Abstract
Background
Pachysolen tannophilus is a non-conventional yeast, which can metabolize many of the carbon sources found in low cost feedstocks including glycerol and xylose. The xylose utilisation pathways have been extensively studied in this organism. However, the mechanism behind glycerol metabolism is poorly understood. Using the recently published genome sequence of P. tannophilus CBS4044, we searched for genes with functions in glycerol transport and metabolism by performing a BLAST search using the sequences of the relevant genes from Saccharomyces cerevisiae as queries.
Results
Quantitative real-time PCR was performed to unveil the expression patterns of these genes during growth of P. tannophilus on glycerol and glucose as sole carbon sources. The genes predicted to be involved in glycerol transport in P. tannophilus were expressed in S. cerevisiae to validate their function. The S. cerevisiae strains transformed with heterologous genes showed improved growth and glycerol consumption rates with glycerol as the sole carbon source.
Conclusions
P. tannophilus has characteristics relevant for a microbial cell factory to be applied in a biorefinery setting, i.e. its ability to utilise the carbon sources such as xylose and glycerol. However, the strain is not currently amenable to genetic modification and transformation. Heterologous expression of the glycerol transporters from P. tannophilus, which has a relatively high growth rate on glycerol, could be used as an approach for improving the efficiency of glycerol assimilation in other well characterized and applied cell factories such as S. cerevisiae.
Keywords: P. tannophilus, Glycerol, Transport, S. cerevisiae
Background
Pachysolen tannophilus is known for its ability to ferment D-xylose, one of the major components of hemicellulose plant residues, to ethanol [1]. However, P. tannophilus has also been shown to be capable of converting crude glycerol to ethanol under microaerobic conditions [2]. This ability is interesting since glycerol, a by-product of biodiesel production, has also been considered as a potential alternative carbon source for industrial bioprocesses due to the recent dramatic increase in production of biodiesel.
The whole genome of P. tannophilus CBS4044 has been sequenced [3], and with this, the possibility for understanding and exploiting glycerol transport in this yeast has arisen. A number of studies have investigated glycerol transport, consumption and production by different types of yeast [4-6], but the glycerol transport and metabolic pathways in P. tannophilus have not been studied so far. P. tannophilus has a relatively high growth rate on glycerol (0.3 h-1) [2], but improvement of the strain for industrial applications through engineering the metabolism is hampered due to the lack of tools for genetic modification. However, with glycerol becoming an increasingly abundant substrate, it is highly relevant to characterize strains which are capable of its utilization, and consider the possibility of expressing genes from these strains in industrial cell factories for which genetic engineering strategies are already well defined.
Glycerol transport is the first barrier to glycerol utilization in a microbial cell. In S. cerevisiae, glycerol enters the cell by two different mechanisms: a low affinity transport system (facilitated diffusion) and a high affinity proton symport system (active transport) [7]. When glucose is present, glycerol diffuses into the cells through a glycerol channel by facilitated transport, a process dependent on the FPS1 gene [8,9]. The physiological role of the facilitator Fps1p in S. cerevisiae has been described to be glycerol export rather than uptake during hypo-osmotic shock [9]. However, when only non-fermentable carbon sources (glycerol, acetate, ethanol) are present, an active uptake system driven by a proton motive force ensures the uptake of glycerol [7,10]. Two multi-membrane-spanning proteins encoded by GUP1 and GUP2 (Glycerol Uptake Protein), were first identified as being involved in active glycerol uptake in S. cerevisiae[10]. However, in later studies Gup1p and 2p were proposed to have different roles than glycerol transport [11]. A screen for genes encoding membrane proteins involved in glycerol assimilation in S. cerevisiae identified a gene, STL1, involved in active glycerol uptake. Stl1p is localized in the plasma membrane, is glucose repressed and inactivated by growth in glucose [7,12]. Importantly, it was concluded that the protein is a member of the sugar transporter family and acts as a glycerol proton symporter [12]. Stl1p has also been shown to have a function in glycerol uptake in several other yeasts. It has been shown that in Candida albicans glycerol was actively transported into the cells by a proton symporter encoded by the C. albicans STL1[13]. It has also been reported that S. cerevisiae strains harboring the STL1 gene from D. hansenii slightly improved their growth and doubling times on glycerol [14].
After glycerol is transported into the cells, two different routes have been identified in yeasts for further assimilation: a phosphorylation route and an oxidation route. Within the first pathway, glycerol is dissimilated by glycerol kinase encoded by GUT1 and then by glycerol-3-phosphate dehydrogenase encoded by GUT2, which is located at the outer surface of the mitochondrial membrane [6]. Another pathway is catalyzed by glycerol dehydrogenase encoded by GCY1 followed by dihydroxyacetone kinase encoded by DAK1 and DAK2. In Schizosaccharomyces pombe, it has been reported that glycerol is utilized solely by this pathway [15,16]. In both pathways, glycerol is converted to dihydroxyacetone phosphate, which then enters glycolysis. The pathways involved in glycerol utilization differ in different yeasts. In some yeast strains, all four enzymes in both pathways are present, but only one pathway functions for glycerol dissimilation [4,6]. In S. cerevisiae, it has been shown that glycerol is degraded by the phosphorylation pathway and that mutants lacking one of the two genes are incapable of utilizing glycerol [17]. Although the fermentative pathway was also discovered to be present in S. cerevisiae[18], the function is unknown. The production of glycerol has two functions in S. cerevisiae: redox balance and protection against osmotic stress as a compatible solute (osmolyte) [6,19]. Glycerol is commonly produced in the cytosol of yeasts from the glycolytic intermediate dihydroxyacetone phosphate. This compound is converted into glycerol in two steps that are catalyzed by glycerol-3- phosphate dehydrogenase (Gpd) and glycerol-3-phosphatase (Gpp) respectively. Each of these enzymatic activities has two isoenzymes, Gpd1p and Gpd2p, Gpp1p and Gpp2p [20].
