Abstract
The mouse, as a genetically defined and easily manipulated model organism, has played a critical role in unraveling the mechanisms of craniofacial development and dysmorphology. While numerous gene knockout strains that display craniofacial abnormalities and essential recombinase tool strains with craniofacial-specific expression have been generated, many are absent from public repositories. Large-scale, international resource-generating initiatives promise to address this concern, providing a comprehensive set of targeted mutations and a suite of new Cre driver strains. In addition, panels of genetically defined strains provide tools to dissect the multigenic, complex nature of craniofacial development, adding to the foundation of information gained from single gene studies. Continued progress will require awareness and access to these essential mouse resources. In this review, current mouse resources, large-scale efforts and potential future directions will be outlined and discussed.
Keywords: Knockout, Cre, KOMP, FaceBase
Introduction
The mouse has played a pivotal role in dissecting the genetic pathways that control craniofacial development and dysmorphology. As a small mammal with a short generation time and simple husbandry, the mouse offers the advantages of other simple model organisms, such as zebrafish, Xenopus and chick, combined with a closer phylogenetic relationship to humans and an easily manipulated genome. As such, the mouse has contributed to craniofacial research for many decades, originally through the identification and mapping of spontaneous mutations that arise in inbred strains. The variation seen among individual inbred strains can be used to model human variation in craniofacial morphology and also can provide a genetic tool for understanding susceptibility to environmental causes of craniofacial dysmorphology. Finally, our ability to genetically manipulate the mouse has and will continue to provide rich ground for identification of new genes and pathways important to craniofacial development. Our understanding of the genes that play an important role in craniofacial development is clearly incomplete, as new genes continue to be identified as part of forward genetic screens (Bjork et al., 2010; Stottmann et al., 2010). Therefore, recent efforts to expand the number and availability of mouse resources promise to reveal new genes and pathways important to craniofacial development.
The intent of this review is to identify and describe useful mouse resources available to craniofacial investigators in the scientific community, with a particular emphasis on large-scale efforts to create new tools and models. I will also review information resources that will help users find the mouse resources they need. Details about individual animal models of craniofacial defects have been reviewed extensively elsewhere (Gritli-Linde, 2008; Jiang et al., 2006), and will not be addressed in detail here.
Knockout and Conditional Allele Resources
Gene targeting has provided a wealth of information about the genetic pathways that govern craniofacial development. Over 2100 spontaneous, knockout and/or conditionally targeted alleles, alone or in combination, result in craniofacial malformations of some type (from the Phenotypes Database at MGI: informatics.jax.org) (Blake et al., 2010). A number of mouse Repositories around the world currently distribute these lines, including The Jackson Laboratory (JAX) (www.jax.org) (Davisson and Taft, 2006), the Mouse Mutant Regional Resource Centers (MMRRC) (www.mmrrc.org) (Grieder, 2002), the European Mutant Mouse Archive (EMMA) (Wilkinson et al., 2010) and others (Summarized in Table 1). Strains, embryonic stem (ES) cells and cryopreserved material currently residing in Repositories around the world are all captured in a single database, the International Mouse Strain Resource (IMSR) (www.findmice.org), providing a centralized access point for finding strains of interest (Eppig and Strivens, 1999).
Table 1. Major Mouse Model Repositories.
| Repository Name | Website | Location | Types of Resources |
|---|---|---|---|
| Mutant Mouse Regional Resource Center (MMRRC) |
www.mmrrc.org | UNC-Chapel Hill; University of Missouri; UC-Davis; JAX (USA) |
Live mice, cryopreserved germplasm, ES cells |
| The Jackson Laboratory (JAX) |
www.jax.org | Bar Harbor ME, USA |
Live mice, cryopreserved germplasm, ES cells |
| European Mutant Mouse Archive |
www.emmanet.org | CNR, Moterotondo, Italy; GSF, Munich, Germany; CNRS, Orleans, France; MRC, Harwell, UK; Karolinska Inst., Huddinge, Sweden; Inst. Gulbenkian, Oeiras, Portugal; EMBL-EBI, Hinxton, UK |
Live mice, cryopreserved germplasm |
| European Mouse Mutant Cell Repository (EuMMCR) |
www.eummcr.org | Munich, Germany | ES Cells |
| KOMP Repository | www.komp.org | UC-Davis, USA | Live mice, cryopreserved germplasm, ES cells |
| Canadian Mouse Mutant Repository CMMR |
www.cmmr.ca | Toronto ON, Canada |
Live mice, cryopreserved germplasm, ES cells |
| Taconic | www.taconic.com | Hudson NY, USA | Live mice |
| Texas A&M Institute for Genomic Medicine (TIGM) |
www.tigm.org | Houston TX, USA | ES cells |
| RIKEN BioResource Center (RBRC) |
http://www.brc.riken.jp/lab/animal/en/ | Japan | Live mice, cryopreserved germplasm, ES cells |
High demand strains are frequently distributed from live colonies, while others can be accessed as cryopreserved sperm or embryos. For the investigator, there are many advantages to using a centralized Repository, such as high health status to minimize quarantine, simplified and standardized ordering and shipping procedures, robust and efficient cryorecovery procedures, validated genotyping assays, strain husbandry information, genetic quality control (e.g., strain background), and technical support resources. Despite these advantages and significant efforts by funding agencies urging individual scientists to donate their strains to a Repository, many critical models are not publically available, leading to frequent duplication of effort and the generation of multiple strains of similar or equivalent utility. Large-scale resource generation efforts, such as those discussed below, have begun to address this challenge by providing access to a wide array of targeted mutations.
