Abstract
Borrelia burgdorferi, the etiological agent of Lyme disease, exploits an array of strategies to establish infection and to overcome host innate and adaptive immune responses. One key borrelial immune escape mechanism involves the inactivation of host complement attack through acquisition of human immune regulators factor H (CFH), factor H-like protein 1 (FHL1), factor H-related protein 1 (CFHR1), CFHR2, and/or CFHR5. Binding of these host proteins is primarily mediated by bacterial surface-exposed proteins that have been collectively referred to as complement regulator-acquiring surface proteins, or CRASPs. Different strains of B. burgdorferi produce as many as 5 different CRASP molecules that comprise 3 distinct, genetically unrelated groups. Depending on bacterial genetic composition, different combinations of these proteins can be found on the borrelial outer surface. The 3 groups differ in their gene location, gene regulatory mechanisms, expression patterns during the tick-mammal infection cycle, protein sequence and structure as well as binding affinity for complement regulators and other serum proteins. These attributes influence the proteins’ abilities to contribute to complement resistance of this emerging human pathogen. In this review, we focus on the current knowledge on structure, function, and gene regulation of these B. burgdorferi infection-associated proteins.
Keywords: Borrelia burgdorferi, Spirochetes, Immune evasion, Complement, Complement regulators
Introduction
Lyme borreliosis/Lyme disease is the most commonly reported vector-borne infectious disease in Eurasia and the United States. It is caused by species of the Borrelia burgdorferi sensu lato complex, which includes B. burgdorferi sensu stricto (s.s.) (hereafter referred to as B. burgdorferi), B. garinii, B. afzelii, B. spielmanii, and B. bavariensis (Margos et al., 2009; Richter et al., 2004; Stanek and Reiter, 2011; Steere, 1989; Wang et al., 1999). The ability of Lyme disease spirochetes to perpetuate their natural vertebrate-tick infectious cycle requires an array of strategies to survive in different host environments and necessitates mechanisms to evade innate and adaptive immune responses of their reservoir hosts. Most Lyme disease spirochetes associated with human infection, members of the species B. burgdorferi, B. afzelii, B. spielmanii, and B. bavariensis, are resistant to killing by human complement (Bhide et al., 2005; Brade et al., 1992; Breitner-Ruddock et al., 1997; Herzberger et al., 2007; Kraiczy et al., 2000; Patarakul et al., 1999; van Dam et al., 1997). This is accomplished, at least in part, by the bacteria camouflaging themselves with the host-derived complement regulators factor H (CFH), factor H-like protein 1 (FHL1), and the factor H-related proteins CFHR1, CFHR2, and CFHR5 (Alitalo et al., 2001; Hammerschmidt et al., 2012; Hellwage et al., 2001; Kraiczy et al., 2001a, 2001b; McDowell et al., 2003; Siegel et al., 2010). Binding of those host complement regulators is mediated by surface-exposed lipoproteins that were initially termed CRASPs (complement regulator-acquiring surface proteins) (Kraiczy et al., 2001a, 2001b, 2001c, 2003, 2004b; Wallich et al., 2005). While isolating and characterizing the 5 CRASPs of B. burgdorferi type strain B31, we identified BbCRASP-1 as being encoded by cspA (ORF BBA68), BbCRASP-2 as being encoded by cspZ (ORF BBH06), and BbCRASP-3, BbCRASP-4, and BbCRASP-5 as being identical to the previously named ErpP, ErpC, and ErpA proteins, respectively (Casjens et al., 2000; Hartmann et al., 2006; Kraiczy et al., 2003, 2004a, 2004b; Kraiczy et al., 2003; Stevenson et al., 1996).
Within the last decade, distinct CRASPs interacting with human CFH have been identified among additional borrelial species including B. afzelii, B. spielmanii, B. garinii, B. bavariensis, B. lusitaniae, B. valaisiana, B. bissettii, B. andersonii, B. turdi, B. tanukii, and B. japonica (Alitalo et al., 2001, 2005; Bhide et al., 2009; Dieterich et al., 2010; Herzberger et al., 2007; Kraiczy et al., 2001a; McDowell et al., 2003; Metts et al., 2003; Stevenson et al., 2002; van Burgel et al., 2010; Wallich et al., 2005). Some of these molecules can bind different animal CFH molecules, suggesting that these proteins share identical or similar mechanisms to interact with the key inhibitor of the alternative pathway of diverse hosts, e.g. mouse, rat, dog, sheep, cattle, horse, cat, pig, goat, and chicken (Alitalo et al., 2004; Bhide et al., 2009; Haupt et al., 2007; Hovis et al., 2006; Rogers and Marconi, 2007; Stevenson et al., 2002; van Burgel et al., 2010).
CFH and FHL1 are the key fluid-phase regulatory proteins of the alternative pathway of complement. Both glycoproteins control complement activation at the level of C3b by competing with factor B for binding to C3b, accelerating the decay of the C3 convertase (decay-accelerating activity), and acting as cofactors for factor I-mediated degradation of C3b (Zipfel and Skerka, 2009). The CFH protein family also consists of additional 6 factor H-related proteins (CFHR): CFHR1, CFHR2, CFHR3, CFHR4A, CFHR4B, and CFHR5. All share high degrees of similarity at their carboxy-termini with the carboxy-terminal short consensus repeats (SCRs) 18–20 of CFH (Józsi and Zipfel, 2008; Zipfel et al., 1999; Zipfel and Skerka, 2009). CFHR1 regulates complement at the level of C5 by inhibiting C5 convertase activity and assembly of the terminal membrane attack complex (Heinen et al., 2009). The biological function(s) of CFHR2 is (are) as yet unclear. Like CFH, CFHR5 displays cofactor activity for factor I-mediated inactivation of C3b, thereby inhibiting activity of the fluid-phase C3 convertase (McRae et al., 2001, 2005).
A variety of gene and protein names have been used for CRASP proteins in the literature, leading to some confusion about their identities and functions. One objective of this review is to clarify the nomenclature of these particular molecules.
Characteristics of CspA
The CspA protein (in the literature also referred to as BbCRASP-1, CRASP-1, BBA68, or class 2 CFH binding protein, FHBP) is a surface-exposed, 25.9-kDa lipoprotein. The cspA gene is located on the linear lp54 replicon of B. burgdorferi B31 (Table 1) (Kraiczy et al., 2004b). Sequence analysis of the B31 genome revealed that cspA is part of a large paralogous gene family, PFam54. B. burgdorferi type strain B31 contains 11 apparently intact PFam54 genes, located on 4 different linear plasmids (Casjens et al., 2000, 2012; Wywial et al., 2009). The lp54 replicon of strain B31 contains a tandem array of 7 PFam54 genes (ORF numbers bba64 to bba70), including cspA, plus 2 truncated PFam54 ORFs (bba71 and bba72) (Casjens et al., 2000, 2012; Wywial et al., 2009). Other borrelial isolates can carry a distinct PFam54 gene array, as was demonstrated by comparative sequence analyses of 10 B. burgdorferi genomes originated from the US and Europe (Wywial et al., 2009).
Table 1.
Characteristics of CRASP proteins
| CspA | CspZ | ErpA | ErpC | ErpP | |
|---|---|---|---|---|---|
| Synonyms and other | CRASP-1 | CRASP-2 | CRASP-5 | CRASP-4 | CRASP-3 |
| designations | BbCRASP-1 | BbCRASP-2 | BbCRASP-5 | BbCRASP-4 | BbCRASP-3 |
| BBA68 | BBH06 | ErpI | BBN38 | ||
| ZS7.A68 | ErpN | ||||
| FHBP | BBP38 | ||||
| BBL39 | |||||
| OspE | |||||
| Gene name | cspA | cspZ | erpA | erpC | erpP |
| Gene location in B. burgdorferi | lp54 | lp28-3 | cp32-1 | cp32-2 | cp32-9 |
| strain B31 | cp32-5 | ||||
| cp32-8 | |||||
| Gene family | PFam54 | None | PFam162 | PFam162 | PFam162 |
| Gene expression in unfed tick | No | No | No | No | No |
| Gene expression in feeding tick | Yes | No | Yes | Yes | Yes |
| Gene expression in skin at tick | Yes (high | Yes (low | Yes | Yes | Yes |
| feeding site | expression) | expression) | |||
| Gene expression in | No | Yes (high | Yes | Yes | Yes |
| mice/disseminated infection | expression) | ||||
| Susceptibility to proteolytic | Yes | No | Yes | Yes | Yes |
| degradation | |||||
| Confers serum resistance | Yes | Yes | No | No | No |
| Interaction of complement | CFH | CFH | CFHR1 | CFHR1 | CFHR1 |
| regulators with native CRASP | FHL1 | FHL1 | CFHR2 | CFHR2 | CFHR2 |
| CFHR5 | CFHR5 | ||||
| Interaction of complement | CFH | CFH | CFH | CFH | CFH |
| regulators with denaturated | FHL1 | FHL1 | CFHR1 | CFHR1 | CFHR1 |
| CRASP | CFHR2 | CFHR2 | CFHR2 | ||
| CFHR5 | CFHR5 | ||||
| Interaction with plasminogen | Yes | Yes | Yes | Yes | Yes |
CspA binds the human complement regulators CFH and FHL1 via SCR domains 5–7 (CFH and FHL1) and SCR domains 19 and 20 (CFH) (Kraiczy et al., 2001a, 2004b) (Fig. 1; Table 1). Intriguingly, no other tested PFam54 paralog of strain B31 can interact with complement regulators. This suggests that the paralogs possess other, unknown function(s) (Kraiczy et al., 2004b, 2006; McDowell et al., 2005). Furthermore, the high genetic variability and the lack of a clear synteny and orthology of the PFam54 genes among the species as well as between isolates of the same species suggest that the ability to interact with complement regulators may be an incidental rather than the primary function and therefore PFam54 paralogs possess other, as yet unknown function(s) (Wywial et al., 2009).