Although there have been several studies on glycerol metabolism in S. cerevisiae, many industrially used baker’s yeast strains and laboratory strains grow poorly on glycerol. For example, the specific growth rate of the well-studied lab strain S. cerevisiae CBS 8066 under aerobic conditions was found to be very low, 0.010 ± 0.002 h-1, during shake flask cultivations in minimal medium containing glycerol as sole carbon source [21]. Strategies based on adaptive evolution, and overexpression or knock out of the genes involved in glycerol metabolism have been performed with the aim of increasing the host strain’s growth on glycerol and production of different value-added products have been pursued [21,22]. By adaptive evolution, strains with growth rates of up to 0.2 h-1 have been achieved [21]. However, one possible rate-limiting step during glycerol metabolism, glycerol transport, was not addressed in those approaches [23].
In this study, a comparison of the genome sequences of S. cerevisiae and P. tannophilus has been performed to study the genes involved in glycerol transport and glycerol metabolism in these two organisms. Quantitative real-time PCR was performed to compare transcript levels during growth on glycerol. The genes involved in glycerol transport in P. tannophilus were heterologously expressed in a S. cerevisiae STL1 knockout strain to assess their function and to evaluate whether they could possibly contribute as transporter genes to improve the growth of S. cerevisiae on glycerol.
Results
Identification of genes in glycerol metabolism and analysis of glycerol transporters in P. tannophilus
In order to find homologous genes related to glycerol transport and metabolism in P. tannophilus, the genome sequence was searched by BLAST with the sequences of genes known to have these functions in S. cerevisiae as queries. In P. tannophilus, two putative glycerol facilitator and two putative glycerol symporter genes were found and named PtFPS1, PtFPS2, PtSTL1 and PtSTL2, respectively. In the glycerol consumption pathways, the genes with high similarity to S. cerevisiae were found and named PtGUT1, PtGUT2, PtGCY1, PtGCY2, PtDAK (Table 1). In the glycerol production pathways, the genes found were named PtGPD and PtGPP. It was noticed that the genes PtFPS2 and PtGUT1 were located close to each other in the P. tannophilus genome. With the aim of predicting the functions of the putative genes, bioinformatics tools Blastx and Blastp were used, and functional domain predictions were applied. The results are presented in Table 1.
Table 1.
Genes potentially involved in glycerol metabolism in P. tannophilus
| Annotated function | Gene name S. cerevisiae | Gene name P. tannophilus | Size (bp) | Identity/Similarity | Accession no. |
|---|---|---|---|---|---|
| Transport |
|
|
|
|
|
| Glycerol Facilitator |
ScFPS1 |
PtFPS1 |
1170 |
35/49 |
[GenBank:JQ481631] |
| |
|
PtFSP2 |
972 |
32/51 |
[GenBank:JQ481632] |
| Glycerol Symporter |
ScSTL1 |
PtSTL1 |
1728 |
34/51 |
[GenBank:JQ481633] |
| |
|
PtSTL2 |
1905 |
31/50 |
[GenBank:JQ481634] |
| Consumption |
|
|
|
|
|
| Glycerol kinase |
ScGUT1 |
PtGUT1 |
1848 |
53/67 |
[GenBank:JQ481635] |
| G3P dehydrogenase |
ScGUT2 |
PtGUT2 |
1998 |
52/66 |
[GenBank:JQ481636] |
| Glycerol dehydrogenase |
ScGCY0031 |
PtGCY1 |
936 |
54/73 |
[GenBank:JQ481637] |
| |
|
PtGCY2 |
933 |
51/70 |
[GenBank:JQ481638] |
| Dihydroxyacetone kinase |
ScDAK1 |
PtDAK |
1767 |
42/58 |
[GenBank:JQ481639] |
| |
ScDAK2 |
|
|
|
|
| Production |
|
|
|
|
|
| G3P dehydrogenase |
ScGPD1 |
PtGPD |
1314 |
67/80 |
[GenBank:JQ481640] |
| |
ScGPD2 |
|
|
|
|
| G3-phosphatase |
ScGPP1 |
PtGPP |
705 |
33/53 |
[GenBank:JQ481641] |
| ScGPP2 |
The potential genes were identified by performing a BLAST search with known S. cerevisiae genes as queries. Sequences were deposited in Genbank and the accession no. is listed in the table. Sizes given for P. tannophilus genes.
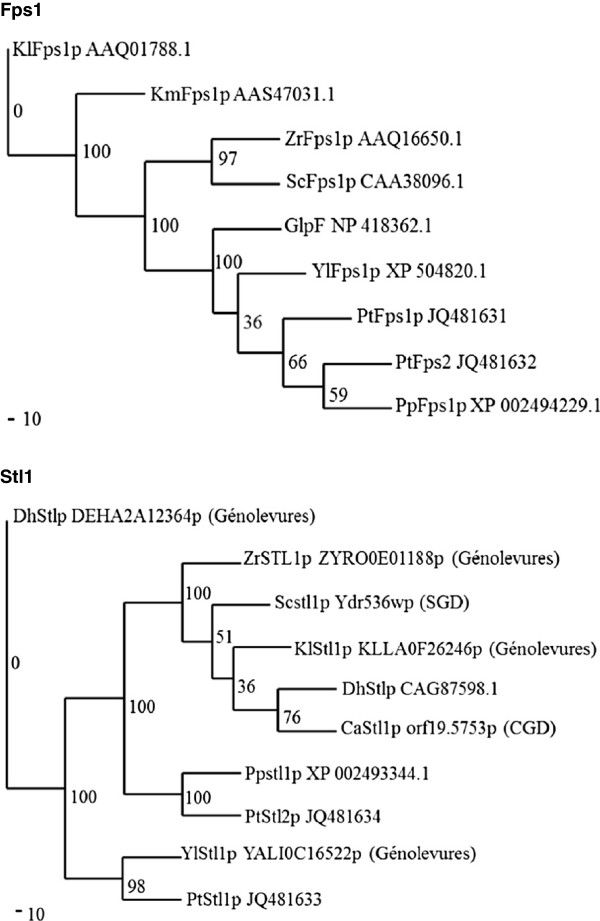
In order to investigate the phylogenetic relationship among putative glycerol transporters in P. tannophilus and their homologues in other yeast strains, an alignment was performed with Ptfps1p, Ptfps2p, Ptstl1p,Ptstl2p as well as with other predicted or published transporter proteins available from GenBank, Génolevures, SGD (Saccharomyces Genome Database) and CGD (Candida Genome Database). Unrooted phylogenetic trees are presented in Figure 1.
Figure 1.