International Knockout Mouse Consortium
The stated goal of the International Knockout Mouse Consortium (IKMC) is to generate targeted mutations in ES cells for every protein-coding gene in the mouse. (Collins et al., 2007b; Gondo, 2008) The consortium is comprised of three coordinated production programs (summarized in Table 2): the NIH-funded Knockout Mouse Project (KOMP), the EU-funded European Conditional Mouse Mutagenesis (EUCOMM) program (Friedel et al., 2007), and the Canadian North American Conditional Mouse Mutagenesis (NorCOMM) program. These efforts are supplemented by the existing Texas Institute for Genomic Medicine (TIGM) gene trap collection, which, between the C57BL6/N and 129S#/SvEvBrd libraries covers 10,689 mouse genes (Collins et al., 2007a). The KOMP project is charged with generating 8,500 targeted mutations, an effort split between the consortium of the Children’s Hospital of Oakland Research Institute (CHORI), the Welcome Trust Sanger Institute (WTSI) and University of California, Davis (together known as CHORI/Sanger/Davis, or CSD), and the Regeneron Corporation in Tarrytown, NY, which is using their Velocigene (LacZ knock-in) platform (Valenzuela et al., 2003) to complete 3,500 of the 8,500 KOMP targets. The EUCOMM program (Friedel et al., 2007) aims to create 8000 “conditional-ready” targeted null alleles (see www.knockoutmouse.org for allele design details) using a high-throughput vector design and cloning platform. Notably, the KOMP program employs the same platform through its CSD consortium, thus increasing the total number of genes slated for conditional-ready design to 11,500. Finally, the NorCOMM program goal is to generate 500 conditional-ready alleles, using an alternate targeting strategy. Both EUCOMM and NorCOMM have additional gene trapping efforts to ensure complete coverage of the mouse genome. With the exception of the TIGM gene trap library and a small number (less than 1%) of EUCOMM alleles using 129S#/SvEvBrd ES cells, all of these efforts will use C57BL/6N ES cell lines to ensure that virtually all IKMC alleles are on a single genetic background. For each program, a small number of live mice are being produced to validate the process and provide animals for pilot phenotyping efforts. The IKMC Data Coordination Center (IKMC-DCC) (Ringwald et al., 2010) at JAX has updated information on project goals and progress at www.knockoutmouse.org.
Table 2. Mouse Knockout Projects and Current Progress.
The stated goal of total genes and current progress towards that goal (as targeted ES cell lines) is indicated. For KOMP clones, the first number indicates the total completed, while the second is the total number currently available from the KOMP Repository. All data are taken from the IKMC coordinating center (www.knockoutmouse.org) and are current for December 2010.
| Name | Website | Goal | Progress | Repository |
|---|---|---|---|---|
| KOMP -Regeneron |
www.knockoutmouse.org | 3500 | 2838 (1885) | www.komp.org |
| KOMP -CSD |
www.knockoutmouse.org | 5000 | 4160 (3899) | www.komp.org |
| EUCOMM | www.eucomm.org | 8000 | 4917 | www.eummcr.org |
| NorCOMM | www.norcomm.org | 500 | 500 | www.cmmr.ca |
| TIGM | www.tigm.org | n/a | 10,689 | www.tigm.org |
Based on information available at the IKMC-DCC at the time of this writing, the IKMC programs are well on their way to accomplishing their goals, with KOMP, EUCOMM and NorCOMM having completed 82%, 62% and 100% of their proposed efforts, respectively. Notably however, the number of conditional-ready alleles is likely to be less than originally planned. A survey of IKMC alleles in MGI reveals that ~45% of KOMP-CSD alleles and ~54% of EUCOMM alleles are actually conditional-ready, with targeted, non-conditional and deletion (KOMP-CSD only) alleles making up the remainder. The number of alleles for each project in MGI exceeds the total deemed “complete” on the IKMC-DCC website, suggesting that in some cases multiple alleles of the same gene have been generated Clearly, however, a substantial proportion of mouse genes are not going to be conditionally targeted by the IKMC. This is not surprising, as many genes will either be difficult or impossible to properly target with a conditional strategy, particularly one that requires a high-throughput design process. Nevertheless, this is a tremendous resource that will greatly benefit the craniofacial research community and the whole of the scientific community as well.
Individual Repositories for the distribution of ES cells (and in some cases mice or cryopreserved material) from IKMC projects have been established and provide a centralized access point to the IKMC resource (Table 2). For KOMP, all ES cells, mice and mouse germplasm are available through the KOMP Repository (www.komp.org), which will also provide an array of services such as genotype confirmation, chromosome count, pathogen testing and microinjection for an additional fee. These quality control measures are critical, whether performed at KOMP or by individual investigators, as up to 35% of KOMP clones have been found to harbor “complex genetic events” at the targeted locus (Dr. Kent Lloyd, personal communication), which could include errors such as duplication of the targeting cassette not easily detected by PCR screening. EUCOMM clones are distributed through the European Mouse Mutant Cell Repository (EuMMCR, www.eummcr.org) and NorCOMM clones are accessible through the Canadian Mouse Mutant Repository (CMMR, www.cmmr.ca).
Each individual program, or in the case of KOMP the KOMP Repository, has its own web-based interface for searching and accessing IKMC materials of interest. However, the best approach is to use the IKMC-DCC (www.knockoutmouse.org), whose purpose is to provide a centralized database for all IKMC projects. Searching by gene, the user can find all IKMC projects underway for a given gene, their current progress, links to the individual project page for each allele and links to the appropriate Repository, if products are complete and available. In addition, the IKMC-DCC provides direct links to information about existing alleles at MGI and publically available strains or ES cell resources listed in the IMSR. Thus, the IKMC-DCC is an excellent entry portal for any investigator who wants to know if a targeted mutation is available for their gene of interest and how to acquire the material when it becomes available.