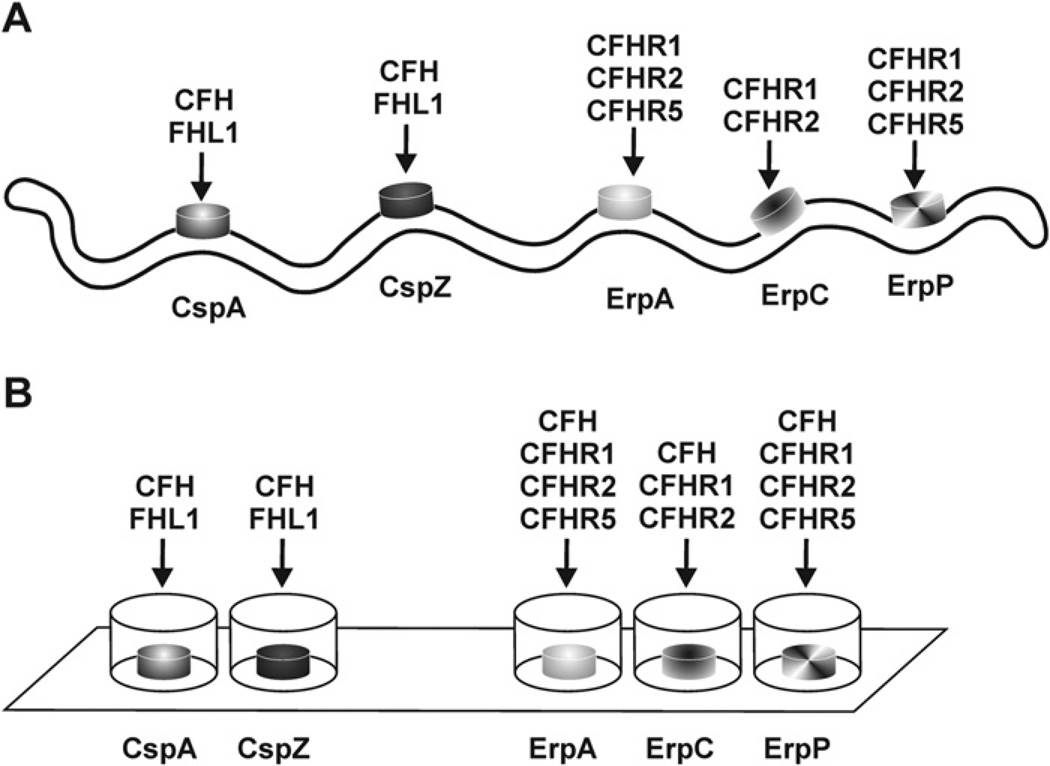
Fig. 1.
Binding capabilities of CRASP proteins of B. burgdorferi to distinct complement regulators. Schematic representation of the binding properties of native CRASP proteins (CspA, CspZ, ErpA, ErpC, and ErpP) exposed to the spirochetal outer surface (A) as well as of recombinant proteins immobilized on a solid, abiotic surface (nitrocellulose membrane or microtiter plate) (B).
Crystals of CspA revealed that it consists of 5 crossing α-helices (A–E), connected by short 310 turns and short loops to form a ‘helical-lollipop’ (Cordes et al., 2005). Based on computer models, it had been hypothesized that CspA (and other CFH-binding proteins) contain coiled-coil elements that form the CFH binding site (McDowell et al., 2005). However, CspA of B. burgdorferi does not contain any coiled-coil structures (Cordes et al., 2005), disproving the coiled-coil hypothesis and demonstrating that such structures are not necessary for CspA to bind CFH. The threedimensional structure of CspA also revealed that the protein is a homodimer. The 2-paired monomers form a large cleft at the dimer interface (Cordes et al., 2005). The architecture of the cleft is suitable for accommodation of a single short consensus repeat (SCR) domain of CFH suggesting that the dimer interface forms the CFH binding site (Cordes et al., 2006). In addition, the dimensions of the cleft would exclude antibodies, suggesting that internal residues are hidden in an immunologically privileged site. This could relieve pressure for variation in the ligand-binding domain. More recently, distinct amino acid residues have been identified that are involved in binding of CFH and FHL1 as well as for sustaining the architecture of the homodimer (Kraiczy et al., 2009). Based on studies with mutagenized proteins, the dimer interface appears to mediate binding of both complement regulators, while the C-terminal region contains a structurally sensitive interaction site. Although some amino acid substitutions introduced in helix E lead to loss of CFH and FHL1 binding, they also cause disruption of the CspA dimer, suggesting that this region is responsible for structure maintenance rather than being involved directly in binding with complement regulators. Differences in the amino acid sequences in the C terminus of PFam54 orthologs encoded on either lp54 (BBA64, BBA65), lp28-4 (BBI36, BBI38, BBI39), or lp38 (BBJ39.1 and BBJ41) may explain the inability of those proteins to bind CFH and FHL1.
Several studies have examined expression levels of the cspA gene during the spirochete’s natural mammal-tick infectious cycle. Transcripts from this gene are largely undetectable in spirochetes residing in the midguts of unfed ticks. However, almost all spirochetes transmitted by feeding ticks produce CspA, indicating rapid gene/protein induction during tick feeding (Bykowski et al., 2007; von Lackum et al., 2005). Two weeks after transmission to mammals, cspA transcripts become undetectable (Bykowski et al., 2007; Lederer et al., 2005; McDowell et al., 2006; Wallich et al., 2003). As a consequence of the short duration of cspA expression during mammalian infection, humans generally do not produce robust levels of antibodies against CspA (McDowell et al., 2006; Rossmann et al., 2006). Noteworthy, detection of anti-CspA antibodies in serum samples obtained from Lyme disease patients has only been successful when non-denaturated CspA protein was utilized. In contrast, reactive antibodies of the same serum samples could not be detected when using conventional Western blotting, which includes denaturation of the protein by SDS-PAGE. Those data indicate that antibodies which develop against CspA during natural infection are largely restricted to natively folded structural determinants (Rossmann et al., 2007). B. burgdorferi again produce CspA during transmission from infected mammals to feeding, naïve ticks (Bykowski et al., 2007; Lederer et al., 2005; McDowell et al., 2006; Wallich et al., 2003). A recent report indicated that expression of cspA orthologous genes of B. burgdorferi, B. afzelii, B. bavariensis, and B. garinii may be induced during incubation of spirochetes with human or certain animal sera (e.g. dog, mouse, and guinea pig or hamster) (Kisova-Vargova et al., 2011) while in contrast, Tokarz et al. (2004) showed that incubation of spirochetes with blood did not change the expression of cspA.
Immune serum generated against CspA is able to prevent infection by needle inoculation of cultured bacteria, but does not efficiently protect infected mice from disseminated spirochetal infection (Wallich et al., 2003). These results are consistent with the known expression profile of CspA, being produced during the initial stage of mammalian infection, but not during later, disseminated stages (von Lackum et al., 2005). Tick challenge experiments with CspA-immunized mice need to be performed to answer the question of whether this particular protein can serve as an effective vaccine to protect humans and domestic animals against B. burgdorferi infection.
The ability of CspA to facilitate complement resistance was demonstrated by the generation of a CspA-deficient B. burgdorferi B31 strain (Brooks et al., 2005; Kenedy et al., 2009). While CspA-deficient spirochetes were highly susceptible to human complement, B. garinii producing surface-exposed CspA displayed a serum-resistant phenotype revealing that CspA alone is sufficient for mediating resistance to human serum.
Studies with B. burgdorferi mutant strains indicated that the cspA gene is transcribed using the housekeeping sigma factor, RpoD (σ70), with neither of the alternative RNA polymerase sigma factors RpoS (σS) and RpoN (NtrA, σ54) playing a significant role (Bykowski et al., 2007; McDowell et al., 2006). Transcription of cspA increased during the mammal-to-tick and tick-to-mammal transmission process, but not during established infection (Bykowski et al., 2007). Expression of cspA was downregulated shortly after transmission of the spirochetes to the mammalian host. Furthermore, wholegenome array analysis using ‘host-adapted’ or ‘blood-treated’ B31 spirochetes also revealed that cspA expression is downregulated in response to different environmental stimuli (Brooks et al., 2003; Tokarz et al., 2004). In contrast to B. burgdorferi, in vitro cultivated B. afzelii spirochetes subjected to environmental conditions simulating the natural mammalian-tick infection cycle produced lower levels of CspA at ambient temperature than at elevated temperatures (Kraiczy et al., 2001a).
Another intriguing feature of the PFam54/cspA paralogous genes comes from in silico RNA secondary structure modeling studies. The sequences immediately downstream of the stop codon carry highly conserved RNA stem-loop structures (Delihas, 2009a). Other borrelial RNA stem-loop motifs are associated with a potential transposon (Delihas, 2009b), suggesting that the PFam54-linked motifs may be involved with expansion of this gene family throughout the borrelial genome.
Characteristics of CspZ
CspZ (in the literature also referred to as BbCRASP-2, CRASP-2, or BBH06) is a surface-exposed 23.2-kDa lipoprotein. It can bind both CFH and FHL1 via SCR domain 7 (Table 1 and Fig. 1) (Hartmann et al., 2006; Siegel et al., 2008b). The cspZ gene is located on the linear lp28-3 replicon of B. burgdorferi B31 and on related lp28-3 plasmids of other borrelial isolates (Hartmann et al., 2006; Siegel et al., 2008b). In contrast to cspA and other members of the PFam54 gene family, there are no other genes within the B. burgdorferi B31 genome that are related to cspZ (Casjens et al., 2000, 2012; Fraser et al., 1997; Hartmann et al., 2006).
Analyses of a number of geographically dispersed borrelial isolates revealed that cspZ sequences are, overall, well conserved among species associated with Lyme disease, e.g. B. burgdorferi, B. afzelii, B. garinii, B. spielmanii, and B. bavariensis as well as B. bissettii (identity ≥97.2%) (Kraiczy et al., 2008; Rogers et al., 2009a; Rogers and Marconi, 2007; Schutzer et al., 2011, 2012). Despite these high degrees of conservation, there are some significant differences among CspZ proteins. There is a species-linked polymorphism in B. afzelii, B. garinii, and B. spielmanii cspZ genes, which encode an N-terminal domain of 44 amino acids that is absent from B. burgdorferi CspZ proteins. CspZ derived from B. burgdorferi bind both CFH and FHL1, while the CspZ orthologs of B. garinii, B. afzelii, and B. spielmanii do not bind these complement regulators (Rogers et al., 2009a; Rogers and Marconi, 2007). The N-terminal extension found in B. garinii CspZ proteins does not prevent binding of CFH or FHL1, indicating that other elements are involved in ligand binding. By investigating a number of additional B. burgdorferi isolates, Rogers et al. (2009a) showed that some natural CspZ orthologs probably lack CFH binding activity due to a 4-amino acid insertion at the N terminus of CspZ and/or point mutations at 3 distinct positions (amino acids 51, 150, and 193). Computer modeling of CspZ suggests that the 4-amino acid insertion is located in a loop between 2 α-helices and thus may destabilize the architecture of the protein and indirectly affect the interactions between CspZ and complement regulators (Kraiczy et al., 2009). In addition, through site-directed mutagenesis and by exogenous expression of mutant CspZ proteins in a borrelial strain that lacks all known CRASPs, a structurally sensitive domain has been identified within the C terminus of CspZ (amino acids 204–211), suggesting that this particular region forms a contiguous binding domain (Siegel et al., 2008b). Changes of amino acid residues within or outside the 4 proposed binding regions may have significant impacts on CFH and FHL1 binding. Those data support a hypothesis of multiple contact sites for the CspZ and its complement regulator ligands. Elucidation of the three-dimensional structure of CspZ will likely yield important clues in our understanding of the molecular interactions between this protein and the host complement regulators of the alternative pathway.
