P. P., Griffin N. W., Goodman A. L. & Gordon J. I. Human nutrition, the gut microbiome and the immune system. Nature 474, 327–36 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrel-Dennis M. et al. TLR-independent type I interferon induction in response to an extracellular bacterial pathogen via intracellular recognition of its DNA. Cell Host Microbe. 4, 543–554 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumo T. et al. Comparison of the immunomodulatory effects of live and heat-killed Lactobacillus pentosus S-PT84. J. Heath Sci. 57, 304–310 (2011). [Google Scholar]
- Chuang L. et al. Heat-killed cells of lactobacilli skew the immune response toward T helper 1 polarization in mouse splenocytes and dendritic cell-treated T cells. J. Agric. Food Chem. 55, 11080–11086 (2007). [DOI] [PubMed] [Google Scholar]
- Ishikawa H. et al. Oral administration of heat-killed Lactobacillus plantarum strain b240 protected mice against Salmonella enterica Serovar Typhimurium. Biosci. Biotechnol. Biochem. 74, 1338–1342 (2010). [DOI] [PubMed] [Google Scholar]
- Sakabe S., Ozawa M., Takano R., Iwatsuki-Horimoto K. & Kawaoka Y. Mutations in PA, NP, and HA of a pandemic (H1N1) 2009 influenza virus contribute to its adaptation to mice. Virus Res. 158, 124–129 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiso M. et al. T-705 (favipiravir) activity against lethal H5N1 influenza A viruses. Proc. Natl. Acad. Sci. U. S. A. 107, 882–887 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information