High-throughput phenotyping
From the IKMC’s inception, the organizers recognized the value of turning the IKMC clones into live mice, performing broad-based phenotyping to interrogate gene function, and providing basic information to potential users who could then perform more detailed hypothesis-driven analyses in their own laboratories (Moore and Committe, 2010). European groups have led the charge, through the development of highthroughput phenotyping pipelines and standard operating procedures (Beckers et al., 2009; Brown et al., 2005; Brown et al., 2009). More recently, the NIH has committed to producing and phenotyping 2500 strains as part of their new KOMP2 program, an extension of the KOMP project currently nearing completion. Funding decisions will be made by June 2011 and the first mice should reach Phenotyping Centers by the end of the year. Of particular interest to the craniofacial development community is the inclusion of a viability screen, in addition to general morphological characterization of knockout (KO) mice at 7 weeks of age. Although only embryonic day 12.5 (E12.5) was defined as a required screen time point, information regarding viability in the perinatal period will likely be captured as well. While it is hard to say how much specific phenotypic data will be captured for lethal KOs, this project still promises to uncover a large set of new potential regulators of craniofacial morphogenesis, and more importantly, easy access to the relevant mouse model.
Cre Recombinase Resources
While the IKMC programs promise to deliver a large number of new conditional alleles to the scientific community, realizing the full potential of this resource will require a large collection of accessible, well-characterized Cre recombinase driver lines. To date, nearly 1300 Cre driver alleles are captured in MGI, representing 543 individual drivers (promoters), of which 262 are publically available in Repositories (as 424 individual strains) and listed in the IMSR. A number of Repositories distribute Cre strains, including JAX, the MMRRC, and EMMA. The total number of strains is constantly increasing thanks to a number of large-scale projects initiated, for the most part, by the neuroscience community, including the Neuroscience Blueprint Cre Driver project, the GENSAT project and new strains from the Allen Brain Institute (http://www.credrivermice.org/)(Gong et al., 2007; Madisen et al.). While the individual strains are not designed to specifically address questions in craniofacial development, there is likely to be significant overlap with regard to individual promoters chosen as drivers of Cre expression. In addition, our group has initiated a project as part of the FaceBase Consortium to produce Cre strains that would be specifically useful for orofacial clefting (see details below).
Until recently, there has been little international coordination of Cre driver efforts, leading to an incomplete catalogue of resources, spotty availability of strains, and unnecessary duplication of driver generation. For example, there are currently 23 versions of the Nes-Cre allele in MGI, including 13 constitutive Cre lines and 10 inducible lines. While a handful represent multiple transgene insertions from the same lab, this set represents the work of a total of 17 independent investigators, highlighting the high rate of duplication of effort. There are several simple explanations for this phenomenon. For one, the availability of a given driver strain is often limited, particularly in the first few years following publication. For example, of the 23 Nes-Cre lines, only 3 are listed as available from a public repository. Another significant explanation is suboptimal functionality and/or fidelity, particularly for first generation transgenic constructs that contain only a fragment of the upstream promoter region and frequently suffer from position effects at the site of integration site. Both of these issues could be addressed through an international consortium to ensure coordination of future driver generation efforts and to establish standards for characterization. The EU-funded CREATE consortium, lead by Dr. Nadia Rosenthal at EMBL-Monterotondo, is the first group to formally address this issue, bringing together groups from around the world that are involved in generating and distributing Cre strain resources (http://dev.creline.org/home). Thus, building on the IKMC model, the CREATE consortium represents an excellent first step towards coordinating existing Cre resources with new efforts to expand the repertoire of Cre driver strains (for a recent review of these international projects, see (Smedley et al., 2010))
JAX Cre Repository
The JAX Cre Repository is a recent effort to integrate the distribution of a large variety of Cre driver strains with the extended characterization necessary to provide users with a full understanding of the functionality of a given line. JAX currently distributes 233 Cre strains, representing 158 individual drivers. Currently, the characterization program comprehensively evaluates Cre activity using a R26R-LacZ reporter (Soriano, 1999) at four time points, E10.5, E15.5, postnatal day 7 (P7) and adult. All staining is carefully annotated using controlled vocabularies and curated along with slide-scanned images on the JAX website (cre.jax.org). The findings have been quite striking, with nearly all Cre strains exhibiting previously unreported activity in one or more “off-target” tissues. These data can then be combined with existing published data to allow the end user to make a more informed choice as to the best Cre strain for their experiment, and make them aware of the potential confounding effects of the off-target Cre activity. We are currently expanding this program to include analysis of potential toxicity in inducible Cre systems, which has been reported in some systems, but is still not widely appreciated. This extra level of characterization is especially important as new large sets of Cre driver strains are produced, frequently with characterization data focused only on the target tissue of interest and without specific data from use in an experiment.
Cre Databases
The final element to providing Cre resources to the scientific community is a database that allows users to search for Cre functionality by either allele/strain or by tissue/cell type of interest. The first resource that addressed this need was developed in Andras Nagy’s lab, now much expanded and known as the Cre-X-mice database (http://www.mshri.on.ca/nagy/) (Nagy and Mar, 2001; Nagy et al., 2009). It currently contains Cre functionality data for over 500 lines with sophisticated search tools that allow the user to search by a number of parameters, including driver promoter, allele type, anatomical area, and cell type. Of particular interest, users are encouraged to register and submit data about new Cre alleles, such as new strains they have developed in their own laboratories, reducing the curation burden on the database administrators. The Cre8 portal (http://dev.creline.org/search_cre_mice), developed as part of the CREATE consortium effort, provides a BioMart interface to integrate data from the different Cre driver databases around the world, including Cre-X-Mice, CreZOO based at the Fleming Institute in Greece, Insitut Clinique de la Souris (ICS) and the MGI Creportal (see below).