Unrooted phylogenetic tree based on alignment of predicted protein sequences. The tree was bootstrapped 1000 times. Accession numbers are presented after protein names. The origin of the protein sequences from databases other than GenBank are given in brackets after accession numbers. Ca, Candida albicans; Dh, Debaryomyces hansenii; Kl, Kluyveromyces lactis; Km, Kluyveromyces marxianus; Pp, Pichia pastoris; Pt, Pachysolen tannophilus; Sc, Sacchromyces cerevisiae; Yl, Yarrowia lipolytica; Zr, Zygosaccharomyces rouxii; GlpF, the aquaglyceroporin protein from. E. coli.
For FPS1, the lengths of PtFps1p, PtFps2p, PpFps1p, YaFps1p and GlpF were relatively short compared to other Fps1p proteins. PtFps1p was shown to be 54% identical to PpFps1p, 45% to YlFps1p, 35% to ScFps1p and only 31% to GlpF. PtFps2p was shown to be 60% identical to PpFps1p, 49% to YlFps1p, 34% to GlpF and only 32% to ScFps1p. PtFps1p and PtFps2p were grouped together with YlFps1p and PpFps1p, while they were in a separate branch than ScFps1p. For STL1, the identity between PtStl1p and ScStl1p was low, 34%, compared to the 56% identity observed between PtStl1p and YlPtStl1p.
Quantitative real time PCR
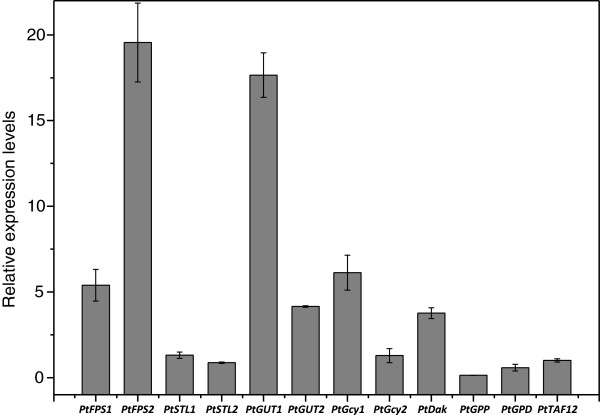
qPCR was used to compare the levels of transcripts during growth on glycerol to those on glucose as the sole carbon source. It was found that the genes involved in glycerol transport and assimilation PtFPS1, PtFPS2, PtGUT1, PtGUT2, PtGCY1, PtDAK were up-regulated on glycerol to different degrees compared to on glucose, while the genes involved with glycerol production PtGPD and PtGPP were down-regulated on glycerol (Figure 2). Among the genes PtFPS2 and PtGUT1 were most up-regulated, by a factor of 19.6 ± 2.3 and 17.6 ± 1.3, respectively, on glycerol compared to glucose. The transcript levels of PtFPS1, another putative facilitator, were up-regulated by a factor of 5.4 ± 0.9 on glycerol compared to glucose, while the putative glycerol symporter genes PtSTL1 and PtSTL2 were expressed almost at the same level on glycerol and glucose. PtGUT2, which is a putative mitochondrial glycerol phosphate ubiquinone oxidoreductase, was also up-regulated 4.2 ± 0.04 fold on glycerol.
Figure 2.
Relative expression levels of glycerol metabolism related genes in P. tannophilus on glycerol compared to that on glucose. The estimation of relative expression levels was based on 2-ΔΔCT, where ΔΔCT = (CT gene of interest - CT internal control) sample A - (CT gene of interest - CT internal control) sample B. CT represents the cycle number at which a sample reaches a predetermined threshold signal value for the specific target gene. All the experiments were performed in triplicate.
Performance of S. cerevisiae stl1Δ harboring different glycerol transporter genes
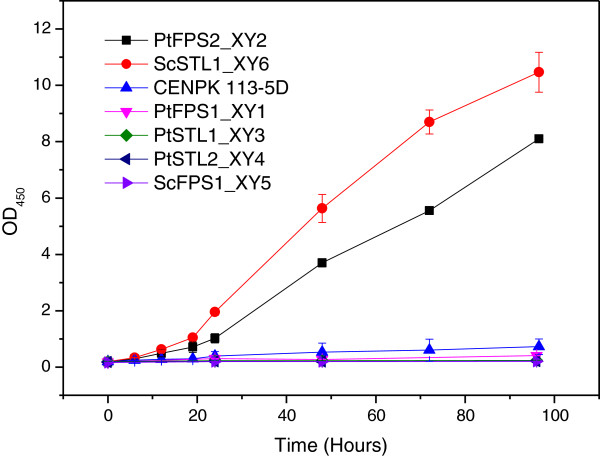
The glycerol symporter gene ScSTL1, which is responsible for glycerol transport in the absence of glucose, was knocked out. In order to validate the function of the glycerol transporters from P. tannophilus, all the predicted transporter genes from P. tannophilus were heterologously expressed in S. cerevisiae stl1Δ. Meanwhile, the glycerol facilitator Fps1p and glycerol symporter Stl1p in S. cerevisiae were also expressed in S. cerevisiae stl1Δ. To analyze the performance of the recombinant strains, growth in defined medium with 1% (v/v) glycerol as the sole carbon source was analyzed. The stl1Δ strain expressing different transporter genes PtFPS1, PtFPS2, PtSTL1, PtSTL2, ScFPS1, ScSTL1 and the S. cerevisiae CENPK 113-5D strain were tested under aerobic conditions in shake flasks.
The stl1Δ recombinant strain expressing the transporter gene PtFPS2 from P. tannophilus and ScSTL1 from S. cerevisiae showed growth on glycerol after 96 hours of cultivation, while the stl1Δ recombinant strain harbouring PtFPS1, PtSTL1, PtSTL2 and ScFPS1 did not grow (Figure 3). The strains expressing PtFPS2 and ScSTL1 genes grew on glycerol with a μmax of 0.07 ± 0.008 and 0.09 ± 0.005 and achieved a final OD of 8.1 and 10.5 at 96 hours cultivation, respectively. In comparison, the S .cerevisiae CENPK 113-5D strain grew very slowly with a μmax of only 0.02 ± 0.004 on glycerol.
Figure 3.
