Abstract
Myotonic dystrophy 2 (DM2) is an autosomal dominant, multisystem disease, which primarily affects skeletal muscle. DM2 is caused by CCTGn expansion in the intron 1 of the ZNF9 gene. Expression of the mutant CCUGn RNA changes RNA processing in patients with DM2; however, the role of ZNF9 protein in DM2 pathology has been not elucidated. ZNF9 has been shown to regulate cap-dependent and cap-independent translation. We have examined a possible role of ZNF9 in the regulation of translation in DM2 patients. We found that ZNF9 interacts with the 5′ UTRs of terminal oligopyrimidine (TOP) tract mRNAs encoding human ribosomal protein, RPS17, poly(A)-binding protein 1 (PABP1), and the elongation factors, eEF1A and eEF2. The binding activity of ZNF9 toward these TOP-containing 5′ UTRs is reduced in DM2 muscle. Consistent with the reduction of this activity, the levels of RPS17, PABP, eEF1A, and eEF2 proteins are also diminished in DM2 muscle. The reduction of ZNF9 RNA-binding activity in DM2 correlates with a decrease of ZNF9 protein levels in cytoplasm of DM2 muscle cells. We found that the reduction of ZNF9 is caused by expression of the mutant CCUG repeats. This decrease of proteins of translational apparatus in DM2 correlates with a reduction of a rate of protein synthesis in myoblasts from DM2 patients. We found that the ectopic expression of ZNF9 in DM2 myoblasts corrects rate of protein synthesis, suggesting that the alterations in CCUG-ZNF9-TOP mRNAs pathway are responsible for the reduction of the rate of protein translation in DM2 muscle cells.
Introduction
Myotonic dystrophy 2 (DM2) is a multisystemic, skeletal muscle disease caused by an expansion of intronic CCTG repeats in the gene encoding Zinc finger factor 9 [ZNF9, also known as cellular nucleic acid binding protein (CNBP)] (Liquori et al., 2001; Day et al., 2003). DM2 mutation is similar to the untranslated CTG expansion causing Myotonic dystrophy 1 (DM1). Similarities of these mutations imply that both diseases might be caused by a disruption of RNA metabolism in patients' tissues through accumulation of untranslated CUGn (for DM1) and CCUGn (for DM2) RNAs (Timchenko, 1999; Ranum and Day, 2004). However, there is an important difference in the locations of DM1 and DM2 mutations: CTG expansion is located within the 3′ UTR of DMPK gene (Aslanidis et al., 1992); whereas the CCTG repeat expansion is located in intron of ZNF9 gene (Liquori et al., 2001), which codes for a protein with DNA- and RNA-binding activities (Rajavashisth et al., 1989; Warden et al., 1994; Yasuda et al., 1995). Given the RNA binding activity of ZNF9, this protein might contribute to the disruption of RNA metabolism in DM2 cells.
Little is known about ZNF9 biological functions in normal and in DM2 cells. ZNF9 structure is highly conserved suggesting that this protein plays a basic biological role (Warden et al., 1994; Yasuda et al., 1995; Flink et al., 1998). ZNF9 contains seven conserve Zinc-finger (CCHC) repeats which are common in transcription factors, ribosomal proteins, and proteins involved in the processing of mRNAs coding for ribosomal proteins. It has been shown that ZNF9 binds to the regulatory elements of several genes (Rajavashisth et al., 1989; Edwards et al., 1992; Flink and Morkin, 1995; Michelotti et al., 1995). Several reports implicated ZNF9 in the regulation of cap-dependent and cap-independent translation (Pellizzoni et al., 1997; Gerbasi and Link, 2007). In Xenopus laevis, Znf9 binds to the 5′ UTRs of mRNAs containing the highly conserved oligopyrimidine element, terminal oligopyrimidine tract (TOP) (Pellizzoni et al., 1997). TOP-containing mRNAs program translation of proteins associated with translational apparatus [ribosomal proteins, elongation factors eEF1A and eEF2, and poly(A)-binding protein] (Meyuhas, 2000).