Creportal
MGI has recently unveiled its Creportal (www.creportal.org) (Blake et al., 2010), which builds upon MGI and Gene Expression Database (GXD) (Finger et al., 2010) infrastructure to provide a comprehensive catalogue of all Cre alleles in the public domain. The database can be searched by anatomical structure or driver name, and the queries themselves can be easily refined and sorted according to the needs of the user. Importantly, each individual allele page extensively cross-references many of the resources available through MGI, including IMSR strain availability. Reporter expression data from the published literature is annotated, and when possible, actual images are included. Direct data submission from the JAX Cre Repository and other large-scale Cre driver projects also contribute to the dataset, although curation and ensuring large unpublished datasets are part of the database is ongoing. Overall, the Creportal provides the first truly comprehensive approach to capturing and disseminating detailed information about Cre driver strains and their functionality.
Craniofacial Mutant Resource
The Craniofacial Mutant Resource (CMR) was established at JAX in 2003 as an extension of the Mouse Mutant Resource (MMR) (Bauschatz et al., 2003), which has been identifying, characterizing and genetically mapping spontaneous mouse mutations for over 50 years. Its primary focus is to provide more detailed analysis of craniofacial phenodeviants that arise in the JAX production colonies, including standardized morphometric analysis of head and face shape, in addition to thorough investigation of dental, ear and eye abnormalities. More recently, developmental phenotyping has been added to the project, with a particular focus on identifying more severe (and lethal) homozygous phenotypes which many times appear as dominantly-inherited, viable phenotypes in heterozygotes. This approach has been quite fruitful, informing gene identification efforts of mapped mutants and revealing interesting, severe phenotypes in otherwise subtly affected strains. Over the past 8 years, the program has identified over 286 deviants, of which 72 were proven heritable. From this group, 23 have been genetically mapped to at least a single chromosome, with the likely causative gene having been identified for 14 mutants (Donahue et al., 2003; Lorenz-Depiereux et al., 2004; Mao et al., 2009; Odgren et al., 2010). Strains are made available to the public after genetic and phenotypic characterization through the primary literature or through a specific web portal (craniofacial.jax.org) providing detailed information in the form of a short “web paper”. The rate of discovery has accelerated in the last year with the development of array-based capture techniques (D'Ascenzo et al., 2009), including exome capture, and next-generation sequencing. These approaches greatly reduce the mapping resolution required and the time needed to identify putative genetic lesions. Of particular interest, CMR mutants can arise on any genetic background and in many cases the penetrance is reduced or the phenotype lost when bred to a “standard” strain such as C57BL/6J. Together with the fact that spontaneous mutations frequently harbor allele types other than null, CMR mutants represent a potential pool of craniofacial models that will not be identified through traditional gene targeting, including the IKMC program.
Mouse genetic panels
Given the multigenic nature of craniofacial malformations and number of susceptibility loci being mapped as part of genome wide association studies (GWAS) in humans (Beaty et al., 2010; Mangold et al., 2010), there has been increasing interest in using mouse strain panels as a tool to understand craniofacial dysmorphology as a complex trait. JAX supports a number of mapping panels through the Special Mouse Strains Resource (SMSR), including the AXB and BXA Recombinant Inbred (RI) strain sets, and the B.A Chromosome Substitution Strain (CSS) panel (Peters et al., 2007). Both of these may be particular useful due to the presence of cleft lip susceptibility alleles fixed in the A/J inbred strain background (Juriloff et al., 2006). In addition, a subset of strains selected from among the AXB and BXA sets has been used to identify a number of teratogen susceptibility loci, demonstrating the utility of these sets (Diehl and Erickson, 1997) in developmental studies.
A new resource promises to bring even greater power to map complex traits, and may stimulate a wave of new interest in complex trait analysis in the craniofacial development community. The Collaborative Cross is a large set (up to 500) of RI strains derived from 8 progenitor strains and being developed for distribution from UNC Chapel Hill (Chesler et al., 2008; Churchill et al., 2004). The process of inbreeding the strains is underway and the first lines should be available in early 2011, with a tentative completion in 2013. When inbreeding is complete, each RI strain will be SNP genotyped at high resolution to identify segments from each progenitor strain, providing a perpetual mapping resource that will not require additional genotyping. The power of this resource comes from the genetic diversity of the progenitor strains, which includes 3 wild-derived strains, coupled with the large size of the set which will greatly increase mapping power and resolution. Like the existing RI sets, these strains can be used by craniofacial investigators to study complex gene by environment interactions, map teratogen susceptibility loci and/or map genes that control midfacial morphometry (see FaceBase project below).
FaceBase Consortium
The FaceBase Consortium is a collection of inter-related research and resource development projects aimed at improving our understanding of the causes of orofacial clefting. The consortium structure allows for greater interaction between projects and closer coordination of effort, adding greater value than that which could be achieved through individual research projects. A central coordination “hub” archives and distributes data through a public interface (www.facebase.org). There is a particularly strong focus on projects that develop resources for the consortium and for the scientific community in general. Many of these projects employ the mouse as a model organism, and thus promise to deliver a set of new resources to the craniofacial research community.
Table 3 lists the different FaceBase projects that use the mouse. Although detailed descriptions of each project are available at the FaceBase website (www.facebase.org), here, I will focus on our efforts within the FaceBase Cre Driver project. The goal of this project is to deliver at least 15 new Cre driver strains to the scientific community; strains that will provide new functionality beyond what is currently available for orofacial clefting research. The design of seven Cre strains has already been established in coordination with a working group of FaceBase members and external experts, while the remaining eight will be defined as the project proceeds. This second set of drivers will use data generated by other members of the FaceBase Consortium to guide the selection of driver genes or enhancer sequences. The consortium structure helps to facilitate interaction and enables collaboration that might not have existed otherwise. Critically, all of these strains will be made available to the public through the JAX Repository as soon as basic characterization is complete.
Table 3. FaceBase Projects, Principle Investigators and Resources.