Growth of recombinant S. cerevisiae strains expressing different glycerol transporter genes. The growth was tested in defined medium containing 1% (v/v) glycerol as the sole carbon source at 30°C in agitated flasks. Results represent the mean of at least duplicate experiments.
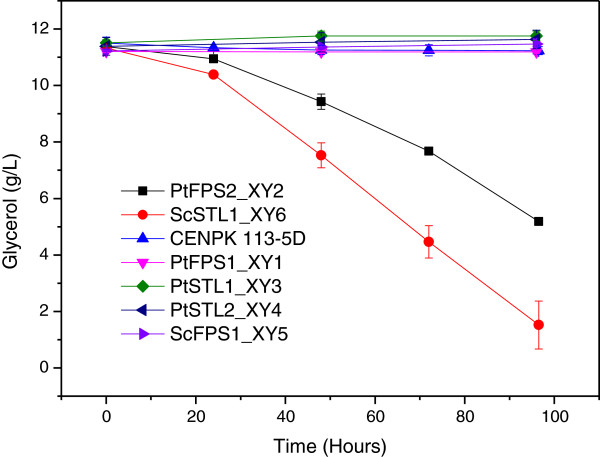
In terms of glycerol consumption, the recombinant stl1Δ strains harboring the genes PtFPS1, PtSTL1, PtSTL2 and ScFPS1 did not consume glycerol even after 96 hours of cultivation. The S .cerevisiae CENPK 113-5D consumed only around 0.1 g glycerol. However, the recombinant stl1Δ strains harboring the genes PtFPS2 and ScSTL1 assimilated glycerol much faster than the other strains and the final glycerol consumed for the two strains was 6.2 ± 0.15 g/L and 9.8 ± 0.6 g/L glycerol, respectively (Figure 4). Although succinate was chosen as a buffer in all the experiments, it was not consumed by the strains.
Figure 4.
Glycerol consumption of recombinant S. cerevisiae strains expressing different glycerol transporter genes. The growth was tested in defined medium containing 1% (v/v) glycerol as the sole carbon source at 30°C in agitated shake flasks. Results represent the means of at least duplicate experiments.
Discussion
With regard to glycerol transport, two glycerol facilitator homologues PtFPS1, PtFPS2 and two glycerol symporter homologues PtSTL1, PtSTL2 were found in the genome of P. tannophilus. Based on qPCR results, all the gene homologues identified to be involved in glycerol transport and assimilation were induced on glycerol as carbon source relative to the levels observed on glucose. The exceptions were the predicted glycerol symporters PtSTL1 and PtSTL2, which were constitutively expressed on both glycerol and glucose as carbon sources. PtFPS2 and PtGUT1 (19.6 fold and 17.6 fold) were the two most strongly up-regulated genes, which might lead to the hypothesis that they are most relevant for glycerol assimilation in P. tannophilus. Moreover, the two genes are located closely together in the genome in agreement with a model where the two genes are co-regulated and contribute to the same overall process, i.e. glycerol assimilation. Next, we focused our efforts on understanding the function of the genes involved in glycerol transport as this function may be the rate limiting step for glycerol metabolism. In this paper the functions of the two types of glycerol transporters from P. tannophilus were addressed by transferring them individually to the well characterized yeast S. cerevisiae.
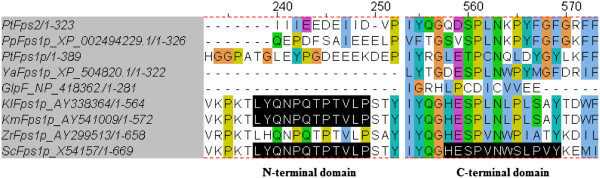
The low affinity transporter Fps1p is a glycerol facilitator protein and belongs to the major intrinsic protein (MIP) family of channel proteins with six putative transmembrane domains (TMDs). Fps1p is responsible for transporting water, small molecules like glycerol, urea, NH3, CO2 or ions without consuming energy. The physiological role of the facilitator Fps1p in S. cerevisiae was previously described to be glycerol export rather than uptake during hypo-osmotic shock and the Fps1p channel closed and retained the glycerol inside the cells in response to hyperosmostic shock [9]. An N-terminal domain 225LYQNPQTPTVLP236 and a C-terminal domain 535HESPVNWSLPVY546 were found to have important roles in controlling Fps1p function. The N-terminus was required for closing of the channels and restricted transport through Fps1p. It was found that the rate of glycerol efflux was higher than that for uptake [9]. In agreement with this, we find in the present study that overexpression of ScFPS1 does not suppress the glycerol transport defect of stl1Δ strains as they do not grow on glycerol. Surprisingly, overexpression of PtFPS1 in stl1Δ strains allowed growth and assimilation of glycerol indicating that this homologous transporter is involved in glycerol uptake and recovers the stl1Δ strain growth defect. The similarities of PtFps1p and PtFps2p to ScFps1p are 35% and 32% respectively, but the homology is only restricted to the core of the protein with the six putative TMDs. ScFps1p (669 amino acid residues) is much longer than PtFps1p (389 residues) and PtFps2p (323 residues), the size of which is more similar to the E. coli GlpF (281 amino acid residues). The size difference is mainly due to the long hydrophilic N- and C-terminal domains of ScFps1p, which are absent in PtFps1p, PtFps2p and GlpF (Figure 5). By expressing PtFPS2 in S. cerevisiae stl1Δ strains, the facilitators from P. tannophilus increased the glycerol influx and glycerol consumption, presumably due to improved glycerol transport ability. Previously it was also reported that the glycerol transport was increased approximately 2.5-fold in S. cerevisiae by introduction of the bacterial gene GlpF [9]. Therefore, we propose that in P. tannophilus the glycerol facilitator might function for glycerol influx rather than efflux.
Figure 5.
Multiple sequence alignments with Fps1p from different yeast strains and E. coli. The alignments were done by CLUSTALW2, showing the N-terminal and C-terminal extension domains of Fps1p.