Two papers have examined the levels of ZNF9 mRNA and protein in total protein extracts from DM2 cells and have not identified significant changes (Botta et al., 2006; Margolis et al., 2006). However, the analyses of the mouse model with deleted ZNF9 showed that these mice have developed DM2-like symptoms: myotonia, muscular dystrophy, and cardiac defects (Chen et al., 2007). These data prompted us to investigate biological functions of ZNF9, and examine whether these functions are altered in muscle cells from DM2 patients. We found that ZNF9 binds to the 5′ UTR of mRNA-encoding ribosomal protein, RPS17. Our data show that RNA-binding activity and protein levels of ZNF9 are reduced in cytoplasm of DM2 myoblasts and in cytoplasm of muscle biopsies from DM2 patients. Consistent with the reduction of ZNF9, the protein levels of RPS17 are also diminished in DM2 muscle cells. In addition to RPS17, ZNF9 interacts with the 5′ UTRs of three other TOP-containing mRNAs: PABP1, eEF1A, and eEF2. These proteins are reduced in DM2 muscle precursors and in muscle biopsies from DM2 patients. In agreement with the decrease of the proteins of translational apparatus, the rate of global protein synthesis is diminished in DM2 myoblasts.
Materials and Methods
Patients, muscle biopsies, and primary human myoblasts.
Primary myoblast cell lines were established from muscle biopsies taken from the vastus lateralis or from the biceps brachii of control patients (free of neurological or musculoskeletal diseases) and patients with the typical late-onset proximal myotonic myopathy (DM2) (Schara and Schoser, 2006). Normal specimens did not show any myopathological and biochemical alterations. The group of DM2 patients, biopsies of which were used for generation of primary myoblast lines, included two females and two males, aged between 39 and 66 years. Examination of ZNF9 and its targets was also performed in muscle biopsies from three DM2 patients (one male and two females, aged between 60 and 70 years) and age-matching biopsies from control (unaffected) patients and two patients (female and male, aged 79 and 70 years) affected with myopathy and neurogenic atrophy, respectively. Genetic analysis of DNA from blood of DM2 patients showed that the CCTG repeat length ranged between 3000 and >15,000 CCTG repeats. One of the DM2 patients was homozygous for DM2 mutation; this patient was described in detail previously (Schoser et al., 2004). Other patients were heterozygous for the DM2 mutation. Primary muscle cultures were established and propagated according to standard protocols (Schoser et al., 2004). All procedures were approved by the Institutional Ethics Committee and were conducted according to the Declaration of Helsinki.
Cloning of human ZNF9 and purification of the recombinant ZNF9 protein.
Human ZNF9 cDNA was generated by reverse transcriptase (RT)-PCR with the specific primers covering the open reading frame of human ZNF9. The sequence of the forward primer was as follows: 5′-ATG AGC AGC AAT GAG TGC TTC AAG TGT GGA-3′. The sequence of the reverse primer was as follows: 5′-TTA GGC TGT AGC CTC AAT TGT GCA TTC CCG-3. The RT-PCR product was cloned into StuI site of pMAL vector, and the resulting plasmid was verified by sequencing. ZNF9 protein fused to maltose-binding protein (MBP) was induced in Escherichia coli with isopropyl-β-d-1-thiogalactopyranoside, and protein was purified by affinity chromatography using amylose resin. The chromatography fractions containing purified MBP-ZNF9 were analyzed by gel electrophoresis.
Generation of polyclonal antibodies to ZNF9.
Rabbit polyclonal antibodies to the ZNF9 specific peptide (CYRCGESGHLARECTIEATA) were generated in Bethyl. This C-terminal conserved peptide has been previously applied for the production of antibodies to ZNF9 (Warden et al., 1994). The antibodies to ZNF9 were further purified by affinity chromatography with the peptide used for the production of antibodies. The titer and specificity of the purified antibodies to ZNF9 were tested with the purified ZNF9 fused to maltose-binding protein, MBP, and with cytoplasmic and nuclear protein extracts from C2C12 myoblasts as described in the text.
Western blotting.