There appears to be a connection between ability/inability of a given strain’s CspZ to bind CFH and that bacterium’s ribosomal spacer sequence type (RST). The RST grouping is also linked to a strain’s tendency toward hematogenous dissemination and invasiveness (Iyer et al., 2001; Rogers et al., 2009a; Wormser et al., 2008). This relationship is linked to 3 CspZ phylogenetic clusters (groups 1–3). Isolates belonging to the highly invasive RST1 and moderate invasive RST3 genogroups fall into groups 1 and 3, which are able to bind CFH. In contrast, group 2 CspZ proteins do not bind CFH and are found in bacteria of the minimally invasive RST2 genogroup. These correlations suggest that CspZ may help B. burgdorferi to disseminate in the mammalian host, as would be expected of a protein involved in the resistance to the host innate immune system.
The cspZ gene is not transcribed by borreliae within the midguts of unfed nymphal ticks (Bykowski et al., 2007; Coleman et al., 2008). During bacterial transmission from feeding ticks to mammalian hosts, only a low percentage of spirochetes produce CspZ. However, within 2 weeks post infection, cspZ transcript levels increase significantly, reaching levels that are considerably higher than those observed in cultured bacteria. Note that the expression profile of CspZ is opposite that of CspA, which is produced at high levels during transmission, then quickly repressed as mammalian infection is established (Bykowski et al., 2007; von Lackum et al., 2005). Human patients with early or late manifestation of Lyme disease generally exhibit a strong antibody response to CspZ, also indicating substantial production of this protein during natural infection (Bykowski et al., 2007; Kraiczy et al., 2009; Rogers et al., 2009a). Moreover, the antibody titers tend to be higher in sera from patients with late-stage disease than with sera obtained from early Lyme disease patients. These observations indicate that spirochetes produce CspZ throughout disseminated and persistent infection of humans and experimental animals. However, for establishment of murine infection, CspZ appears to be not absolutely required (Coleman et al., 2008). Thus, it is not yet known how much CspZ contributes to the ability of Lyme spirochetes to infect their mammalian hosts.
CspZ can provide spirochetes with resistance to host complement. When a strain of B. garinii that normally lacks CRASPs was transformed such that it produced CspZ on its surface, the bacterium acquired ability to bind CFH and became resistant to complement-mediated killing (Siegel et al., 2008a). Contradictory data have been reported from a CspZ-deficient B. burgdorferi strain that resists complement-mediated killing by human serum (Coleman et al., 2008). However, that observation might be due to CspA compensating for the loss of CspZ in the studied B. burgdorferi strain. As noted in the preceding paragraph, wild-type B. burgdorferi produce substantially greater levels of cspZ transcripts during mammalian infection than they do when grown in culture medium (Bykowski et al., 2007). Moreover, CspZ protein is often undetectable in cultured spirochetes. This is a likely explanation for an observation that a B. burgdorferi strain specifically deleted of cspA (cspZ+, cspA−) was unable to bind CFH (Coleman et al., 2008). The low level of CspZ expression by cultured wild-type B. burgdorferi may also explain the inability of CspZ-directed antibodies to protect mice from challenge with cultured B. burgdorferi (Rogers et al., 2009a).
As are all the other borrelial CRASP-encoding genes, cspZ transcription is directed by the housekeeping sigma factor, RpoD (Bykowski et al., 2007, 2008). Mutant spirochetes, deficient of either the alternative sigma factors, RpoN and RpoS, express cspZ at levels comparable to wild-type B. burgdorferi (Bykowski et al., 2007, 2008). Expression of cspZ is influenced by Rrp1 (BB0419) and Rrp2 (BB0763), parts of 2 distinct two-component sensory transduction systems (Rogers et al., 2009b; Yang et al., 2003). Rrp1 produces cyclic di-GMP in response to unknown signals, which appears to influence expression levels of a wide variety of borrelial genes, many of which are involved in mammalian infection (Rogers et al., 2009b).
Immunofluorescence microscopy of intact spirochetes indicated surface exposure of CspZ (Hartmann et al., 2006; Siegel et al., 2008a). The majority of surface-exposed proteins of B. burgdorferi are susceptible to protease treatment in situ, in particular to proteolytic enzymes exhibiting broad substrate specificity. An uncommon feature of native CspZ is its insensitivity to proteolytic degradation by proteinase K and trypsin (Coleman et al., 2008; Hartmann et al., 2006). However, several other Borrelia spp. outer membrane proteins that have been confirmed to be surface-exposed, such as the B. burgdorferi OspA, Oms66, ErpB, ErpM, ErpK, ErpX, and B. turicatae Vsp proteins, are also innately resistant to protease degradation in situ (El-Hage et al., 2001; Exner et al., 2000; Zückert et al., 2001). CspZ and these other surface proteins may naturally fold such that protease target sequences are hidden, might be glycosylated or otherwise modified, or may interact with other surface components that shield them from proteases (Benz and Schmidt, 2002; El-Hage et al., 2001; Exner et al., 2000; Zückert et al., 2001). Alternatively, resistance to proteolytic degradation might also be due to CspZ not being exposed at the outer surface (Coleman et al., 2008).
Characteristics of ErpA, ErpC, and ErpP
B. burgdorferi type strain B31 encodes three 17- to 20-kDa surface-exposed lipoproteins that have high affinities for CFH and some CFHRs (Fig. 1) (Hammerschmidt et al., 2012; Haupt et al., 2007; Hellwage et al., 2001; Siegel et al., 2010). These are ErpA, ErpC, and ErpP (also collectively described as OspE, although each protein has a distinct sequence) (Table 1) (Casjens et al., 1997b, 2000; Stevenson et al., 1996). Several additional alternative names are found in publications: ErpA has also been described as BbCRASP-5, ErpI, ErpN, BBP38, and BBL39; ErpC has also been called BbCRASP-4; and ErpP has been called BbCRASP-3 or BBN38 (Hovis et al., 2006; Kraiczy et al., 2004a; McDowell et al., 2004; Metts et al., 2003). Type strain B31 carries 3 separate but identical erpA genes, one each on cp32-1, cp32-5, and cp23-8, and an erp gene redundancy that has also been found in some other B. burgdorferi isolates (Stevenson and Miller, 2003; Stevenson et al., 2000b). Of note, the erpC gene was not identified in the sequenced subculture of type strain B31 (B31-MI), due to its lack of cp32-2, although that plasmid is maintained by other subcultures of strain B31 (Casjens et al., 2000; Stevenson et al., 1996, 2006). Orthologs that have been characterized in other B. burgdorferi strains were given various names. Adding to confusion, some nomenclatures give the same name to distinct proteins/genes, such as the very different proteins of B. burgdorferi strains N40 and 297 that have both been named ‘P21’. As noted above, the approximately 20-kDa CRASPs have been collectively named OspE. However, as we describe below, the proteins in the OspE group do not all possess the same functions. In the Erp nomenclature, each distinct protein has a unique name, e.g. ErpA, ErpC, Erp63, Erp64, etc.
Almost all of the examined approximately 20-kDa Erp/CRASPs bind human CFH through that complement regulator’s SCR20 domain (Kraiczy et al., 2001a). They also bind CFHR-1, CFHR-2, and CFHR-5 through those proteins’ C-terminal SCR domains (which are >90% identical to the C-terminal SCR20 of CFH). None of these molecules, however, bind complement regulator FHL1. Despite their high degrees of sequence similarity, the strain B31 ErpA, ErpC, and ErpP proteins differ in their affinities for CFH, CFHR-1, CFHR-2, and CFHR-5 (Hammerschmidt et al., 2012; Haupt et al., 2007; Siegel et al., 2010). There are several lines of evidence, indicating that native ErpA, ErpC, and ErpP do not confer resistance of B. burgdorferi to complement-mediated lysis in vitro under conditions where high concentrations of human serum (up to 50%) were applied. A CspA-deficient B31 strain as well as B313, a derivative of B31, lacked CFH binding and were highly sensitive to human complement despite the production of ErpA and ErpN or ErpA, respectively (Brooks et al., 2005; Hartmann et al., 2006; Rossmann et al., 2007; Schott et al., 2010). Moreover, heterologous production of ErpA, ErpC, or ErpP in a strain of B. garinii that lacks naturally occurring CRASPs did not provide resistance to complement-mediating bacteriolysis (Hammerschmidt et al., 2012; Siegel et al., 2010), suggesting that these particular Erp proteins may act in concert with CspA and/or CspZ to help the bacteria evade complement-mediated killing, confirming earlier findings with CRASP-deficient spirochetes. However, simultaneous overproduction of ErpP and ErpA in the serum-sensitive CspA-deficient B31 strain imparts serum resistance, thus under the expression of an artificial promoter, Erp proteins are sufficient to restore serum resistance (Kenedy and Akins, 2011).
Type strain B31 encodes 13 unique Erp proteins, most of which have highly divergent sequences (Casjens et al., 2000; Stevenson et al., 1998a). Other members of the Erp protein family, such as ErpX or ErpY of type strain B31 and Erp proteins of B. lusitaniae can also bind CFH under certain conditions, although it is not clear whether those interactions are of biological significance (Dieterich et al., 2010; Stevenson et al., 2002).