The high-affinity transporter Stl1p was identified as the glycerol proton symporter in S. cerevisiae, which is a member of the sugar permease family of the major facilitator superfamily (MFS) [24]. It was demonstrated that the transcription of STL1 gene was significantly induced with glycerol as the sole carbon source, and the STL1 gene was subject to glucose repression based on microarray-based transcriptome analysis [25,26]. However, glycerol uptake by STL1 from C. albicans was not affected by the carbon source and salt stress [13]. In D. hansenii, it was shown that the active glycerol transport system was constitutively expressed and not subject to glucose repression [27]. In agreement with these experiments, we showed that the glycerol symporter genes PtSTL1 and PtSTL2 in P. tannophilus are constitutively expressed on glycerol and glucose based on qPCR expression analysis. However, the presence of the PtSTL1 and PtSTL2 genes had no obvious effect on the physiology of S. cerevisiae, while the glycerol consumption and growth in strains that overexpressed ScSTL1 was improved compared to stl1Δ strains. The symporter PtStl1p showed a low degree of sequence identities to ScStl1p with 34%. However, the PtStl1p from P. tannophilus exhibited 52% identity to the Stl1p from D. hansenii (DEHA2A12364p) and 56% to the Stl1p from Yarrowia lipolytica (YALI0C16522p). It has been reported that Y. lipolytica can grow on glycerol with a μmax around 0.3 h-1[28] and Pichia pastoris can grow on glycerol with a μmax 0.26 h-1[29]. P. tannophilus can grow on glycerol with μmax around 0.29 h-1[2], while S. cerevisiae grows relatively slowly on glycerol with μmax of 0.02 h-1 (CEN.PK 113-5D). Both the facilitator and symporter similarities among P. tannophilus, Y. lipolytica and P. pastoris were higher than that compared to S. cerevisiae. However, more protein sequence data with verified functions are needed to provide a definitive conclusion.
Since glycerol transport might be the rate-limiting step for glycerol utilization, heterologous expression of the glycerol transporters from yeasts which have relatively high growth rates on glycerol could be used as the approach for improving the efficiency of glycerol assimilation in S. cerevisiae. Improved glycerol transport has been demonstrated here with an increased glycerol consumption rate and growth rate under aerobic conditions with S. cerevisiae.
Conclusions
The current study demonstrates the function of the glycerol transporters from P. tannophilus. Our studies open new possibilities for further improvement of glycerol fermentation in industrial yeast strains with heterologous expression of glycerol transporters from the glycerol utilizing P. tannophilus. This study thus proposes a possible route to development of glycerol-based bioprocesses in S. cerevisiae.
Methods
Strains and plasmids
The P. tannophilus strain used in this study was CBS4044. S. cerevisiae CEN.PK 113-5D was used for the construction of the STL1 knockout strain. The plasmid PUG6 [30] was utilized as the template for amplifying of the loxP-kanMX-loxP cassette. The integrative USER vector pXI-5 [31] was used in this study for constructing the expression vectors. Plasmid pSP-G1 [32] was used as a template for amplifying the bidirectional promoter TEF1/PGK1. Plasmid pSH47 [30] was used for excision of the kanMX marker gene. All plasmids were propagated in Escherichia coli strain DH5α. All the plasmids and S. cerevisiae strains used in this study are listed in Table 2.
Table 2.
Strains and plasmids used in this study
| Plasmid or strain | Relevant characteristics | Source or reference |
|---|---|---|
| Plasmids |
|
|
| PUG6 |
Plasmid with loxP-kanMX-loxP disruption cassette |
[23] |
| pSP-G1 |
2μ-based URA3 plasmid with TEF1/PGK1 promoter |
[25] |
| pXI-5 |
Integrative USER vector, with reusable URA3 Marker |
[24] |
| pSH47 |
Centromeric plasmid, URA3, PGAL1-Cre-TCYC1 |
[23] |
| pXI-5-PtFPS1 |
pXI-5 with TEF1/PGK1 promoter and PtFPS1 gene |
This study |
| pXI-5-PtFPS2 |
pXI-5 with TEF1/PGK1 promoter and PtFPS2 gene |
This study |
| pXI-5-PtSTL1 |
pXI-5 with TEF1/PGK1 promoter and PtSTL1 gene |
This study |
| pXI-5-PtSTL2 |
pXI-5 with TEF1/PGK1 promoter and PtSTL2 gene |
This study |
| pXI-5-ScFPS1 |
pXI-5 with TEF1/PGK1 promoter and ScFPS1 gene |
This study |
| pXI-5- ScSTL1 |
pXI-5 with TEF1/PGK1 promoter and ScSTL1 gene |
This study |
|
S.cerevisiae Strains |
|
|
| CEN.PK 113-5D |
MATaMAL2-8cSUC2 ura3-52 |
Peter Kötter |
|
stl1Δ |
CEN.PK 113-5D Stl1Δ (490,1279 )::loxP |
This study |
| PtFPS1_XY1 |
stl1Δ with plasmid pXI-5-PtFPS1 |
This study |
| PtFPS2_XY2 |
stl1Δ with plasmid pXI-5-PtFPS2 |
This study |
| PtSTL1_XY3 |
stl1Δ with plasmid pXI-5-PtSTL1 |
This study |
| PtSTL2_XY4 |
stl1Δ with plasmid pXI-5-PtSTL2 |
This study |
| ScFPS1_XY5 |
stl1Δ with plasmid pXI-5-ScFPS1 |
This study |
| ScSTL1_XY6 | stl1Δ with plasmid pXI-5- ScSTL1 | This study |
Medium and culture conditions
For quantitative real-time PCR (qPCR) experiments, P. tannophilus was grown at 30°C in defined medium, containing (per liter) 0.67 g yeast nitrogen base w/o amino acids, 10 g succinic acid and 6 g NaOH as buffer system (initial pH around 5.6), 2% (v/v) glycerol or 2% (w/v) glucose as the carbon source. Cells were harvested at mid exponential growth phase at the same optical density by centrifugation at 5000 g, 4°C for 5mins, resuspended in 1 ml ice cold ddH2O, and the cell pellet was stored at −80°C. S. cerevisiae stl1Δ harbouring different glycerol transporter genes were cultivated at 30°C for 96 hours in agitated flasks with defined medium, containing (per liter) 7.25 g synthetic complete (SC) powder, 10 g succinic acid and 6 g NaOH as buffer system, 1% (v/v) glycerol as the sole carbon source.