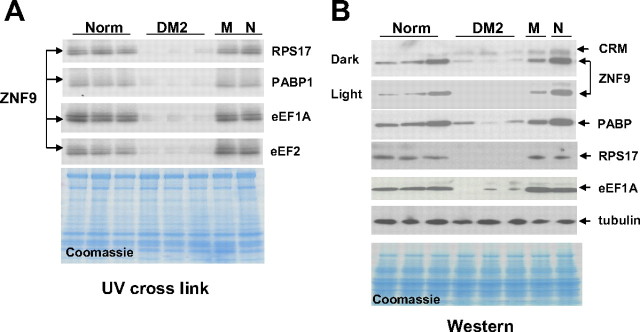
Primary human myoblasts were grown in 5% CO2 until 60% density and maintained for no more than 10 passages. Cytoplasmic and nuclear protein extracts were prepared from mouse C2C12 myoblasts and from human normal and DM2 myoblasts as described previously (Timchenko et al., 1996). Human muscle samples were sequentially homogenized on ice with loose and tight glass homogenizers, and cytoplasmic proteins were extracted with the low salt buffer (20 mm Tris-HCl pH 7.5, 30 mm KCl, 5 mm DDT, 10% glycerol and phosphatase and protease inhibitors), which does not disrupt nuclear membrane (Timchenko et al., 1996). Prior to Western blot analyses, cytoplasmic proteins from muscle samples were examined by gel electrophoresis and staining with Coomassie blue to ensure identical patterns of protein composition from different muscle samples. Electrophoretic analysis of cytoplasmic proteins from muscle biopsies derived from normal, DM2 patients and patients with other muscle diseases showed that all analyzed samples had similar patterns of major proteins suggesting that there was no degradation of proteins during homogenization or cross-contamination with major nuclear proteins (see Fig. 5).
Figure 5.
Expression of ZNF9 and its targets is reduced in skeletal muscle of DM2 patients. A, ZNF9 RNA-binding activity toward mRNAs (shown on the right) was examined by UV cross-link assay. Cytoplasmic extracts of muscle from three normal patients and three patients affected with DM2 were used. Muscle biopsies from patients with other muscle diseases, myopathy (M) and neurogenic atrophy (N), were used as additional control. A parallel gel with identical loadings of proteins was stained with Coomassie blue to verify protein loading (bottom). B, Protein levels of ZNF9 and its targets are reduced in cytoplasm of DM2 muscle. Western blot analysis was performed with antibodies shown on the right. The membrane was sequentially probed with antibodies to ZNF9, RPS17, PABP, and eEF1A. As the control, Western assay with anti-tubulin was performed. The membrane was stained with Coomassie blue (bottom). CRM, A cross-reactive material detected after longer exposure on the membrane with ZNF9 Western blotting. Norm, Normal.
Twenty to 50 μg of protein from human myoblasts and human muscle biopsies were subjected to electrophoresis on 12% or 8–16% polyacrylamide gels, transferred on nitrocellulose and probed with the affinity-purified antibodies to ZNF9 (1:1000). Fifteen to 60 μg of proteins from C2C12 myoblasts were resolved by 12% gel-electrophoresis, transferred on membrane, and incubated with anti-ZNF9 (1:500 to 1:2000). Antibodies to RPS17, poly (A)-binding protein (PABP; 10E10), eEF1A (all from Santa Cruz Biotechnology), eEF2 (Millipore), and β-actin (Sigma) were used according to the manufactures' protocols. There are several protein isoforms of human PABP with close molecular weights. Monoclonal anti-PABP (10E10) recognizes one protein with molecular weight 70 kDa in the cytoplasm of human myoblasts and skeletal muscle tissue after separation on 8–16% polyacrylamide gel. Thus, we have referred to the protein product, recognized by these antibodies, as “PABP” without specification of PABP isoforms.
Ultraviolet cross-link assay.
The sequences of RNA probes used for ultraviolet (UV) cross-linking were as follows: (1) 5′-guuuccucuuuuaccaaggaccccccgccaacaugggc-3′(RPS17); (2) 5′-gcgcucuacuccuguaacggaaag- gucgcggcuugugugccugcgggcagccgugccgagaaug-3′ (PABP isoform 3); (3) 5′-cccuucuccccggcgguuagug cugagagu-3′ (PABP isoform 1); (4) 5′-cucuuccg-ccgucgucgccgccauccucggcgcgacucgcuucuuucgguucuaccugggagaauccaccgc-cauccgccaccaug-3′ (eEF2); 5′-cuuuuucgcaacggguuugccgccagaacacag- gugucgugaaaacuaccccuaaaagccaaaaug-3′ (eEF1A). The sequences representing the TOP elements in these 5′ UTRs are shown in italics. The purified ZNF9-MBP and cytoplasmic protein extracts from control and DM2 myoblasts were incubated with the RNA probes labeled with γ32P-ATP, under conditions described in our previous paper (Salisbury et al., 2008). RNA-protein mixtures were treated with UV light, and RNA-protein complexes were separated by electrophoresis in 4–20% polyacrylamide gel. After electrophoresis, RNA-protein complexes were transferred on nitrocellulose membrane and exposed to x-ray film. After exposure, membranes were stained with Coomassie blue to verify protein loading. Where indicated, 10–100 ng of specific competitor (unlabeled RNA of the same sequence) were added prior to the radioactive probe addition. All UV cross-link reaction mixtures contained 0.1 μg of nonspecific competitor (tRNA).