Projects listed here represent individual component projects of the FaceBase Consortium that will generate mouse resources and/or data. More detailed descriptions of these and other FaceBase projects are available at www.facebase.org.
| Project Title | PI(s) | Resource |
|---|---|---|
| Genetic Determinants of Orofacial Shape and Relationship to Cleft Lip/Palate |
Richard Spritz, | Morphometrics, face shape QTL |
| Research on Functional Genomics, Image Analysis and Rescue of Cleft Palate |
Yang Chai Scott Fraser Marianne Bronner- Fraser Joseph Hacia |
Basic Research, Image data |
| Genome-Wide Atlas of Craniofacial Transcriptional Enhancers |
Axel Visel | Enhancer sequence map |
| Functional Analysis of Neural Crest and Palate: Imaging Craniofacial Development |
Scott Fraser Marianne Bronner- Fraser Yang Chai |
Image Data and Technology |
| Global Gene Expression | Steve Potter | Gene Expression Data |
| Atlas of Craniofacial | Bruce Arronow | |
| Development | Paul Trainor | |
| Identification of miRNAs Involved in Midfacial Development and Clefting |
David Clouthier, Kristin Artinger and John Postlethwait |
miRNA Expression Data |
| Genetic Tools and Resources for Orofacial Clefting Research |
Leah Rae Donahue, Steve Murray |
Animal models and Cre tool strains |
The FaceBase Cre Driver project will initially employ two basic strategies to create new Cre driver lines. The first is to use a standard BAC transgenic approach (Gong et al., 2007; Warming et al., 2005), wherein a CreIRESmCherry cassette will be targeted to the ATG of the first exon of the driver gene, replacing the entire exon with the Cre cassette (Figure 1A). This strategy provides a balance between the advantages of a knock-in approach, which would ensure the presence of all regulatory elements, and a standard transgenic approach, which does not cause haploinsufficiency at the driver locus. This is an important consideration, because many genes that have useful expression patterns for driver construction also play a critical role in craniofacial development. Four of these transgenic strains are currently under development, with the first few expected to be released to the public by June 2011. The remainder of the drivers will be chosen in coordination with the FaceBase working group, and will use FaceBase data sets, such as the Global Gene Expression Atlas of Craniofacial Development, as a source for new drivers whenever possible. We are also exploring alternate techniques, such as developing 3’ knock-ins, which target the Cre cassette directly to the target locus, thus retaining endogenous gene function, but avoiding issues related to BAC transgenesis.
Figure 1. Design scheme for FaceBase project Cre driver strains.
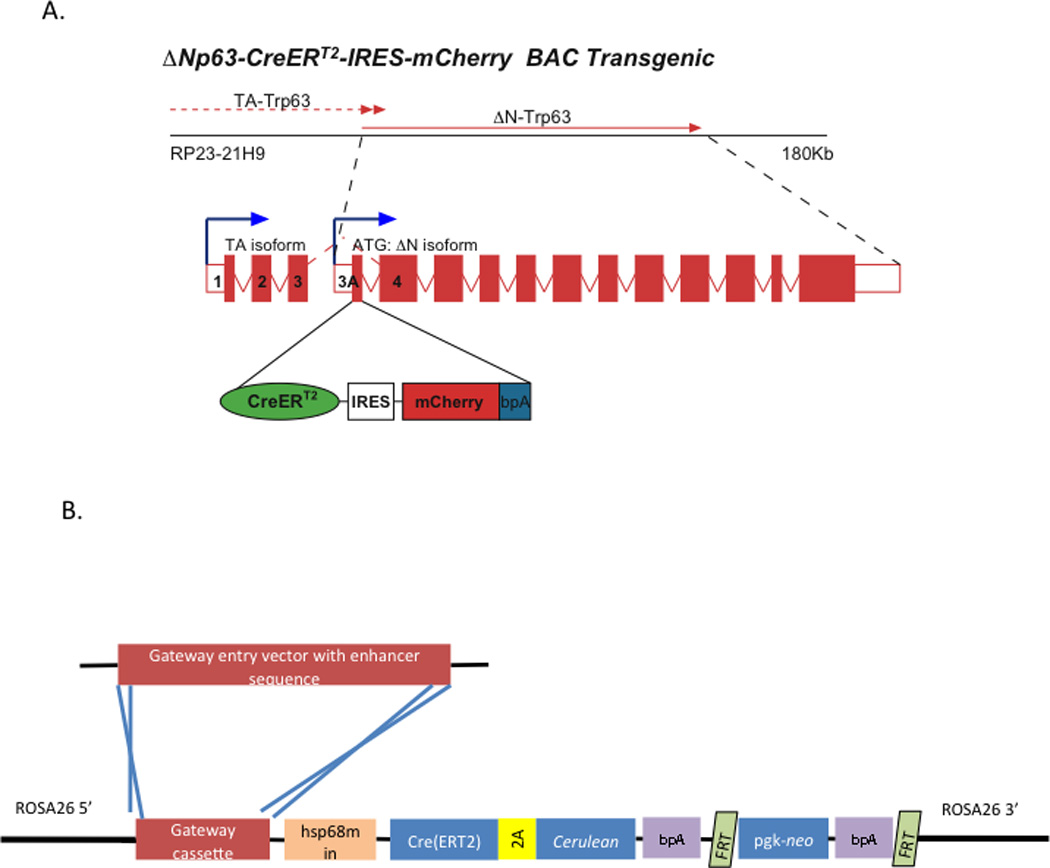
A) An example of the BAC transgenic design using deltaNp63-CreERT2 as a model. A CreERT2-IRESmCherry cassette is cloned directly into the ATG of the target gene, deleting the entire first exon. In this case, a BAC that excluded the 5’ exons was deliberately chosen to ensure expression from the deltaN isoform promoter only. CreERT2 was specifically required in this case so that expression could be induced in the basal epithelium after formation of the periderm layer. B) Overall strategy for creation of Cre driver strains using highly conserved enhancer elements. Individual elements are cloned via Gateway system 5’ of an hsp68 minimal promoter. This drives expression of a Cre or CreERT2 gene linked by a 2A self-cleaving peptide to a Cerulean (blue) fluorescent marker. This entire construct is then targeted to the ROSA26 locus in C57BL/6 ES cells.