Sequence retrieval and analysis
The genome sequence from P. tannophilus CBS4044 was obtained from previous work and is available in the EMBL database [3]. The sequences of genes involved in glycerol transport and metabolism in S. cerevisiae FPS1, STL1, GUT1, GUT2, GCY1, Dak1/2, GPD1/2, GPP1/2 were used as queries in a BLAST search against the genome sequence of P. tannophilus. The genes with high similarities and high identities were listed as potential orthologous genes. Gene sequences were registered in GenBank at NCBI. The multiple sequence alignments with the amino acid sequences of FPS1 and STL1 transporters from different yeast strains and E. coli were performed by using ClustalW2 free program at PDBe. Phylogenetic analyses were performed by using PAUP* 4.0b10 [33]. Unweighted parsimony analysis was performed. Trees were inferred using the heuristic search option with tree bisection-reconnection branch swapping and 1000 random sequence additions. Maxtrees were unlimited, branches of zero length were collapsed and all multiple parsimonious trees were saved. Clade stability was assessed in a bootstrap analysis with 1000 replicates. Trees were visualized in Treeview [34].
Quantitative real-time PCR
Total RNA isolation and cDNA synthesis
Total RNA was isolated from frozen cells using an RNeasy Mini Kit (QIAGEN, USA) according to the manufacturer’s protocol. The quantity and quality of the isolated RNA were measured by NanoDrop ND-1000. The total RNA was treated with DNaseI (Fermentas) prior to cDNA synthesis. Five μg total RNA were used to synthesize cDNA employing the RevertAid™ First Strand cDNA Synthesis kit (Fermentas) following the manufacturer’s recommendations using oligo(dT)18 primer. The cDNA was used as the template for quantitative real-time PCR for determining the transcript levels of the target genes under different growth conditions.
qPCR was performed on a Stratagene Mx3005P using the SYBR Green technology. The qPCR reaction mixture was prepared with 5 μl of 5 times dilution of cDNA as template, 10 μl SYBR Green master mix, 2 μl of 1 μM forward primer and 2 μl of 1 μM reverse primer and ddH2O to 20 μl. The PCR program for qPCR was as follows: 10 min of incubation at 95°C, followed by 40 cycles of 95°C for 30 s, 58°C for 30 s and 72°C for 30 s, and finally the temperature was increased from 55°C to 94°C to check for unspecific products. The number of fluorescence threshold cycles (Ct) was calculated with the set threshold value by using Mx3005P software. Results presented are mean values of three independent experiments. Suitable primer pairs for all the genes investigated were designed using the Primer Express software v3.0 (Applied Biosystems) software with the following parameters: product size 140 - 180 bp and melting temperature (Tm) 57-59°C. The primers used in this work are listed in Additional file 1. In addition, control reactions which include all components for qPCR, except for the reverse transcriptase, were performed to detect the genomic DNA contamination. The absence of products under these conditions meant the absence of enough genomic DNA for successful amplification.
The relative expression levels were calculated approximately based on 2-ΔΔCT, where ΔΔCT = (CT gene of interest - CT internal control) sample A - (CT gene of interest - CT internal control) sample B. CT represents the cycle number at which a sample reaches a predetermined threshold signal value for the specific target gene. Relative expression data were normalized to the relative expression value of the housekeeping gene TAF12 in each sample, thus giving normalized relative expression for a target gene.
S. cerevisiae STL1 knockout strain construction
The construction of the STL1 knockout strain was done through the loxP-kanMX-loxP /Cre recombinase system [30]. The primers for amplification of loxP-kanMX-loxP fragment from pUG6 were: KanMX_fw_STL1 (sequence 5′- AAAGCTGAAAATAGAGGGTTGCTGGTCAATTTAGAAGGTTCCACAATTGCCTGAAGCTTCGTACGCTG -3′) and KanMX_re_STL1 (sequence 5′- AAGCGTTTGTTGATGCACGAACTTTCATTGATGCAATTTCTGGTGGGTATCTAGTGGATCTGATATCAC -3′). The loxP-kanMX-loxP fragment with 50 bp homologous sequences in both ends to the gene STL1was transformed into CENPK-113-5D [35] for obtaining the knockout strain by selection colonies on YPD supplemented with 200 μg/ml Geneticin (G418). The resistant cells were transformed with pSH47 and selected on SC medium lacking uracil. Positive clones were purified and grown in liquid YPD overnight. The next day, the cells were washed once with distilled water and then incubated in YPG medium for 2 h in order to induce expression of the Cre recombinase. The culture medium was diluted and spread on YPD plates. The colonies were picked and restreaked on YPD plates supplemented with and without G418. Clones that were not able to grow on the G418 containing medium were the knockout strains. The knockout strains were streaked on 5-FOA (740 mg/L) plates to eliminate the pSH47 plasmid. Yeast extract-peptone-dextrose (YPD) medium, synthetic complete (SC) medium, and SC lacking specific amino acids were prepared as described previously [36].
Constructs harboring different glycerol transporters
In this study, six expression plasmids carrying the glycerol transporter genes from P. tannophilus and S.cerevisiae were constructed. The USER vector pXI-5 was digested with AsiSI and then with the nicking endonuclease Nb.BsmI for making the AsiSI/Nb.BsmI USER cassettes [37]. The proofreading PfuX7 [38] was used for amplification of fragments PtFPS1, PtFPS2, PtSTL1, PtSTL2, ScFPS1, ScSTL1 and bidirectional promoter TEF1/PGK1 with appropriate USER tails for insertion into the designated USER cassette AsiSI/Nb.BsmI. Primers used are listed in Additional file 1. PCR was performed with PfuX7 DNA polymerase according to manufacturer’s instructions. DNA mixtures were prepared from purified digested vector, glycerol transporter gene, and bidirectional promoter TEF1/PGK1, 5× Phusion HF buffer (Fermentas), and 1 U of USER enzyme (Fermentas), adjusted to 10 μl by adding ddH2O. The mixture was incubated at 37°C for 20 min, followed by 25°C for 20 min. The 10 μl reaction mix was used directly to transform chemically competent E. coli DH5α cells. All the glycerol transporter genes were cloned under control of the TEF1 promoter. Constructs were named as: pXI-5-PtFPS1, pXI-5-PtFPS2, pXI-5-PtSTL1, pXI-5-PtSTL2, pXI-5-ScFPS1, pXI-5-ScSTL1 (Table 2). The plasmids were verified by sequencing and digested with NotI (Fermentas) for 1 h at 37°C and then subjected to gel purification. Each linearized construct was homologously integrated into the S. cerevisiae stl1Δ genome by transformation; six yeast strain lines were constructed as summarized in Table 2. Transformants were plated onto SC-Ura for selection. The S. cerevisiae transformants were restreaked on SC-Ura plates for single colonies, and the desired homologous integration was checked by colony PCR. Sequences and descriptions of primers are presented in Additional file 1.