Immunofluorescence analysis.
Normal and DM2 primary myoblasts were grown on 4× chambers at 60% density. Cells were washed consequently with PBS and cytoskeleton (CSK) buffer and then incubated in the 0.5% solution of Triton X-100 in CSK buffer. The composition of CSK buffer was 10 mm Tris-HCl, pH 7.0; 300 mm sucrose, 3 mm MgCl2; 10 μg/ml leupeptin and 10 μg/ml pepstatin. Cells were washed several times with CSK buffer and PBS and then fixed in 3,7% formaldehyde in 1× PBS for 1 h with change of solution in 30 min. Where indicated, cells were treated with RNase A (1 mg/ml). Nonspecific binding was blocked with the solution containing goat serum (1:40) and 1% bovine serum albumin in PBS for 1 h. Cells were incubated with the affinity-purified antibodies to ZNF9 (1:20) for 1 h and with goat anti-rabbit antibodies labeled with FITC (1:200) from Santa Cruz Biotechnology for 1 h. Nuclei were stained with DAPI. Cells were covered with mounting medium and analyzed on the fluorescent microscope Nikon. To quantify the intensities of ZNF9 signals, fluorescent images on normal and DM2 myoblasts were taken under identical settings of time of exposure and brightness. Images of normal and DM2 myoblasts were taken at 100 × to observe the appearance of ZNF9 in diffused and/or aggregated form. The percentage of cells with weak and strong intensities was calculated based on analyses of 121 normal cells and 109 DM2 cells. Calculations of three independent experiments are presented in this paper. To count the number of ZNF9-positive aggregates with and without RNase treatment, the digital immunofluorescence (IF) images, taken at 100×, were enlarged and the number of ZNF9 aggregates in cytoplasm of normal and DM2 myoblasts per cell was counted. An average number of ZNF9 aggregates in cytoplasm per cell was calculated based on three independent experiments.
Transient transfections.
To inhibit expression of ZNF9, normal human myoblasts were transfected with siRNAs to ZNF9 (purchased from Qiagen) using nucleofection protocol (Amaxa). Initially, siRNA, targeting the sequence 5′-AAGAGTTGAGTCAGACCAGTA-3′ in the 3′ UTR of ZNF9, was used. Three additional siRNAs to ZNF9 (siRNA #1, targeting the sequence 5′-CAGCAAGACAAGTGAAGTCAA-3′, located in the coding region of ZNF9; and siRNAs #2 and #3, targeting the sequences 5′-CTGACCTCAGTAGCTATTAAA-3′ and 5′-CACATAGATGGTGATGGCAAA-3′ in the 3′ UTR of ZNF9) were used to confirm the role of ZNF9 in the inhibition of the rate of protein synthesis. Thus, total four different siRNAs to ZNF9 were applied. To visualize transfected cells and evaluate the efficiency of transfection, siRNA to ZNF9 was cotransfected with wild-type green fluorescent protein (GFP; 1 μg/10 cm plate).
Where indicated, normal and DM2 myoblasts were transfected with plasmids expressing ZNF9 or long CCUG repeats cloned in pcDNA vector. Long CCTG repeats (CCTG300) were synthesized as described previously (Ordway and Detloff, 1996). The medium was changed the day after transfection and protein extracts were isolated in 48 h after transfection.
Examination of the rate of protein synthesis.
Normal and DM2 myoblasts were grown on two 10-cm-diameter plates. Cells were washed with PBS and maintained in the Met-free medium supplemented with 35S-Met (50 μCi/plate) for 2 h. Cells were washed several times with PBS and protein extracts were isolated as described (Timchenko et al., 1996). Radioactive proteins were analyzed by gel electrophoresis and X-radiography. To quantify the rate of protein synthesis, radioactive proteins were precipitated with 10% TCA, proteins were washed with 70% acetone, and separated into two portions. One portion was used to determine protein concentration and the second portion was used to determine radioactivity using a scintillation counter.