Multi-label immunofluorescence analysis confirmed that a bacterium’s entire repertoire of Erp proteins is simultaneously co-expressed (El-Hage et al., 2001; El-Hage and Stevenson, 2002). All erp genes are carried on circular replicons of the cp32 family and all examined B. burgdorferi isolates naturally carry multiple, distinct cp32 plasmids (Table 1) (Casjens et al., 2012; Schutzer et al., 2012; Stevenson and Miller, 2003; Stevenson et al., 2001). In addition, numerous related cp32 replicons have been identified in all Lyme disease-associated spirochetes, including B. afzelii, B. garinii, B. bavariensis, and B. spielmanii as well as B. lusitaniae, and other members of the genus Borrelia such as the relapsing fever spirochetes B. hermsii and B. turicatae (Brissette et al., 2008; Casjens et al., 2012; Schutzer et al., 2011, 2012; Stevenson et al., 2000a, 2006). Of note, cp32 plasmids of examined relapsing fever Borrelia species do not contain any erp loci (Stevenson et al., 2000b). Several lines of evidence indicate that cp32 replicons are genomes of lysogenic prophages, which could be responsible for the observed horizontal transmission of erp genes among Borrelia (Casjens et al., 1997a, 2000; Chenail et al., 2012; Eggers et al., 2001; Eggers and Samuels, 1999; Stevenson et al., 1998b; Stevenson and Miller, 2003; Zhang and Marconi, 2005).
All Erp family members contain a typical consensus signal peptidase II cleavage sequence at the N terminus (Stevenson et al., 2000b). A variety of techniques have confirmed that all Erp proteins are located in the borrelial outer membrane and are exposed to the external environment (El-Hage et al., 2001; Kraiczy et al., 2003; Lam et al., 1994; Siegel et al., 2010). An additional feature of Erp proteins is their high content of the charged amino acids lysine and glutamate. Lysine residues, in particular, are proposed to be involved in the interaction of Erp proteins with host CFH (Alitalo et al., 2004, 2005). Truncations of the N- and C terminus of ErpA and ErpP completely disrupt binding of CFH, supporting the hypothesis that discontinuous or conformational domains are required for binding of CFH (Kraiczy et al., 2003; Metts et al., 2003).
Three-dimensional structures have yet to be determined for any Erp protein, so it is not entirely clear whether hypothesized coiled-coil structures are present, and if those or other higher-ordered structures are required for proper ligand binding (Alitalo et al., 2002, 2004, 2005; Kraiczy et al., 2003; McDowell et al., 2004; Seling et al., 2010). The contribution of individual amino acid residues in binding of CFH has been clearly demonstrated for the Erp63 protein of B. spielmanii strain TIsar3 (Seling et al., 2010). Exchange of a single amino acid residue completely abolished ability to bind CFH, yet did not alter the ability of Erp63 to bind plasminogen, suggesting that the entire architecture of this particular molecule was not impaired and that this particular amino acid residue directly interacts with CFH.
Unexpectedly, spirochetes that lack cspA and cspZ, but contain erpA, erpC, or erpP, do not bind CFH to their surface, although the complement regulators CFHR-1, CFHR-2, and CFHR-5 were bound (Fig. 1) (Hammerschmidt et al., 2012; Siegel et al., 2010). Those results indicate that ErpA, ErpC, and ErpP in situ possess different binding properties than do the isolated recombinant proteins. One possible explanation is that other membrane proteins proximate to these molecules impair interactions between Erps and the large CFH protein, while permitting binding of the smaller CFHR-1, CFHR-2, and CFHR-5 molecules. These findings warrant continuative investigations of Erp protein architecture and protein-protein interactions to define the functions of these ubiquitous, infection-associated surface proteins.
Erp proteins are produced during mammalian infection, but are largely repressed during tick colonization (Akins et al., 1995; Hefty et al., 2001; McDowell et al., 2001a, 2001b; Miller and Stevenson, 2006; Miller et al., 2003; Stevenson et al., 1995, 1998a; Wallich et al., 1995, 2005). The spirochetes continually synthesize Erp proteins throughout mammalian infection, although levels of expression vary over time (Miller and Stevenson, 2006; Miller et al., 2003). Analysis of the mechanisms governing erp gene expression revealed that all erp operons are controlled by the housekeeping sigma factor RpoD, although some are also transcribed to varying extents by the alternative sigma factor RpoS (Caimano et al., 2004; Eggers et al., 2004, 2006). Regulation of erp transcription depends on a DNA region adjacent to the promoter the erp operator, to which 3 cytoplasmic proteins bind. BpaB binds to the erp operator and represses erp transcription. EbfC competes with BpaB and functions as an anti-repressor. BpuR binding enhances the repressive effects of BpaB (Babb et al., 2004, 2006; Burns et al., 2010; Jutras et al., 2012a, 2012b; Riley et al., 2009). The alarmones autoinducer- 2 (AI-2) and cyclic di-GMP also influence erp transcription levels (Rogers et al., 2009b; Stevenson and Babb, 2002; von Lackum et al., 2006). Noteworthy, some inconsistencies in regulation of erp genes have been reported for some B. burgdorferi strains, possibly due to genetic differences that result in different levels of regulatory factors or affinities of DNA binding protein alleles (Das et al., 1997; Miller et al., 2005). Nucleotide differences found in erp promoter and operator regions may also contribute to strain-to-strain variations in erp expression levels.
During mammalian infection, the entire repertoire of Erp proteins produced by the spirochetes are exposed to the external environment, and host adaptive immune systems produce robust antibody responses against these lipoproteins (Akins et al., 1995; Lam et al., 1994; Miller et al., 2003; Nguyen et al., 1994; Stevenson et al., 1998a; Wallich et al., 1995). Due to the large strain-tostrain variability of Erp protein sequences, the suitability of these molecules as vaccine components or for serodiagnostic tests is very restricted (Miller et al., 2000; Stevenson et al., 1998a). Antibodies directed against the identical Erp proteins produced by the challenge strain of bacteria are unable to effectively protect mice from infection (Hefty et al., 2002; Nguyen et al., 1994). It remains to be determined how an extracellular pathogen such as B. burgdorferi is able to produce such highly antigenic proteins on its outer surface during long-term infection of immunocompetent animals.
Conclusions
Independent of the geographical origin, all examined natural B. burgdorferi isolates contain cspA and numerous erp genes. In addition, a large number of investigated Lyme disease borreliae also harbor a cspZ gene. The ubiquity of these genes points toward an important role(s) for their encoded proteins in the Lyme disease spirochete’s natural infectious cycle. The CRASPs’ strong affinities and binding capacities for host complement regulatory proteins strongly supports their hypothesized roles in immune evasion in diverse mammalian hosts. Although the three-dimensional structure of CspA has been determined, and mutagenesis approaches with CspA, CspZ, and ErpA/ErpP have identified probable structural determinants involved in interactions with complement regulators, much remains to be learned about this particular group of molecules. Continued studies of borrelial CRASPs will further clarify the role(s) of these proteins in infection and in the pathogenesis of Lyme disease/Lyme borreliosis.
As with B. burgdorferi, sequence heterogeneity of CRASP-encoding genes is also found in serum-resistant B. afzelii and B. spielmanii. All examined isolates of those species contain a cspA gene located on an lp54 replicon and numerous erp genes (Herzberger et al., 2007, 2009; Kraiczy et al., 2001a; Wallich et al., 2005). Although cspA orthologous genes are presented in the genomes of serum-sensitive B. valaisiana and B. lusitaniae (Schutzer et al., 2012), tested orthologs do not bind CFH or FHL1 (Dieterich et al., 2010, and P. Kraiczy unpublished data). In contrast to CspA, several Erp homologs of B. valaisiana and B. lusitaniae do bind CFH and CFHR1 (Dieterich et al., 2010). Of note, no CRASP-encoding sequences have been identified in genomes of bacteria outside of the genus Borrelia. Moreover, many important human pathogens, e.g. Staphylococcus aureus, Streptococcus pyogenes, S. pneumoniae, Neisseria meningitidis, N. gonorrhoeae, and Haemophilus influenzae, bind host complement regulators through highly diverse bacterial proteins (for review see Lambris et al., 2008). Investigations of Lyme disease spirochetes provide not only insight into the infectious mechanisms of these spirochetes, but also provide helpful comparisons and contrasts with many other infectious agents.
Acknowledgements
We thank all our colleagues and friends whose research has contributed to an increased understanding of CRASP proteins. Research in the Kraiczy laboratory is supported by the Deutsche Forschungs-gemeinschaft grant Kr3383/1-2 and in the Stevenson laboratory by U.S. National Institutes of Health grant R01-AI44254.