The second approach will use findings from another FaceBase project, which is using ChIP-seq to identify discrete enhancer elements that direct gene expression to restricted regions of the developing mid-face and palate. This project builds upon prior work using high conservation and ChIP-seq in other embryonic tissues, which was very successful at identifying such elements (Visel et al., 2009; Visel et al., 2008). The individual elements are validated and specific expression patterns determined through transient transgenesis, which will provide empirical evidence for choosing those best suited for Cre drivers. The elements will be linked to an hsp68 minimal promoter (Gong et al., 2007; Warming et al., 2005) and placed in front of a Cre(ERT2)-2A-Cerulean cassette, and the entire construct knocked into the ROSA26 locus (Figure 1B). The “2A” refers to a Picornaviridae virus family “self-cleaving” peptide (Szymczak et al., 2004), allowing for continued translation following cleavage between the linked proteins and providing an alternative to an IRES element. The use of Cerulean, a blue fluorescent protein (Rizzo et al., 2004), will allow for greater allele combination flexibility, as many reporters constructs employ GFP or red wavelength fluorescent proteins. Like the BAC transgenics, the first set of these lines will be available to the public by June 2011.
Summary
Clearly, the mouse continues to provide an excellent tool for understanding craniofacial development and investigating the causes of dysmorphology. The large-scale targeted mutagenesis programs promise to deliver a complete set of gene mutations to the scientific community, while new technologies are potentially fueling renewed interest in forward mutagenesis approaches for gene discovery. In both cases, awareness of, and access to, mouse resources is essential for continued progress. As rapid growth increases the challenges of providing seamless access to mouse and information resources continued international cooperation and coordination is essential. Despite these challenges, these new mouse resources promise to accelerate our understanding of craniofacial development and disease.
Acknowledgements
Thank you to Leah Rae Donahue and David Bergstrom for their excellent suggestions, comments and careful reading of the manuscript.
NIH support: DE020052 DE019451 RR026117
Literature Cited
- Bauschatz JD, Curtain MM, Davisson MT, Lane PW, Donahue LR. In collaboration: the Jackson Laboratory Craniofacial Resource. Crit Rev Eukaryot Gene Expr. 2003;13:107–108. doi: 10.1615/critreveukaryotgeneexpr.v13.i24.40. [DOI] [PubMed] [Google Scholar]
- Beaty TH, Murray JC, Marazita ML, Munger RG, Ruczinski I, Hetmanski JB, Liang KY, Wu T, Murray T, Fallin MD, Redett RA, Raymond G, Schwender H, Jin SC, Cooper ME, Dunnwald M, Mansilla MA, Leslie E, Bullard S, Lidral AC, Moreno LM, Menezes R, Vieira AR, Petrin A, Wilcox AJ, Lie RT, Jabs EW, Wu-Chou YH, Chen PK, Wang H, Ye X, Huang S, Yeow V, Chong SS, Jee SH, Shi B, Christensen K, Melbye M, Doheny KF, Pugh EW, Ling H, Castilla EE, Czeizel AE, Ma L, Field LL, Brody L, Pangilinan F, Mills JL, Molloy AM, Kirke PN, Scott JM, Arcos-Burgos M, Scott AF. A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nat Genet. 2010;42:525–529. doi: 10.1038/ng.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckers J, Wurst W, de Angelis MH. Towards better mouse models: enhanced genotypes, systemic phenotyping and envirotype modelling. Nat Rev Genet. 2009;10:371–380. doi: 10.1038/nrg2578. [DOI] [PubMed] [Google Scholar]
- Bjork BC, Turbe-Doan A, Prysak M, Herron BJ, Beier DR. Prdm16 is required for normal palatogenesis in mice. Hum Mol Genet. 2010;19:774–789. doi: 10.1093/hmg/ddp543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake JA, Bult CJ, Kadin JA, Richardson JE, Eppig JT. The mouse genome database (MGD): premier model organism resource for mammalian genomics and genetics. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SD, Chambon P, de Angelis MH. EMPReSS: standardized phenotype screens for functional annotation of the mouse genome. Nat Genet. 2005;37:1155. doi: 10.1038/ng1105-1155. [DOI] [PubMed] [Google Scholar]
- Brown SD, Wurst W, Kuhn R, Hancock JM. The functional annotation of mammalian genomes: the challenge of phenotyping. Annu Rev Genet. 2009;43:305–333. doi: 10.1146/annurev-genet-102108-134143. [DOI] [PubMed] [Google Scholar]