Results
ZNF9 binds to the 5′ UTR of RPS17 mRNA
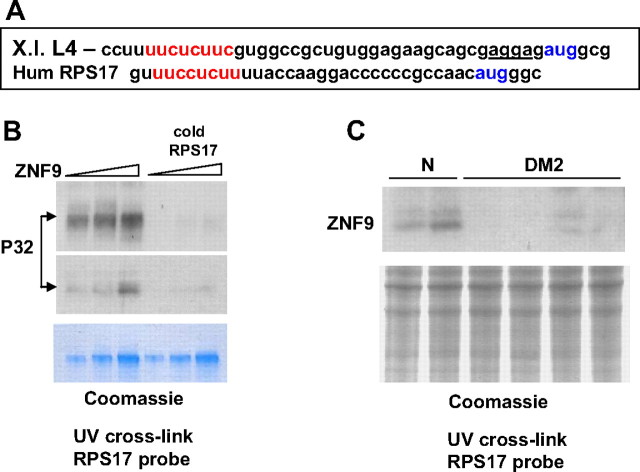
To examine whether human ZNF9 binds to mRNAs containing TOP tract, we have analyzed ZNF9 interaction with the 5′ UTR of mRNA encoding human ribosomal protein RPS17. In Xenopus laevis, Znf9 binds to the 5′ UTRs of ribosomal mRNAs between TOP tract and AUG codon (Pellizzoni et al., 1997). As shown in Figure 1A, the 5′ UTR of RPS17 contains the oligopyrimidine tract (TOP) with the sequence 5′-UUCCUCUU-3′, homologous to the TOP element in the 5′ UTR of L4 mRNA in Xenopus laevis. The 38 nt fragment of the 5′ UTR of RPS17 RNA was synthesized and used for UV cross-link assay with the human ZNF9 fused with MBP. The UV cross-linking analysis showed that MBP-ZNF9 binds to this RNA probe (Fig. 1B). The binding is specific because addition of the cold specific competitor completely inhibits the interaction of MBP-ZNF9 with the 5′ UTR of RPS17. The interaction of ZNF9 with the 5′ UTR of RPS17 mRNA suggested that ZNF9 may regulate translation of RPS17.
Figure 1.
RNA-binding activity of ZNF9 is reduced in cytoplasm of DM2 myoblasts. A, The 5′ UTRs of mRNAs encoding ribosomal protein L4 (Xenopus laevis) and RPS17 (human) contain the pyrimidine tracts (shown in red). AUG codons are shown in blue. Znf9-binding site within the 5′ UTR of rpsL4 is underlined. X.l., Xenopus laevis; Hum, human. B, The fusion MBP-ZNF9 binds to the 5′ UTR of RPS17 mRNA. The increased amounts of the purified MBP-ZNF9 were added to the UV cross-link reactions with RPS17 mRNA probe. Specific cold competitor was added before the probe addition. Short and long exposures of the membrane are shown. The membrane was stained with Coomassie blue to verify protein loading (bottom). C, Reduction of ZNF9 RNA-binding activity in DM2 myoblasts. Equal amounts of cytoplasmic protein extracts from normal (N) and DM2 cultured myoblasts were examined by UV cross-link assay with the RPS17 RNA probe. Coomassie staining shows loading of proteins used for UV-cross linking.
Because ZNF9 knock-out mice showed signs of DM2 phenotype, we asked whether ZNF9 RNA-binding activity toward the 5′ UTR of RPS17 mRNA is affected in DM2 primary myoblasts. UV cross-link assay with the RPS17 RNA probe showed that ZNF9 strongly binds to the 5′ UTR of RPS17 in normal cytoplasm; however, ZNF9 RNA-binding activity toward this RNA was significantly reduced in DM2 myoblasts (Fig. 1C). The Coomassie staining of the membrane after exposure to x-ray film showed that the equal amounts of proteins from normal and DM2 myoblasts were used for UV cross link. Thus, these studies show that the ZNF9 RNA-binding activity toward the 5′ UTR of RPS17 is decreased in cytoplasm of myoblasts derived from DM2 patients. Since previous reports did not identify changes in the total levels of ZNF9 (Margolis et al., 2006) or in cytoplasmic extracts from lymphoblastoid DM2 cell lines (Botta et al., 2006), we suggested that such reduction of ZNF9-binding activity might be caused by inhibition of its activity without alterations of protein levels or by a reduction of ZNF9 protein levels in cytoplasm of DM2 myoblasts.