Abbreviations
- CFH
factor H
- FHL1
factor H-like protein 1
- CFHR
human factor H-related proteins
- CRASP
complement regulator-acquiring surface protein
- PF
paralogous family
- CspA
complement regulator-acquiring surface protein 1
- CspZ
complement regulator-acquiring surface protein 2
- Erp
OspE/F-related surface protein
References
- Akins DR, Porcella SF, Popova TG, Shevchenko D, Baker SI, Li M, Norgard MV, Radolf JD. Evidence for in vivo but not in vitro expression of a Borrelia burgdorferi outer surface protein F (OspF) homologue. Mol. Microbiol. 1995;18:507–520. doi: 10.1111/j.1365-2958.1995.mmi_18030507.x. [DOI] [PubMed] [Google Scholar]
- Alitalo A, Meri T, Chen T, Lankinen H, Cheng Z-Z, Jokiranta TS, Seppälä IJT, Lahdenne P, Hefty PS, Akins DR, Meri S. Lysine-dependent multipoint binding of the Borrelia burgdorferivirulence factor outer surface protein E to the C terminus of factor H. J. Immunol. 2004;172:6195–6201. doi: 10.4049/jimmunol.172.10.6195. [DOI] [PubMed] [Google Scholar]
- Alitalo A, Meri T, Comstedt P, Jeffery L, Tornberg J, Strandin T, Lankinen H, Bergström S, Cinco M, Vuppala SR, Akins DR, Meri S. Expression of complement factor H binding immunoevasion proteins in Borrelia garinii isolated from patients with neuroborreliosis. Eur. J. Immunol. 2005;35:3043–3053. doi: 10.1002/eji.200526354. [DOI] [PubMed] [Google Scholar]
- Alitalo A, Meri T, Lankinen H, Seppälä I, Lahdenne P, Hefty PS, Akins D, Meri S. Complement inhibitor factor H binding to Lyme disease spirochetes is mediated by inducible expression of multiple plasmid-encoded outer surface protein E paralogs. J. Immunol. 2002;169:3847–3853. doi: 10.4049/jimmunol.169.7.3847. [DOI] [PubMed] [Google Scholar]
- Alitalo A, Meri T, Rämö L, Jokiranta TS, Heikkilä T, Seppälä IJT, Oksi J, Viljanen M, Meri S. Complement evasion by Borrelia burgdorferi: serum-resistant strains promote C3b inactivation. Infect. Immun. 2001;69:3685–3691. doi: 10.1128/IAI.69.6.3685-3691.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babb K, Bykowski T, Riley SP, Miller MC, Demoll E, Stevenson B. Borrelia burgdorferi EbfC, a novel, chromosomally encoded protein, binds specific DNA sequences adjacent to erp loci on the spirochete’s resident cp32 prophages. J. Bacteriol. 2006;188:4331–4339. doi: 10.1128/JB.00005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babb K, McAlister JD, Miller JC, Stevenson B. Molecular characterization of Borrelia burgdorferi erp promoter/operator elements. J. Bacteriol. 2004;186:2745–2756. doi: 10.1128/JB.186.9.2745-2756.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benz I, Schmidt MA. Never say never again: protein glycosylation in pathogenic bacteria. Microbiol. Mol. 2002;45:267–276. doi: 10.1046/j.1365-2958.2002.03030.x. [DOI] [PubMed] [Google Scholar]
- Bhide MR, Escudero R, Camafeita E, Gil H, Jado I, Anda P. Complement factor H binding by different Lyme disease and relapsing fever Borrelia in animals and human. BMC Res. Notes. 2009;2:134. doi: 10.1186/1756-0500-2-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhide MR, Travnicek M, Levkutova M, Curlik J, Revajova V, Levkut M. Sensitivity of Borrelia genospecies to serum complement from different animals and human: a host–pathogen relationship. FEMS Immunol. Med. Microbiol. 2005;43:165–172. doi: 10.1016/j.femsim.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Brade V, Kleber I, Acker G. Differences of two Borrelia burgdorferistrains in complement activation and serum resistance. Immunobiology. 1992;185:453–465. doi: 10.1016/S0171-2985(11)80087-2. [DOI] [PubMed] [Google Scholar]
- Breitner-Ruddock S, Würzner R, Schulze J, Brade V. Heterogeneity in the complement-dependent bacteriolysis within the species of Borrelia burgdorferi . Med. Microbiol. Immunol. 1997;185:253–260. doi: 10.1007/s004300050038. [DOI] [PubMed] [Google Scholar]
- Brissette CA, Cooley AE, Burns LH, Riley SP, Verma A, Woodman ME, Bykowski T, Stevenson B. Lyme borreliosis spirochete Erp proteins, their known host ligands, and potential roles in mammalian infection. Int. J. Med. Microbiol. 2008;298(Suppl. 1):257–267. doi: 10.1016/j.ijmm.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks CS, Hefty PS, Jolliff SE, Akins DR. Global analysis of Borrelia burgdorferi genes regulated by mammalian host-specific signals. Infect. Immun. 2003;71:3371–3383. doi: 10.1128/IAI.71.6.3371-3383.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks CS, Vuppala SR, Jett AM, Alitalo A, Meri S, Akins DR. Complement regulator-acquiring surface protein 1 imparts resistance to human serum in Borrelia burgdorferi . J. Immunol. 2005;175:3299–3308. doi: 10.4049/jimmunol.175.5.3299. [DOI] [PubMed] [Google Scholar]
- Burns LH, Adams CA, Riley SP, Jutras BL, Bowman A, Chenail AM, Cooley AE, Haselhorst LA, Moore AM, Babb K, Fried MG, Stevenson B. BpaB, a novel protein encoded by the Lyme disease spirochete’s cp32 prophages, binds to erp operator 2 DNA. Nucleic Acids Res. 2010;38:5443–5455. doi: 10.1093/nar/gkq284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bykowski T, Woodman ME, Cooley AE, Brissette CA, Brade V, Wallich R, Kraiczy P, Stevenson B. Coordinated expression of Borrelia burgdorferi complement regulator-acquiring surface proteins during the Lyme disease spirochete’s mammal-tick infection cycle. Infect. Immun. 2007;75:4227–4236. doi: 10.1128/IAI.00604-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bykowski T, Woodman ME, Cooley AE, Brissette CA, Wallich R, Brade V, Kraiczy P, Stevenson B. Borrelia burgdorfericomplement regulator-acquiring surface proteins (BbCRASPs): expression patterns during the mammal-tick infection cycle. Int. J. Med. Microbiol. 2008;298(Suppl 1):249–256. doi: 10.1016/j.ijmm.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caimano MJ, Eggers CH, Hazlett KR, Radolf JD. RpoS is not central to the general stress response in Borrelia burgdorferi but does control expression of one or more essential virulence determinants. Infect. Immun. 2004;72:6433–6445. doi: 10.1128/IAI.72.11.6433-6445.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S, Murphy M, DeLange M, Sampson L, van Vugt R, Huang WM. Telomeres of the linear chromosomes of Lyme disease spirochaetes: nucleotide sequence and possible exchange with linear plasmid telomeres. Mol. Microbiol. 1997a;26:581–596. doi: 10.1046/j.1365-2958.1997.6051963.x. [DOI] [PubMed] [Google Scholar]
- Casjens S, Palmer N, van Vugt R, Huang WM, Stevenson B, Rosa P, Lathigra R, Sutton G, Peterson J, Dodson RJ, Haft D, Hickey E, Gwinn M, White O, Fraser CM. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi . Mol. Microbiol. 2000;35:490–516. doi: 10.1046/j.1365-2958.2000.01698.x. [DOI] [PubMed] [Google Scholar]
- Casjens S, van Vugt R, Tilly K, Rosa PA, Stevenson B. Homology throughout the multiple 32-kilobase circular plasmids present in Lyme disease spirochetes. J. Bacteriol. 1997b;179:217–227. doi: 10.1128/jb.179.1.217-227.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens SR, Mongodin EF, Qiu WG, Luft BJ, Schutzer SE, Gilcrease EB, Huang WM, Vujadinovic M, Aron JK, Vargas LC, Freeman S, Radune D, Weidman JF, Dimitrov GI, Khouri HM, Sosa JE, Halpin RA, Dunn JJ, Fraser CM. Genome stability of Lyme disease spirochetes: comparative genomics of Borrelia burgdorferi plasmids. PLoS ONE. 2012;7:e33280. doi: 10.1371/journal.pone.0033280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenail AM, Jutras BL, Adams CA, Burns LH, Bowman A, Verma A, Stevenson B. Borrelia burgdorferi cp32 BpaB modulates expression of the prophage NucP nuclease and SsbP single-stranded DNA-binding protein. J. Bacteriol. 2012;194:4570–4578. doi: 10.1128/JB.00661-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman AS, Yang X, Kumar M, Zhang X, Promnares K, Shroder D, Kenedy MR, Anderson JF, Akins DR, Pal U. Borrelia burgdorferi complement regulator-acquiring surface protein 2 does not contribute to complement resistance or host infectivity. PLoS ONE. 2008;3:3010e. doi: 10.1371/journal.pone.0003010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes FS, Kraiczy P, Roversi P, Simon MM, Brade V, Jahraus O, Wallis R, Goodstadt L, Ponting CP, Skerka C, Zipfel PF, Wallich R, Lea SM. Structure–function mapping of BbCRASP-1, the key complement factor H and FHL-1 binding protein of Borrelia burgdorferi . Int. J. Med. Microbiol. 2006;296(Suppl. 40):177–184. doi: 10.1016/j.ijmm.2006.01.011. [DOI] [PubMed] [Google Scholar]
- Cordes FS, Roversi P, Kraiczy P, Simon MM, Brade V, Jahraus O, Wallis R, Skerka C, Zipfel PF, Wallich R, Lea SM. A novel fold for the factor H-binding protein BbCRASP-1 of Borrelia burgdorferi . Nat. Struct. Mol. Biol. 2005;12:276–277. doi: 10.1038/nsmb902. [DOI] [PubMed] [Google Scholar]
- Das S, Barthold SW, Giles SS, Montgomery RR, Telford SR, 3rd, Fikrig E. Temporal pattern of Borrelia burgdorferi p21 expression in ticks and the mammalian host. J. Clin. Invest. 1997;99:987–995. doi: 10.1172/JCI119264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delihas N. Intergenic regions of Borrelia plasmids contain phylogenetically conserved RNA secondary structure motifs. BMC Genomics. 2009a;10:101. doi: 10.1186/1471-2164-10-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delihas N. Stem loop sequences specific to transposable element IS605 are found linked to lipoprotein genes in Borrelia plasmids. PLoS ONE. 2009b;4:e7941. doi: 10.1371/journal.pone.0007941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich R, Hammerschmidt C, Richter D, Skerka C, Wallich R, Matuschka FR, Zipfel PF, Kraiczy P. Inadequate binding of immune regulator factor H is associated with sensitivity of Borrelia lusitaniae to human complement. Infect. Immun. 2010;78:4467–4476. doi: 10.1128/IAI.00138-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggers CH, Caimano MJ, Radolf JD. Analysis of promoter elements involved in the transcriptional initiation of RpoS-dependent Borrelia burgdorferi genes. J. Bacteriol. 2004;186:7390–7402. doi: 10.1128/JB.186.21.7390-7402.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggers CH, Caimano MJ, Radolf JD. Sigma factor selectivity in Borrelia burgdorferi: RpoS recognition of the ospE/ospF/elp promoters is dependent on the sequence of the −10 region. Mol. Microbiol. 2006;59:1859–1875. doi: 10.1111/j.1365-2958.2006.05066.x. [DOI] [PubMed] [Google Scholar]