- Chesler EJ, Miller DR, Branstetter LR, Galloway LD, Jackson BL, Philip VM, Voy BH, Culiat CT, Threadgill DW, Williams RW, Churchill GA, Johnson DK, Manly KF. The Collaborative Cross at Oak Ridge National Laboratory: developing a powerful resource for systems genetics. Mamm Genome. 2008;19:382–389. doi: 10.1007/s00335-008-9135-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill GA, Airey DC, Allayee H, Angel JM, Attie AD, Beatty J, Beavis WD, Belknap JK, Bennett B, Berrettini W, Bleich A, Bogue M, Broman KW, Buck KJ, Buckler E, Burmeister M, Chesler EJ, Cheverud JM, Clapcote S, Cook MN, Cox RD, Crabbe JC, Crusio WE, Darvasi A, Deschepper CF, Doerge RW, Farber CR, Forejt J, Gaile D, Garlow SJ, Geiger H, Gershenfeld H, Gordon T, Gu J, Gu W, de Haan G, Hayes NL, Heller C, Himmelbauer H, Hitzemann R, Hunter K, Hsu HC, Iraqi FA, Ivandic B, Jacob HJ, Jansen RC, Jepsen KJ, Johnson DK, Johnson TE, Kempermann G, Kendziorski C, Kotb M, Kooy RF, Llamas B, Lammert F, Lassalle JM, Lowenstein PR, Lu L, Lusis A, Manly KF, Marcucio R, Matthews D, Medrano JF, Miller DR, Mittleman G, Mock BA, Mogil JS, Montagutelli X, Morahan G, Morris DG, Mott R, Nadeau JH, Nagase H, Nowakowski RS, O'Hara BF, Osadchuk AV, Page GP, Paigen B, Paigen K, Palmer AA, Pan HJ, Peltonen-Palotie L, Peirce J, Pomp D, Pravenec M, Prows DR, Qi Z, Reeves RH, Roder J, Rosen GD, Schadt EE, Schalkwyk LC, Seltzer Z, Shimomura K, Shou S, Sillanpaa MJ, Siracusa LD, Snoeck HW, Spearow JL, Svenson K, Tarantino LM, Threadgill D, Toth LA, Valdar W, de Villena FP, Warden C, Whatley S, Williams RW, Wiltshire T, Yi N, Zhang D, Zhang M, Zou F. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat Genet. 2004;36:1133–1137. doi: 10.1038/ng1104-1133. [DOI] [PubMed] [Google Scholar]
- Collins FS, Finnell RH, Rossant J, Wurst W. A new partner for the international knockout mouse consortium. Cell. 2007a;129:235. doi: 10.1016/j.cell.2007.04.007. [DOI] [PubMed] [Google Scholar]
- Collins FS, Rossant J, Wurst W. A mouse for all reasons. Cell. 2007b;128:9–13. doi: 10.1016/j.cell.2006.12.018. [DOI] [PubMed] [Google Scholar]
- D'Ascenzo M, Meacham C, Kitzman J, Middle C, Knight J, Winer R, Kukricar M, Richmond T, Albert TJ, Czechanski A, Donahue LR, Affourtit J, Jeddeloh JA, Reinholdt L. Mutation discovery in the mouse using genetically guided array capture and resequencing. Mamm Genome. 2009;20:424–436. doi: 10.1007/s00335-009-9200-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davisson MT, Taft RA. Strategies for managing an ever increasing mutant mouse repository. Brain Res. 2006;1091:255–257. doi: 10.1016/j.brainres.2006.02.031. [DOI] [PubMed] [Google Scholar]
- Diehl SR, Erickson RP. Genome scan for teratogen-induced clefting susceptibility loci in the mouse: evidence of both allelic and locus heterogeneity distinguishing cleft lip and cleft palate. Proc Natl Acad Sci U S A. 1997;94:5231–5236. doi: 10.1073/pnas.94.10.5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue LR, Chang B, Mohan S, Miyakoshi N, Wergedal JE, Baylink DJ, Hawes NL, Rosen CJ, Ward-Bailey P, Zheng QY, Bronson RT, Johnson KR, Davisson MT. A missense mutation in the mouse Col2a1 gene causes spondyloepiphyseal dysplasia congenita, hearing loss, and retinoschisis. J Bone Miner Res. 2003;18:1612–1621. doi: 10.1359/jbmr.2003.18.9.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppig JT, Strivens M. Finding a mouse: the International Mouse Strain Resource (IMSR) Trends Genet. 1999;15:81–82. doi: 10.1016/s0168-9525(98)01665-5. [DOI] [PubMed] [Google Scholar]
- Finger JH, Smith CM, Hayamizu TF, McCright IJ, Eppig JT, Kadin JA, Richardson JE, Ringwald M. The mouse Gene Expression Database (GXD): 2011 update. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedel RH, Seisenberger C, Kaloff C, Wurst W. EUCOMM--the European conditional mouse mutagenesis program. Brief Funct Genomic Proteomic. 2007;6:180–185. doi: 10.1093/bfgp/elm022. [DOI] [PubMed] [Google Scholar]
- Gondo Y. Trends in large-scale mouse mutagenesis: from genetics to functional genomics. Nat Rev Genet. 2008;9:803–810. doi: 10.1038/nrg2431. [DOI] [PubMed] [Google Scholar]
- Gong S, Doughty M, Harbaugh CR, Cummins A, Hatten ME, Heintz N, Gerfen CR. Targeting Cre recombinase to specific neuron populations with bacterial artificial chromosome constructs. J Neurosci. 2007;27:9817–9823. doi: 10.1523/JNEUROSCI.2707-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieder FB. Mutant Mouse Regional Resource Center Program: a resource for distribution of mouse models for biomedical research. Comp Med. 2002;52:203. [PubMed] [Google Scholar]
- Gritli-Linde A. The etiopathogenesis of cleft lip and cleft palate: usefulness and caveats of mouse models. Curr Top Dev Biol. 2008;84:37–138. doi: 10.1016/S0070-2153(08)00602-9. [DOI] [PubMed] [Google Scholar]
- Jiang R, Bush JO, Lidral AC. Development of the upper lip: morphogenetic and molecular mechanisms. Dev Dyn. 2006;235:1152–1166. doi: 10.1002/dvdy.20646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juriloff DM, Harris MJ, McMahon AP, Carroll TJ, Lidral AC. Wnt9b is the mutated gene involved in multifactorial nonsyndromic cleft lip with or without cleft palate in A/WySn mice, as confirmed by a genetic complementation test. Birth Defects Res A Clin Mol Teratol. 2006;76:574–579. doi: 10.1002/bdra.20302. [DOI] [PubMed] [Google Scholar]