Protein levels and RNA binding activity of ZNF9 are reduced in cytoplasm of DM2 myoblasts
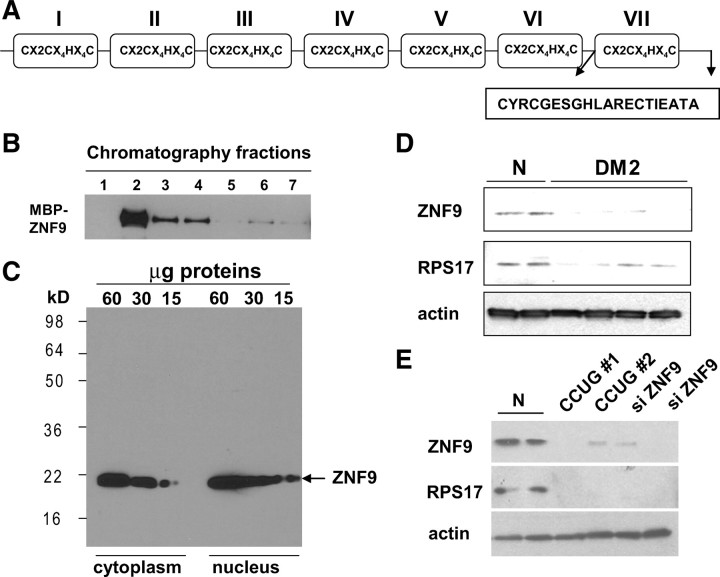
To examine whether DM2 myoblasts have reduced amounts of ZNF9 protein, we have generated antibodies to the specific peptide located in the domain VII of ZNF9 (Fig. 2A). Initial experiments showed that the generated antibodies specifically recognized the purified MBP-ZNF9 protein (Fig. 2B), but they also cross reacted with several other proteins of higher molecular weights (data not shown). To increase the specificity of these antibodies, they have been subjected to affinity chromatography. To determine the interactions of these antibodies with cellular proteins, we have performed Western blotting using cytoplasmic and nuclear proteins from C2C12 myoblasts. This analysis showed that the antibodies interact with a single 20 kDa protein in C2C12 cytoplasm and nucleus (Fig. 2C). Given the generation of these antibodies, we next examined the levels of ZNF9 in cytoplasm from normal and DM2 myoblasts by Western blotting. As seen in Figure 2D, ZNF9 levels are reduced in DM2 cytoplasm. The reprobe of the filter with Abs to β-actin showed equal loading of proteins from normal and DM2 myoblasts. Since ZNF9 binds to the 5′ UTR of RPS17 mRNA (Fig. 1), we have examined RPS17 in DM2 myoblasts. Our data demonstrated that RPS17 is reduced in DM2 cytoplasm consistent with the decrease of ZNF9 protein (Fig. 2D). To examine the causal role of ZNF9 in the reduction of RPS17, we inhibited ZNF9 in normal cells by siRNA. In these experiments, we have used Amaxa Transfector system which gives up to 70–89% efficiency of transfections. As seen in Figure 2E, the inhibition of ZNF9 leads to a reduction of RPS17. These studies showed that ZNF9 positively regulates RPS17, and that the decline of ZNF9 in cytoplasm of DM2 myoblasts reduces the levels of RPS17. It has been suggested that the accumulation of noncoding CCUG repeats resulted in the formation of nuclear foci that cause DM2 pathology (Margolis et al., 2006). Thus, we have examined whether the reduction of ZNF9 in cytoplasm of DM2 myoblasts is caused by pure RNA CCUG repeats. Normal myoblasts were transiently transfected with a plasmid expressing long CCUG repeats. As shown in Figure 2E, expression of RNA CCUG repeats reduces the levels of ZNF9 and the levels of its target, RPS17. Together, these data suggest that the decrease of ZNF9 in cytoplasm of DM2 myoblasts is caused by RNA CCUG repeats.
Figure 2.