- Eggers CH, Kimmel BJ, Bono JL, Elias AF, Rosa P, Samuels DS. Transduction by phiBB-1, a bacteriophage of Borrelia burgdorferi . J. Bacteriol. 2001;183:4771–4778. doi: 10.1128/JB.183.16.4771-4778.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggers CH, Samuels DS. Molecular evidence for a new bacteriophage of Borrelia burgdorferi . J. Bacteriol. 1999;181:7308–7313. doi: 10.1128/jb.181.23.7308-7313.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Hage N, Babb K, Carroll JA, Lindstrom N, Fischer ER, Miller JC, Gilmore RD, Jr, Mbow ML, Stevenson B. Surface exposure and protease insensitivity of Borrelia burgdorferi Erp (OspEF-related) lipoproteins. Microbiology. 2001;147:821–830. doi: 10.1099/00221287-147-4-821. [DOI] [PubMed] [Google Scholar]
- El-Hage N, Stevenson B. Simultaneous coexpression of Borrelia burgdorferiErp proteins occurs through a specific, erp locus-directed regulatory mechanism. J. Bacteriol. 2002;184:4536–4543. doi: 10.1128/JB.184.16.4536-4543.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exner MM, Wu X, Blanco DR, Miller JN, Lovett MA. Protection elicited by native outer membrane protein Oms66 (p66) against host-adapted Borrelia burgdorferi: conformational nature of bactericidal epitopes. Infect. Immun. 2000;68:2647–2654. doi: 10.1128/iai.68.5.2647-2654.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, Gwinn M, Dougherty B, Tomb JF, Fleischmann RD, Richardson D, Peterson J, Kerlavage AR, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams MD, Gocayne J, Weidman J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fujii C, Cotton MD, Horst K, Roberts K, Hatch B, Smith HO, Venter JC. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi . Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- Hammerschmidt C, Hallström T, Skerka C, Wallich R, Stevenson B, Zipfel PF, Kraiczy P. Contribution of the infection-associated complement regulator-acquiring surface protein 4 (ErpC) to complement resistance of Borrelia burgdorferi . Clin. Dev. Immunol. 2012;2012:349657. doi: 10.1155/2012/349657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann K, Corvey C, Skerka C, Kirschfink M, Karas M, Brade V, Miller JC, Stevenson B, Wallich R, Zipfel PF, Kraiczy P. Functional characterization of BbCRASP-2, a distinct outer membrane protein of Borrelia burgdorferithat binds host complement regulators factor H and FHL-1. Mol. Microbiol. 2006;61:1220–1236. doi: 10.1111/j.1365-2958.2006.05318.x. [DOI] [PubMed] [Google Scholar]
- Haupt K, Kraiczy P, Wallich R, Brade V, Skerka C, Zipfel PF. Binding of human factor H-related protein 1 to serum-resistant Borrelia burgdorferiis mediated by borrelial complement regulator-acquiring surface proteins. J. Infect. Dis. 2007;196:124–133. doi: 10.1086/518509. [DOI] [PubMed] [Google Scholar]
- Hefty PS, Brooks CS, Jett AM, White GL, Wikel SK, Kennedy RC, Akins DR. OspE-related, OspF-related, and Elp lipoproteins are immunogenic in baboons experimentally infected with Borrelia burgdorferiand in human lyme disease patients. J. Clin. Microbiol. 2002;40:4256–4265. doi: 10.1128/JCM.40.11.4256-4265.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hefty PS, Jolliff SE, Caimano MJ, Wikel SK, Radolf JD, Akins DR. Regulation of OspE-related, OspF-related, and Elp lipoproteins of Borrelia burgdorferi strain 297 by mammalian host-specific signals. Infect. Immun. 2001;69:3618–3627. doi: 10.1128/IAI.69.6.3618-3627.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinen S, Hartmann A, Lauer N, Wiehl U, Dahse HM, Schirmer S, Gropp K, Enghardt T, Wallich R, Halbich S, Mihlan M, Schlotzer-Schrehardt U, Zipfel PF, Skerka C. Factor H-related protein 1 (CFHR-1) inhibits complement C5 convertase activity and terminal complex formation. Blood. 2009;114:2439–2447. doi: 10.1182/blood-2009-02-205641. [DOI] [PubMed] [Google Scholar]
- Hellwage J, Meri T, Heikkilä T, Alitalo A, Panelius J, Lahdenne P, Seppälä IJT, Meri S. The complement regulator factor H binds to the surface protein OspE of Borrelia burgdorferi . J. Biol. Chem. 2001;276:8427–8435. doi: 10.1074/jbc.M007994200. [DOI] [PubMed] [Google Scholar]
- Herzberger P, Siegel C, Skerka C, Fingerle V, Schulte-Spechtel U, van Dam A, Wilske B, Brade V, Zipfel PF, Wallich R, Kraiczy P. Human pathogenic Borrelia spielmanii sp. nov. resists complement-mediated killing by direct binding of immune regulators factor H and factor H-like protein 1. Infect. Immun. 2007;75:4817–4825. doi: 10.1128/IAI.00532-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzberger P, Siegel C, Skerka C, Fingerle V, Schulte-Spechtel U, Wilske B, Brade V, Zipfel PF, Wallich R, Kraiczy P. Identification and characterization of the factor H and FHL-1 binding complement regulator-acquiring surface protein 1 of the Lyme disease spirochete Borrelia spielmanii sp. nov. Int. J. Med. Microbiol. 2009;299:141–154. doi: 10.1016/j.ijmm.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Hovis KM, Tran E, Sundy CM, Buckles E, McDowell JV, Marconi RT. Selective binding of Borrelia burgdorferiOspE paralogs to factor H and serum proteins from diverse animals: possible expansion of the role of OspE in Lyme disease pathogenesis. Infect. Immun. 2006;74:1967–1972. doi: 10.1128/IAI.74.3.1967-1972.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer R, Liveris D, Adams A, Nowakowski J, McKenna D, Bittker S, Cooper D, Wormser GP, Schwartz I. Characterization of Borrelia burgdorferiisolated from erythema migrans lesions: interrelationship of three molecular typing methods. J. Clin. Microbiol. 2001;39:2954–2957. doi: 10.1128/JCM.39.8.2954-2957.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Józsi M, Zipfel PF. Factor H family proteins and human diseases. Trends Immunol. 2008;29:380–387. doi: 10.1016/j.it.2008.04.008. [DOI] [PubMed] [Google Scholar]
- Jutras BL, Bowman A, Brissette CA, Adams CA, Verma A, Chenail AM, Stevenson B. EbfC (YbaB) is a new type of bacterial nucleoid-associated protein, and a global regulator of gene expression in the Lyme disease spirochete. J. Bacteriol. 2012a;194:3395–3406. doi: 10.1128/JB.00252-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jutras BL, Verma A, Adams CA, Brissette CA, Burns LH, Whetstine CR, Bowman A, Chenail AM, Zückert WR, Stevenson B. BpaB and EbfC DNA-binding proteins regulate production of the Lyme disease spirochete’s infection-associated Erp surface proteins. J. Bacteriol. 2012b;194:778–786. doi: 10.1128/JB.06394-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenedy MR, Akins DR. The OspE-related proteins inhibit complement deposition and enhance serum resistance of Borrelia burgdorferi the Lyme disease spirochete. Infect. Immun. 2011;79:1451–1457. doi: 10.1128/IAI.01274-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenedy MR, Vuppala SR, Siegel C, Kraiczy P, Akins DR. CspA-mediated binding of human factor H inhibits complement deposition and confers serum resistance in Borrelia burgdorferi Infect. Immun. 2009;77:2773–2782. doi: 10.1128/IAI.00318-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisova-Vargova L, Mucha R, Cernanka D, Bhide M. Host-dependent differential expression of factor H binding proteins, their affinity to factor H and complement evasion by Lyme and relapsing fever borreliae. Vet. Microbiol. 2011;148:341–347. doi: 10.1016/j.vetmic.2010.09.026. [DOI] [PubMed] [Google Scholar]
- Kraiczy P, Hanssen-Hubner C, Kitiratschky V, Brenner C, Besier S, Brade V, Simon MM, Skerka C, Roversi P, Lea SM, Stevenson B, Wallich R, Zipfel PF. Mutational analyses of the BbCRASP-1 protein of Borrelia burgdorferi identify residues relevant for the architecture and binding of host complement regulators FHL-1 and factor H. Int. J. Med. Microbiol. 2009;299:255–268. doi: 10.1016/j.ijmm.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Kraiczy P, Hartmann K, Hellwage J, Skerka C, Kirschfink M, Brade V, Zipfel PF, Wallich R, Stevenson B. Immunological characterization of the complement regulator factor H-binding CRASP and Erp proteins of Borrelia burgdorferi . Int. J. Med. Microbiol. 2004a;293:152–157. doi: 10.1016/s1433-1128(04)80029-9. [DOI] [PubMed] [Google Scholar]
- Kraiczy P, Hellwage J, Skerka C, Becker H, Kirschfink M, Simon MM, Brade V, Zipfel PF, Wallich R. Complement resistance of Borrelia burgdorferi correlates with the expression of BbCRASP-1, a novel linear plasmid-encoded surface protein that interacts with human factor H and FHL-1 and is unrelated to Erp proteins. J. Biol. Chem. 2004b;279:2421–2429. doi: 10.1074/jbc.M308343200. [DOI] [PubMed] [Google Scholar]
- Kraiczy P, Hellwage J, Skerka C, Kirschfink M, Brade V, Zipfel PF, Wallich R. Immune evasion of Borrelia burgdorferi: mapping of a complement-inhibitor factor H-binding site of BbCRASP-3, a novel member of the Erp protein family. Eur. J. Immunol. 2003;33:697–707. doi: 10.1002/eji.200323571. [DOI] [PubMed] [Google Scholar]
- Kraiczy P, Hunfeld KP, Breitner-Ruddock S, Würzner R, Acker G, Brade V. Comparison of two laboratory methods for the determination of serum resistance in Borrelia burgdorferi isolates. Immunobiology. 2000;201:406–419. doi: 10.1016/S0171-2985(00)80094-7. [DOI] [PubMed] [Google Scholar]
- Kraiczy P, Rossmann E, Brade V, Simon MM, Skerka C, Zipfel PF, Wallich R. Binding of human complement regulators FHL-1 and factor H to CRASP-1 orthologs of Borrelia burgdorferi Wien. Klin. Wochenschr. 2006;118:669–676. doi: 10.1007/s00508-006-0691-1. [DOI] [PubMed] [Google Scholar]