- Lorenz-Depiereux B, Guido VE, Johnson KR, Zheng QY, Gagnon LH, Bauschatz JD, Davisson MT, Washburn LL, Donahue LR, Strom TM, Eicher EM. New intragenic deletions in the Phex gene clarify X-linked hypophosphatemia-related abnormalities in mice. Mamm Genome. 2004;15:151–161. doi: 10.1007/s00335-003-2310-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangold E, Ludwig KU, Birnbaum S, Baluardo C, Ferrian M, Herms S, Reutter H, de Assis NA, Chawa TA, Mattheisen M, Steffens M, Barth S, Kluck N, Paul A, Becker J, Lauster C, Schmidt G, Braumann B, Scheer M, Reich RH, Hemprich A, Potzsch S, Blaumeiser B, Moebus S, Krawczak M, Schreiber S, Meitinger T, Wichmann HE, Steegers-Theunissen RP, Kramer FJ, Cichon S, Propping P, Wienker TF, Knapp M, Rubini M, Mossey PA, Hoffmann P, Nothen MM. Genome-wide association study identifies two susceptibility loci for nonsyndromic cleft lip with or without cleft palate. Nat Genet. 2010;42:24–26. doi: 10.1038/ng.506. [DOI] [PubMed] [Google Scholar]
- Mao M, Thedens DR, Chang B, Harris BS, Zheng QY, Johnson KR, Donahue LR, Anderson MG. The podosomal-adaptor protein SH3PXD2B is essential for normal postnatal development. Mamm Genome. 2009;20:462–475. doi: 10.1007/s00335-009-9210-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M, Committe IS. The International Mouse Phenotyping Consortium: Initial Business Plan. 2010 In. [Google Scholar]
- Nagy A, Mar L. Creation and use of a Cre recombinase transgenic database. Methods Mol Biol. 2001;158:95–106. doi: 10.1385/1-59259-220-1:95. [DOI] [PubMed] [Google Scholar]
- Nagy A, Mar L, Watts G. Creation and use of a cre recombinase transgenic database. Methods Mol Biol. 2009;530:365–378. doi: 10.1007/978-1-59745-471-1_19. [DOI] [PubMed] [Google Scholar]
- Odgren PR, Pratt CH, Mackay CA, Mason-Savas A, Curtain M, Shopland L, Ichicki T, Sundberg JP, Donahue LR. Disheveled hair and ear (Dhe), a spontaneous mouse Lmna mutation modeling human laminopathies. PLoS One. 2010;5:e9959. doi: 10.1371/journal.pone.0009959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters LL, Robledo RF, Bult CJ, Churchill GA, Paigen BJ, Svenson KL. The mouse as a model for human biology: a resource guide for complex trait analysis. Nat Rev Genet. 2007;8:58–69. doi: 10.1038/nrg2025. [DOI] [PubMed] [Google Scholar]
- Ringwald M, Iyer V, Mason JC, Stone KR, Tadepally HD, Kadin JA, Bult CJ, Eppig JT, Oakley DJ, Briois S, Stupka E, Maselli V, Smedley D, Liu S, Hansen J, Baldock R, Hicks GG, Skarnes WC. The IKMC web portal: a central point of entry to data and resources from the International Knockout Mouse Consortium. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo MA, Springer GH, Granada B, Piston DW. An improved cyan fluorescent protein variant useful for FRET. Nat Biotechnol. 2004;22:445–449. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- Smedley D, Salimova E, Rosenthal N. Cre recombinase resources for conditional mouse mutagenesis. Methods. 2010 doi: 10.1016/j.ymeth.2010.12.027. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Stottmann RW, Bjork BC, Doyle JB, Beier DR. Identification of a Van der Woude syndrome mutation in the cleft palate 1 mutant mouse. Genesis. 2010;48:303–308. doi: 10.1002/dvg.20618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymczak AL, Workman CJ, Wang Y, Vignali KM, Dilioglou S, Vanin EF, Vignali DA. Correction of multi-gene deficiency in vivo using a single 'self-cleaving' 2A peptide-based retroviral vector. Nat Biotechnol. 2004;22:589–594. doi: 10.1038/nbt957. [DOI] [PubMed] [Google Scholar]
- Valenzuela DM, Murphy AJ, Frendewey D, Gale NW, Economides AN, Auerbach W, Poueymirou WT, Adams NC, Rojas J, Yasenchak J, Chernomorsky R, Boucher M, Elsasser AL, Esau L, Zheng J, Griffiths JA, Wang X, Su H, Xue Y, Dominguez MG, Noguera I, Torres R, Macdonald LE, Stewart AF, DeChiara TM, Yancopoulos GD. High-throughput engineering of the mouse genome coupled with high-resolution expression analysis. Nat Biotechnol. 2003;21:652–659. doi: 10.1038/nbt822. [DOI] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Ren B, Rubin EM, Pennacchio LA. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Prabhakar S, Akiyama JA, Shoukry M, Lewis KD, Holt A, Plajzer-Frick I, Afzal V, Rubin EM, Pennacchio LA. Ultraconservation identifies a small subset of extremely constrained developmental enhancers. Nat Genet. 2008;40:158–160. doi: 10.1038/ng.2007.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005;33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson P, Sengerova J, Matteoni R, Chen CK, Soulat G, Ureta-Vidal A, Fessele S, Hagn M, Massimi M, Pickford K, Butler RH, Marschall S, Mallon AM, Pickard A, Raspa M, Scavizzi F, Fray M, Larrigaldie V, Leyritz J, Birney E, Tocchini-Valentini GP, Brown S, Herault Y, Montoliu L, de Angelis MH, Smedley D. EMMA--mouse mutant resources for the international scientific community. Nucleic Acids Res. 2010;38:D570–D576. doi: 10.1093/nar/gkp799. [DOI] [PMC free article] [PubMed] [Google Scholar]