Protein levels of ZNF9 are reduced in cytoplasm of DM2 myoblasts. A, The schematic structure of ZNF9 protein and the position of peptide used for production of antibodies to ZNF9 are shown. B, The generated anti-peptide antibodies to ZNF9 recognize purified MBP-ZNF9. Different fractions of the purification of MBP-ZNF9 by affinity column (shown on the top) were probed by Western blotting with anti-ZNF9. C, Interactions of the generated antibodies with endogenous ZNF9 in cytoplasmic and nuclear extracts from C2C12 myoblasts. D, Protein levels of ZNF9 and its target, RPS17, are reduced in cytoplasm of DM2 myoblasts. Western blotting was performed with two normal and four DM2 myoblast lines. The membrane was reprobed with antibodies to β-actin to verify protein loading. E, Ectopic expression of the mutant CCUG RNA or siRNA to ZNF9 inhibits ZNF9 in normal myoblasts and causes the reduction of RPS17. Western blotting of cytoplasmic proteins from normal myoblasts transfected with mutant CCUG RNA and with siRNA to ZNF9 was performed with antibodies to ZNF9. The membrane was reprobed with antibodies to RPS17 and β-actin. N, Normal.
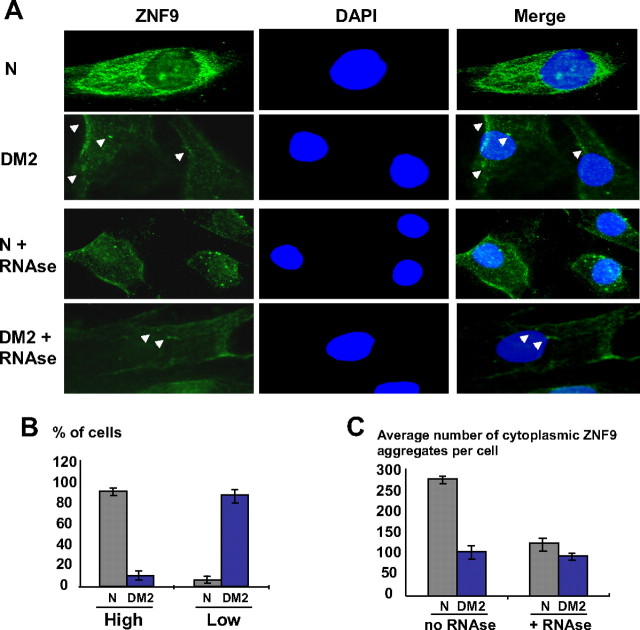
To further characterize intracellular distribution of ZNF9 in human cells, we performed IF analysis of normal and DM2 myoblasts. As shown in Figure 3A, ZNF9 is detected in both cytoplasm and nuclei of normal myoblasts with larger amounts of ZNF9 in cytoplasm. IF at high magnification (100 ×) showed that ZNF9 is detected in dispersed and aggregated forms (Fig. 3A). To compare the levels of ZNF9 in normal and DM2 myoblasts, we have used myoblasts derived from a patient which is homozygous for DM2 mutation (Schoser et al., 2004). The intensity of ZNF9 signals in the cytoplasm of myoblasts from this patient is lower compare with normal myoblasts (Fig. 3A). Whereas the signals of ZNF9 vary from cell-to-cell in normal and DM2 cycling myoblasts, ∼91% of DM2 myoblasts have weaker ZNF9 signals in cytoplasm compared with those in cytoplasm of normal myoblasts (Fig. 3B). Thus, three independent approaches (Western blotting, UV-cross linking, and IF analyses) have shown the reduction of ZNF9 protein and its RNA binding activity in cytoplasm of DM2 myoblasts. IF analysis also showed that, similar to normal myoblasts, ZNF9 is detected in dispersed and aggregated forms in DM2 myoblasts (Fig. 3A) with the variable number of ZNF9-positive aggregates in normal and DM2 myoblasts. To test whether ZNF9-positive aggregates represent ZNF9-RNA complexes, normal and DM2 myoblasts were treated with RNase A prior to detection of ZNF9 by IF analysis. The RNase A treatment of normal myoblasts reduced ∼2-fold the number of ZNF9-positive aggregates (Fig. 3A, C). We found that RNase treatment did not destroy ZNF9-positive aggregates in DM2 cytoplasm (Fig. 3C), but it has reduced their brightness (Fig. 3A). The majority of the ZNF9 aggregates in DM2 have lost their sharpness and become more diffused after RNase A treatments (Fig. 3B). These results suggest that ZNF9 interacts with RNAs and perhaps with some other proteins. The lesser effect of RNase A on the aggregated ZNF9 in DM2 cytoplasm relatively to normal cells perhaps is a result of reduced ZNF9 binding to some mRNAs (Fig. 4).
Figure 3.