- Kraiczy P, Seling A, Brissette CA, Rossmann E, Hunfeld KP, Bykowski T, Burns LH, Troese MJ, Cooley AE, Miller JC, Brade V, Wallich R, Casjens S, Stevenson B. Borrelia burgdorfericomplement regulator-acquiring surface protein 2 (CspZ) as a serological marker of human Lyme disease. Clin. Vaccine Immunol. 2008;15:484–491. doi: 10.1128/CVI.00415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraiczy P, Skerka C, Brade V, Zipfel PF. Further characterization of complement regulator-acquiring surface proteins of Borrelia burgdorferi . Infect. Immun. 2001a;69:7800–7809. doi: 10.1128/IAI.69.12.7800-7809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraiczy P, Skerka C, Kirschfink M, Brade V, Zipfel PF. Immune evasion of Borrelia burgdorferiby acquisition of human complement regulators FHL-1/reconectin and factor-H. Eur. J. Immunol. 2001b;31:1674–1684. doi: 10.1002/1521-4141(200106)31:6<1674::aid-immu1674>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Kraiczy P, Skerka C, Kirschfink M, Zipfel PF, Brade V. Mechanism of complement resistance of pathogenic Borrelia burgdorferiisolates. Intern. Immunopharmacol. 2001c;1:393–401. doi: 10.1016/s1567-5769(00)00041-2. [DOI] [PubMed] [Google Scholar]
- Lam TT, Nguyen TP, Montgomery RR, Kantor FS, Fikrig E, Flavell RA. Outer surface proteins E and F of Borrelia burgdorferithe agent of Lyme disease. Infect. Immun. 1994;62:290–298. doi: 10.1128/iai.62.1.290-298.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambris JD, Ricklin D, Geisbrecht BV. Complement evasion by human pathogens. Nat. Rev. Microbiol. 2008;6:132–142. doi: 10.1038/nrmicro1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederer S, Brenner C, Stehle T, Gern L, Wallich R, Simon MM. Quantitative analysis of Borrelia burgdorferigene expression in naturally (tick) infected mouse strains. Med. Microbiol. Immunol. 2005;194:81–90. doi: 10.1007/s00430-004-0218-1. [DOI] [PubMed] [Google Scholar]
- Margos G, Vollmer SA, Cornet M, Garnier M, Fingerle V, Wilske B, Bormane A, Vitorino L, Collares-Pereira M, Drancourt M, Kurtenbach K. A new Borrelia species defined by multilocus sequence analysis of housekeeping genes. Appl. Environ. Microbiol. 2009;75:5410–5416. doi: 10.1128/AEM.00116-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell JV, Harlin ME, Rogers EA, Marconi RT. Putative coiled-coil structural elements of the BBA68 protein of Lyme disease spirochetes are required for formation of its factor H binding site. J. Bacteriol. 2005;187:1317–1323. doi: 10.1128/JB.187.4.1317-1323.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell JV, Hovis KM, Zhang H, Tran E, Lankford J, Marconi RT. Evidence that the BBA68 protein (BbCRASP-1) of the Lyme disease spirochetes does not contribute to factor H-mediated immune evasion in humans and other animals. Infect. Immun. 2006;74:3030–3034. doi: 10.1128/IAI.74.5.3030-3034.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell JV, Sung SY, Labandeira-Rey M, Skare JT, Marconi RT. Analysis of mechanisms associated with loss of infectivity of clonal populations of Borrelia burgdorferi B31MI. Infect. Immun. 2001a;69:3670–3677. doi: 10.1128/IAI.69.6.3670-3677.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell JV, Sung SY, Price G, Marconi RT. Demonstration of the genetic stability and temporal expression of select members of the lyme disease spirochete OspF protein family during infection in mice. Infect. Immun. 2001b;69:4831–4838. doi: 10.1128/IAI.69.8.4831-4838.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell JV, Wolfgang J, Senty L, Sundy CM, Noto MJ, Marconi RT. Demonstration of the involvement of outer surface protein E coiled coil structural domains and higher order structural elements in the binding of infection-induced antibody and the complement-regulatory protein, factor H. J. Immunol. 2004;173:7471–7480. doi: 10.4049/jimmunol.173.12.7471. [DOI] [PubMed] [Google Scholar]
- McDowell JV, Wolfgang J, Tran E, Metts MS, Hamilton D, Marconi RT. Comprehensive analysis of the factor H binding capabilities of Borrelia species associated with Lyme disease: delineation of two distinct classes of factor H binding proteins. Infect. Immun. 2003;71:3597–3602. doi: 10.1128/IAI.71.6.3597-3602.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRae JL, Cowan PJ, Power DA, Mitchelhill KI, Kemp BE, Morgan BP, Murphy BF. Human factor H-related protein 5 (FHR-5). A new complement-associated protein. J.Biol. Chem. 2001;276:6747–6754. doi: 10.1074/jbc.M007495200. [DOI] [PubMed] [Google Scholar]
- McRae JL, Duthy TG, Griggs KM, Ormsby RJ, Cowan PJ, Cromer BA, McKinstry WJ, Parker MW, Murphy BF, Gordon DL. Human factor H-related protein 5 has cofactor activity, inhibits C3 convertase activity, binds heparin and C-reactive protein, and associates with lipoprotein. J. Immunol. 2005;174:6250–6256. doi: 10.4049/jimmunol.174.10.6250. [DOI] [PubMed] [Google Scholar]
- Metts MS, McDowell JV, Theisen M, Hansen PR, Marconi RT. Analysis of the OspE determinants involved in binding of factor H and OspE-targeting antibodies elicited during Borrelia burgdorferi infection in mice. Infect. Immun. 2003;71:3587–3596. doi: 10.1128/IAI.71.6.3587-3596.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, El-Hage N, Babb K, Stevenson B. Borrelia burgdorferi B31 Erp proteins that are dominant immunoblot antigens of animals infected with isolate B31 are recognized by only a subset of human Lyme disease patient sera. J. Clin. Microbiol. 2000;38:1569–1574. doi: 10.1128/jcm.38.4.1569-1574.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Narayan K, Stevenson B, Pachner AR. Expression of Borrelia burgdorferi erp genes during infection of non-human primates. Microb. Pathog. 2005;39:27–33. doi: 10.1016/j.micpath.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Miller JC, Stevenson B. Borrelia burgdorferi erp genes are expressed at different levels within tissues of chronically infected mammalian hosts. Int. J. Med. Microbiol. 2006;296(Suppl. 401):185–194. doi: 10.1016/j.ijmm.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Miller JC, von Lackum K, Babb K, McAlister JD, Stevenson B. Temporal analysis of Borrelia burgdorferiErp protein expression throughout the mammaltick infectious cycle. Infect. Immun. 2003;71:6943–6952. doi: 10.1128/IAI.71.12.6943-6952.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TP, Lam TT, Barthold SW, Telford SR, 3rd, Flavell RA, Fikrig E. Partial destruction of Borrelia burgdorferi within ticks that engorged on OspE-or OspF-immunized mice. Infect. Immun. 1994;62:2079–2084. doi: 10.1128/iai.62.5.2079-2084.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patarakul K, Cole MF, Hughes CAN. Complement resistance in Borrelia burgdorferistrain 297: outer membrane proteins prevent MAC formation at lysis susceptible sites. Microb. Pathog. 1999;27:25–41. doi: 10.1006/mpat.1999.0280. [DOI] [PubMed] [Google Scholar]
- Richter D, Schlee DB, Allgöwer R, Matuschka F-R. Relationships of a novel Lyme disease spirochete, Borrelia spielmani sp. nov. with its hosts in Central Europe. Appl. Environ. Microbiol. 2004;70:6414–6419. doi: 10.1128/AEM.70.11.6414-6419.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley SP, Bykowski T, Cooley AE, Burns LH, Babb K, Brissette CA, Bowman A, Rotondi M, Miller MC, DeMoll E, Lim K, Fried MG, Stevenson B. Borrelia burgdorferiEbfC defines a newly-identified, widespread family of bacterial DNA-binding proteins. Nucleic Acids Res. 2009;37:1973–1983. doi: 10.1093/nar/gkp027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers EA, Abdunnur SV, McDowell JV, Marconi RT. Comparative analysis of the properties and ligand binding characteristics of CspZ, a factor H binding protein, derived from Borrelia burgdorferiisolates of human origin. Infect. Immun. 2009a;77:4396–4405. doi: 10.1128/IAI.00393-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers EA, Marconi RT. Delineation of species-specific binding properties of the CspZ protein (BBH06) of Lyme disease spirochetes: evidence for new contributions to the pathogenesis of Borrelia spp. Infect. Immun. 2007;75:5272–5281. doi: 10.1128/IAI.00850-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers EA, Terekhova D, Zhang HM, Hovis KM, Schwartz I, Marconi RT. Rrp1, a cyclic-di-GMP-producing response regulator, is an important regulator of Borrelia burgdorferi core cellular functions. Mol. Microbiol. 2009b;71:1551–1573. doi: 10.1111/j.1365-2958.2009.06621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann E, Kitiratschky V, Hofmann H, Kraiczy P, Simon MM, Wallich R. Borrelia burgdorfericomplement regulator-acquiring surface protein 1 of the Lyme disease spirochetes is expressed in humans and induces antibody responses restricted to nondenatured structural determinants. Infect. Immun. 2006;74:7024–7028. doi: 10.1128/IAI.01028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann E, Kraiczy P, Herzberger P, Skerka C, Kirschfink M, Simon MM, Zipfel PF, Wallich R. Dual binding specificity of a Borrelia hermsii associated complement regulator-acquiring surface protein for factor H and plasminogen discloses a putative virulence factor of relapsing fever spirochetes. J. Immunol. 2007;178:7292–7301. doi: 10.4049/jimmunol.178.11.7292. [DOI] [PubMed] [Google Scholar]