Analytical methods
Growth was monitored by measuring OD450nm with a Shimadzu UV mini-1240 spectrophotometer (Shimadzu, Japan). Samples were taken periodically from the flasks and filtered through a 0.22 μM syringe filter, and supernatants were preserved at −20°C for later HPLC analysis. Concentrations of the substrate glycerol in supernatants were measured by HPLC refractive index detector RID-10A using an Aminex87H column (Bio-Rad, USA). Separations were performed at 60°C, flow rate of 0.6 ml/min and 5 mM H2SO4 as mobile phase.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
XL carried out the experimental work, analysis and drafted the manuscript. MW, supervised the experimental work and drafting of the manuscript, and has critically reviewed the text. UHM participated in the design and discussion of the experimental work and critical reviewing of the manuscript. All authors have read and approved the manuscript.
Supplementary Material
Primers used for Semi-quantitative RT-PCR and USER Cloning.
Contributor Information
Xiaoying Liu, Email: xiali@bio.dtu.dk.
Uffe Hasbro Mortensen, Email: um@bio.dtu.dk.
Mhairi Workman, Email: mwo@bio.dtu.dk.
Acknowledgements
This work was funded by the European Community’s 7th Framework Research Programme under Grant Agreement Number 213506 (Project GLYFINERY) and the EU European Regional Development Fund InterReg IVA project Øresund Biorefinery, providing financial support to XL and MW.
References
- Kurtzman CP. Biology and physiology of the D-xylose fermenting yeast Pachysolen tannophilus. Springer Berlin/ Heidelberg: Advances in Biochemical Engineering/Biotechnology; 1983. pp. 73–83. (Pentoses and Lignin. Volume 27). [Google Scholar]
- Liu X, Jensen PR, Workman M. Bioconversion of crude glycerol feedstocks into ethanol by Pachysolen tannophilus. Bioresour Technol. 2012;104:579–586. doi: 10.1016/j.biortech.2011.10.065. [DOI] [PubMed] [Google Scholar]
- Liu X, Kaas RS, Jensen PR, Workman M. Draft Genome Sequence of the Yeast Pachysolen tannophilus CBS 4044/NRRL Y-2460. Eukaryot Cell. 2012;11:827. doi: 10.1128/EC.00114-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler L, Blomberg A, Nilsson A. Glycerol metabolism and osmoregulation in the salt-tolerant yeast Debaryomyces hansenii. J Bacteriol. 1985;162:300–306. doi: 10.1128/jb.162.1.300-306.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lages F, Lucas C. Characterization of a glycerol/H + symport in the halotolerant yeast Pichia sorbitophila. Yeast. 1995;11:111–119. doi: 10.1002/yea.320110203. [DOI] [PubMed] [Google Scholar]
- Nevoigt E, Stahl U. Osmoregulation and glycerol metabolism in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev. 1997;21:231–241. doi: 10.1111/j.1574-6976.1997.tb00352.x. [DOI] [PubMed] [Google Scholar]
- Lages F, Lucas C. Contribution to the physiological characterization of glycerol active uptake in Saccharomyces cerevisiae. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 1997;1322:8–18. doi: 10.1016/S0005-2728(97)00062-5. [DOI] [PubMed] [Google Scholar]
- Sutherland FC, Lages F, Lucas C, Luyten K, Albertyn J, Hohmann S, Prior BA, Kilian SG. Characteristics of Fps1-dependent and -independent glycerol transport in Saccharomyces cerevisiae. J Bacteriol. 1997;179:7790–7795. doi: 10.1128/jb.179.24.7790-7795.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamás MJ, Luyten K, Sutherland FCW, Hernandez A, Albertyn J, Valadi H, Li H, Prior BA, Kilian SG, Ramos J. Fps1p controls the accumulation and release of the compatible solute glycerol in yeast osmoregulation. Mol Microbiol. 1999;31:1087–1104. doi: 10.1046/j.1365-2958.1999.01248.x. [DOI] [PubMed] [Google Scholar]
- Holst B, Lunde C, Lages F, Oliveira R, Lucas C, Kielland-Brandt MC. GUP1 and its close homologue GUP2, encoding multimembrane-spanning proteins involved in active glycerol uptake in Saccharomyces cerevisiae. Mol Microbiol. 2000;37:108–124. doi: 10.1046/j.1365-2958.2000.01968.x. [DOI] [PubMed] [Google Scholar]
- Neves L, Lages F, Lucas C. New insights on glycerol transport in Saccharomyces cerevisiae. FEBS Lett. 2004;565:160–162. doi: 10.1016/j.febslet.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Ferreira C, van Voorst F, Martins A, Neves L, Oliveira R, Kielland-Brandt MC, Lucas C, Brandt A. A Member of the Sugar Transporter Family, Stl1p Is the Glycerol/H + Symporter in Saccharomyces cerevisiae. Mol Biol Cell. 2005;16:2068–2076. doi: 10.1091/mbc.E04-10-0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayingo G, Martins A, Andrie R, Neves L, Lucas C, Wong B. A permease encoded by STL1 is required for active glycerol uptake by Candida albicans. Microbiology. 2009;155:1547–1557. doi: 10.1099/mic.0.023457-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan Carlos G-H. Molecular cloning and characterization of STL1 gene of Debaryomyces hansenii. Journal of Yeast and Fungal Research. 2010;1(4):62–72. [Google Scholar]
- May JW, SLOAN J. Glycerol Utilization by Schizosaccharomyces pombe: Dehydrogenation as the Initial Step. J Gen Microbiol. 1981;123:183–185. [Google Scholar]
- May JW, Marshall JH, Sloan J. Glycerol Utilization by Schizosaccharomyces pombe: Phosphorylation of Dihydroxyacetone by a Specific Kinase as the Second Step. J Gen Microbiol. 1982;128:1763–1766. [Google Scholar]