- Schott M, Grosskinsky S, Brenner C, Kraiczy P, Wallich R. Molecular characterization of the interaction of Borrelia parkeri and Borrelia turicatae with human complement regulators. Infect. Immun. 2010;78:2199–2208. doi: 10.1128/IAI.00089-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutzer SE, Fraser-Liggett CM, Casjens SR, Qiu WG, Dunn JJ, Mongodin EF, Luft BJ. Whole-genome sequences of thirteen isolates of Borrelia burgdorferi . J. Bacteriol. 2011;193:1018–1020. doi: 10.1128/JB.01158-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutzer SE, Fraser-Liggett CM, Qiu WG, Kraiczy P, Mongodin EF, Dunn JJ, Luft BJ, Casjens SR. Whole-genome sequences of Borrelia bissettii, Borrelia valaisiana and Borrelia spielmanii . J. Bacteriol. 2012;194:545–546. doi: 10.1128/JB.06263-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seling A, Siegel C, Fingerle V, Jutras BL, Brissette CA, Skerka C, Wallich R, Zipfel PF, Stevenson B, Kraiczy P. Functional characterization of Borrelia spielmanii outer surface proteins that interact with distinct members of the human factor H protein family and with plasminogen. Infect. Immun. 2010;78:39–48. doi: 10.1128/IAI.00691-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel C, Hallström T, Skerka C, Eberhardt H, Uzonyi B, Beckhaus T, Karas M, Wallich R, Stevenson B, Zipfel PF, Kraiczy P. Complement factor H-related proteins CFHR2 and CFHR5 represent novel ligands for the infection-associated CRASP proteins of Borrelia burgdorferi . PLoS ONE. 2010;5:e13519. doi: 10.1371/journal.pone.0013519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel C, Herzberger P, Skerka C, Brade V, Fingerle V, Schulte-Spechtel U, Wilske B, Zipfel PF, Wallich R, Kraiczy P. Binding of complement regulatory protein factor H enhances serum resistance of Borrelia spielmanii sp. nov. Int. J. Med. Microbiol. 2008a;298(S1):292–294. doi: 10.1016/j.ijmm.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Siegel C, Schreiber J, Haupt K, Skerka C, Brade V, Simon MM, Stevenson B, Wallich R, Zipfel PF, Kraiczy P. Deciphering the ligand-binding sites in the Borrelia burgdorferi complement regulator-acquiring surface protein 2 required for interactions with the human immune regulators factor H and factor H-like protein 1. J Biol. Chem. 2008b;283:34855–34863. doi: 10.1074/jbc.M805844200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanek G, Reiter M. The expanding Lyme Borrelia complex – clinical significance of genomic species? Clin. Microbiol. Infect. 2011;17:487–493. doi: 10.1111/j.1469-0691.2011.03492.x. [DOI] [PubMed] [Google Scholar]
- Steere AC. Lyme disease. N. Engl. J. Med. 1989:586–596. doi: 10.1056/NEJM198908313210906. [DOI] [PubMed] [Google Scholar]
- Stevenson B, Babb K. LuxS-mediated quorum sensing in Borrelia burgdorferi the lyme disease spirochete. Infect. Immun. 2002;70:4099–4105. doi: 10.1128/IAI.70.8.4099-4105.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson B, Bono JL, Schwan TG, Rosa P. Borrelia burgdorferiErp proteins are immunogenic in mammals infected by tick bite, and their synthesis is inducible in cultured bacteria. Infect. Immun. 1998a;66:2648–2654. doi: 10.1128/iai.66.6.2648-2654.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson B, Bykowski T, Cooley AE, Babb K, Miller JC, Woodman ME, von Lakum K, Riley SP. The Lyme disease spirochetes Erp protein family: structure, function, and regulation of gene expression. In: Cabello FC, Godfrey HP, Hulinska D, editors. Molecular Biology of Spirochetes. Amsterdam: IOS Press; 2006. pp. 352–372. [Google Scholar]
- Stevenson B, Casjens S, Rosa P. Evidence of past recombination events among the genes encoding the Erp antigens of Borrelia burgdorferi . Microbiology. 1998b;144((Pt 7)):1869–1879. doi: 10.1099/00221287-144-7-1869. [DOI] [PubMed] [Google Scholar]
- Stevenson B, El-Hage N, Hines MA, Miller JC, Babb K. Differential binding of host complement inhibitor factor H by Borrelia burgdorferi Erp surface proteins: a possible mechanism underlying the expansive host range of Lyme disease spirochetes. Infect. Immun. 2002;70:491–497. doi: 10.1128/IAI.70.2.491-497.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson B, Miller JC. Intra- and interbacterial genetic exchange of Lyme disease spirochete erp genes generates sequence identity amidst diversity. J. Mol. Evol. 2003;57:309–324. doi: 10.1007/s00239-003-2482-x. [DOI] [PubMed] [Google Scholar]
- Stevenson B, Porcella SF, Oie KL, Fitzpatrick CA, Raffel SJ, Lubke L, Schrumpf ME, Schwan TG. The relapsing fever spirochete Borrelia hermsii contains multiple, antigen-encoding circular plasmids that are homologous to the cp32 plasmids of Lyme disease spirochetes. Infect. Immun. 2000a;68:3900–3908. doi: 10.1128/iai.68.7.3900-3908.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson B, Schwan TG, Rosa PA. Temperature-related differential expression of antigens in the Lyme disease spirochete, Borrelia burgdorferi . Infect. Immun. 1995;63:4535–4539. doi: 10.1128/iai.63.11.4535-4539.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson B, Tilly K, Rosa PA. A family of genes located on four separate 32-kilobase circular plasmids in Borrelia burgdorferi B31. J. Bacteriol. 1996;178:3508–3516. doi: 10.1128/jb.178.12.3508-3516.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson B, Zückert WR, Akins DR. Repetition, conservation, and variation: the multiple cp32 plasmids of Borrelia species. J. Mol. Microbiol. Biotechnol. 2000b;2:411–422. [PubMed] [Google Scholar]
- Stevenson B, Zückert WR, Akins DR. Repetition, conservation and variation: the multiple cp32 plasmids of Borrelia species. In: Saier MH, Garcia-Lara J, editors. The Spirochetes: Molecular and Cellular Biology. Oxford: Horizon Press; 2001. pp. 87–100. [Google Scholar]
- Tokarz R, Anderton JM, Katona LI, Benach JL. Combined effects of blood and temperature shift on Borrelia burgdorferigene expression as determined by whole genome DNA array. Infect. Immun. 2004;72:5419–5432. doi: 10.1128/IAI.72.9.5419-5432.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Burgel ND, Kraiczy P, Schuijt TJ, Zipfel PF, van Dam AP. Identification and functional characterisation of complement regulator acquiring surface protein-1 of serum resistant Borrelia garinii OspA serotype 4. BMC Microbiol. 2010;10:43. doi: 10.1186/1471-2180-10-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dam AP, Oei A, Jaspars R, Fijen C, Wilske B, Spanjaard L, Dankert J. Complement-mediated serum sensitivity among spirochetes that cause Lyme disease. Infect. Immun. 1997;65:1228–1236. doi: 10.1128/iai.65.4.1228-1236.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Lackum K, Babb K, Riley SP, Wattier RL, Bykowski T, Stevenson B. Functionality of Borrelia burgdorferi LuxS: the Lyme disease spirochete produces and responds to the pheromone autoinducer-2 and lacks a complete activated-methyl cycle. Int. J. Med. Microbiol. 2006;296(Suppl. 40):92–102. doi: 10.1016/j.ijmm.2005.12.011. [DOI] [PubMed] [Google Scholar]
- von Lackum K, Miller JC, Bykowski T, Riley SP, Woodman ME, Brade V, Kraiczy P, Stevenson B, Wallich R. Borrelia burgdorferiregulates expression of complement regulator-acquiring surface protein 1 during the mammal-tick infection cycle. Infect. Immun. 2005;73:7398–7405. doi: 10.1128/IAI.73.11.7398-7405.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallich R, Brenner C, Kramer MD, Simon MM. Molecular cloning and immunological characterization of a novel linear-plasmid-encoded gene, pG, of Borrelia burgdorferi expressed only in vivo. Infect. Immun. 1995;63:3327–3335. doi: 10.1128/iai.63.9.3327-3335.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallich R, Jahraus O, Stehle T, Tran TT, Brenner C, Hofmann H, Gern L, Simon MM. Artificial-infection protocols allow immunodetection of novel Borrelia burgdorferi antigens suitable as vaccine candidates against Lyme disease. Eur. J. Immunol. 2003;33:708–719. doi: 10.1002/eji.200323620. [DOI] [PubMed] [Google Scholar]
- Wallich R, Pattathu J, Kitiratschky V, Brenner C, Zipfel PF, Brade V, Simon MM, Kraiczy P. Identification and functional characterization of complement regulator-acquiring surface protein 1 of the Lyme disease spirochetes Borrelia afzelii and Borrelia garinii . Infect. Immun. 2005;73:2351–2359. doi: 10.1128/IAI.73.4.2351-2359.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, van Dam AP, Dankert J. Phenotypic and genetic characterization of a novel Borrelia burgdorferi sensu lato isolate from a patient with lyme borreliosis. J. Clin. Microbiol. 1999;37:3025–3028. doi: 10.1128/jcm.37.9.3025-3028.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wormser GP, Brisson D, Liveris D, Hanincova K, Sandigursky S, Nowakowski J, Nadelman RB, Ludin S, Schwartz I. Borrelia burgdorferi genotype predicts the capacity for hematogenous dissemination during early Lyme disease. JInfect. Dis. 2008;198:1358–1364. doi: 10.1086/592279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wywial E, Haven J, Casjens SR, Hernandez YA, Singh S, Mongodin EF, Fraser-Liggett CM, Luft BJ, Schutzer SE, Qiu WG. Fast, adaptive evolution at a bacterial host-resistance locus: the PFam54 gene array in Borrelia burgdorferi . Gene. 2009;445:26–37. doi: 10.1016/j.gene.2009.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XF, Alani SM, Norgard MV. The response regulator Rrp2 is essential for the expression of major membrane lipoproteins in Borrelia burgdorferi . Proc. Natl. Acad. Sci. U. S. A. 2003;100:11001–11006. doi: 10.1073/pnas.1834315100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Marconi RT. Demonstration of cotranscription and 1-methyl-3-nitroso-nitroguanidine induction of a 30-gene operon of Borrelia burgdorferi: evidence that the 32-kilobase circular plasmids are prophages. J. Bacteriol. 2005;187:7985–7995. doi: 10.1128/JB.187.23.7985-7995.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel PF, Jokiranta TS, Hellwage J, Koistinen V, Meri S. The factor H protein family. Immunopharmacology. 1999;42:53–60. doi: 10.1016/s0162-3109(99)00015-6. [DOI] [PubMed] [Google Scholar]
- Zipfel PF, Skerka C. Complement regulators and inhibitory proteins. Nat. Rev. Immunol. 2009;9:729–740. doi: 10.1038/nri2620. [DOI] [PubMed] [Google Scholar]
- Zückert WR, Kerentseva TA, Lawson CL, Barbour AG. Structural conservation of neurotropism-associated VspA within the variable Borrelia Vsp-OspC lipoprotein family. J. Biol. Chem. 2001;276:457–463. doi: 10.1074/jbc.M008449200. [DOI] [PubMed] [Google Scholar]