- Sprague GF, Cronan JE. Isolation and characterization of Saccharomyces cerevisiae mutants defective in glycerol catabolism. J Bacteriol. 1977;129:1335–1342. doi: 10.1128/jb.129.3.1335-1342.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norbeck J, Blomberg A. Metabolic and Regulatory Changes Associated with Growth of Saccharomyces cerevisiae in 1.4 M NaCl: EVIDENCE FOR OSMOTIC INDUCTION OF GLYCEROL DISSIMILATION VIA THE DIHYDROXYACETONE PATHWAY. J Biol Chem. 1997;272:5544–5554. doi: 10.1074/jbc.272.9.5544. [DOI] [PubMed] [Google Scholar]
- Albertyn J, Hohmann S, Thevelein JM, Prior BA. GPD1, which encodes glycerol-3-phosphate dehydrogenase, is essential for growth under osmotic stress in Saccharomyces cerevisiae, and its expression is regulated by the high-osmolarity glycerol response pathway. Mol Cell Biol. 1994;14:4135–4144. doi: 10.1128/mcb.14.6.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zhuge J, Fang H, Prior BA. Glycerol production by microbial fermentation: A review. Biotechnol Adv. 2001;19:201–223. doi: 10.1016/s0734-9750(01)00060-x. [DOI] [PubMed] [Google Scholar]
- Ochoa-Estopier A, Lesage J, Gorret N, Guillouet SE. Kinetic analysis of a Saccharomyces cerevisiae strain adapted for improved growth on glycerol: Implications for the development of yeast bioprocesses on glycerol. Bioresour Technol. 2011;102:1521–1527. doi: 10.1016/j.biortech.2010.08.003. [DOI] [PubMed] [Google Scholar]
- Jung J, Yun H, Lee J, Oh M. Production of 1,2-propanediol from glycerol in Saccharomyces cerevisiae. J Microbiol Biotechnol. 2011;21:846–853. doi: 10.4014/jmb.1103.03009. [DOI] [PubMed] [Google Scholar]
- Burd G, Bhattacharyya A. Compositions and methods for enhancing glycerol utilization. 2008. U.S. Patent 2009/0176285 A1.
- Zhao S, Douglas NW, Heine MJS, Williams GM, Winther-Larsen HC, Meaden PG. The STL1 gene of Saccharomyces cerevisiae is predicted to encode a sugar transporter-like protein. Gene. 1994;146:215–219. doi: 10.1016/0378-1119(94)90295-X. [DOI] [PubMed] [Google Scholar]
- Roberts G, Hudson A. Transcriptome profiling of Saccharomyces cerevisiae during a transition from fermentative to glycerol-based respiratory growth reveals extensive metabolic and structural remodeling. Mol Genet Genomics. 2006;276:170–186. doi: 10.1007/s00438-006-0133-9. [DOI] [PubMed] [Google Scholar]
- Roberts Iii GG, Hudson AP. Rsf1p is required for an efficient metabolic shift from fermentative to glycerol-based respiratory growth in S. cerevisiae. Yeast. 2009;26:95–110. doi: 10.1002/yea.1655. [DOI] [PubMed] [Google Scholar]
- Lucas C, Da Costa M, Van Uden N. Osmoregulatory active sodium-glycerol co-transport in the halotolerant yeast Debaryomyces hansenii. Yeast. 1990;6:187–191. doi: 10.1002/yea.320060303. [DOI] [Google Scholar]
- Papanikolaou S, Muniglia L, Chevalot I, Aggelis G, Marc I. Yarrowia lipolytica as a potential producer of citric acid from raw glycerol. J Appl Microbiol. 2002;92:737–744. doi: 10.1046/j.1365-2672.2002.01577.x. [DOI] [PubMed] [Google Scholar]
- Mattanovich D, Graf A, Stadlmann J, Dragosits M, Redl A, Maurer M, Kleinheinz M, Sauer M, Altmann F, Gasser B. Genome, secretome and glucose transport highlight unique features of the protein production host Pichia pastoris. Microb Cell Fact. 2009;8:29. doi: 10.1186/1475-2859-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güldener U, Heck S, Fiedler T, Beinhauer J, Hegemann JH. A New Efficient Gene Disruption Cassette for Repeated Use in Budding Yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen MD, Buron LD, Salomonsen B, Olsen CE, Hansen BG, Mortensen UH, Halkier BA. Microbial production of indolylglucosinolate through engineering of a multi-gene pathway in a versatile yeast expression platform. Metab Eng. 2012;14:104–111. doi: 10.1016/j.ymben.2012.01.006. [DOI] [PubMed] [Google Scholar]
- Partow S, Siewers V, Bjørn S, Nielsen J, Maury J. Characterization of different promoters for designing a new expression vector in Saccharomyces cerevisiae. Yeast. 2010;27:955–964. doi: 10.1002/yea.1806. [DOI] [PubMed] [Google Scholar]
- Swofford DL. DL PAUP* Phylogenetic Analysis Using Parsimony (*And Other Methods). Version 4 2003. Sunderland, MA: Sinauer Associates; 2002. [Google Scholar]
- Page RD. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH, Willems AR, Woods RA. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- Sherman F. In: Methods in Enzymology. Volume Volume 350. Christine G, Gerald RF, editor. New York: Academic Press; 2002. Getting started with yeast; pp. 3–41. [DOI] [PubMed] [Google Scholar]
- Hansen BG, Salomonsen B, Nielsen MT, Nielsen JB, Hansen NB, Nielsen KF, Regueira TB, Nielsen J, Patil KR, Mortensen UH. Versatile Enzyme Expression and Characterization System for Aspergillus nidulans, with the Penicillium brevicompactum Polyketide Synthase Gene from the Mycophenolic Acid Gene Cluster as a Test Case. Appl Environ Microbiol. 2011;77:3044–3051. doi: 10.1128/AEM.01768-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norholm M. A mutant Pfu DNA polymerase designed for advanced uracil-excision DNA engineering. BMC Biotechnol. 2010;10:21. doi: 10.1186/1472-6750-10-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used for Semi-quantitative RT-PCR and USER Cloning.