A, IF analysis of intracellular distribution of ZNF9 in myoblasts derived from normal and DM2 patient homozygous for DM2 mutation with and without treatment with RNase A. Images of normal and DM2 myoblasts are taken under the same conditions (time of exposure, brightness, and contrast). Magnification is 100×. Nuclei were stained with DAPI. Arrows show ZNF9-positive aggregates in DM2 myoblasts with and without treatment by RNase A. RNase treatment of DM2 cells leads to transformation of sharp aggregates into the diffused ones. Only few aggregates (shown by arrows on the bottom panel) remain sharp. B, The majority of DM2 myoblasts have low intensity of ZNF9 in cytoplasm relatively to normal myoblasts. ZNF9 intensity in normal myoblasts (shown in A) was used as a standard for “high” intensity. The intensity of ZNF9 in DM2 myoblast (shown in A) was used as a standard for “low” intensity. C, RNase A treatment reduces the number of ZNF9-positive aggregates in cytoplasm of normal myoblasts. RNase treatment of DM2 cells transforms sharp aggregates to the diffused ones without changing their number. N, Normal.
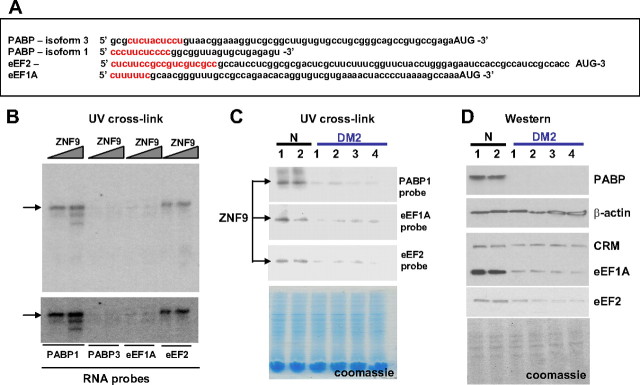
Figure 4.
The levels of the ZNF9 targets PABP, eEF1A, and eEF2 are reduced in DM2 myoblasts. A, Nucleotide sequences of the 5′ UTRs of PABP (isoforms 1 and 3), eEF1A, and eEF2 are shown. The TOP elements are shown in red. B, ZNF9 binds to the 5′ UTRs of PABP1, eEF1A, and eEF2. Increasing amounts of MBP-ZNF9 were incubated with RNA probes and examined by UV cross-link. The position of MBP-ZNF9 is shown by arrow. The signal for eEF1A after 2 h of exposure was weaker than for PABP1 and eEF2, but it was detectable after 24 h of exposure (bottom). C, Reduction of ZNF9 RNA-binding activity toward the 5′ UTRs of PABP1, eEF1A, and eEF2 in cytoplasm of DM2 myoblasts. Top, UV cross-link of cytoplasmic protein extracts from normal and DM2 myoblasts with PABP1, eEF1A, and eEF2 RNA probes is shown. Bottom, The membrane was stained with Coomassie blue to verify protein loading. D, Reduction of protein levels of PABP, eEF1A, and eEF2 in DM2 myoblasts. Western blotting of the cytoplasmic extracts from two normal and four myoblast lines derived from DM2 patients with antibodies to PABP, eEF1A, and eEF2 is shown. Reprobe of the membrane with Abs to β-actin and the cross-reactive molecule (CRM, on the eEF1A membrane) shows equal protein loading. The membrane was also stained with Coomassie blue. N, Normal.
ZNF9 regulates expression of several key proteins of translational apparatus
Given the positive regulation of RPS17 by ZNF9, we suggested that ZNF9 may also control other TOP-containing mRNAs. A group of mRNAs containing TOP includes PABP and elongation factors eEF1A and eEF2. To test whether ZNF9 binds to these mRNAs, the RNA oligomers containing the 5′ UTRs of eEF1A, eEF2, and two isoforms of PABP (isoforms 1 and 3) (Fig. 4A) were synthesized and used for UV cross-linking with the purified ZNF9-MBP. UV cross-link data showed that ZNF9 strongly binds to the 5′ UTRs of PABP1 and eEF2 (Fig. 4B). The binding of ZNF9 to eEF1A was lower, but was detectable after longer exposure (Fig. 4B, bottom). In contrast, PABP3 RNA does not interact with ZNF9 and did not show any signal even after long exposure to x-ray film (24 h) (Fig. 4B, bottom image). This result shows that ZNF9 binds to the 5′ UTRs of mRNAs encoding PABP1, eEF2 and to less extent, to mRNA coding for eEF1A. This binding is specific because ZNF9 does not interact with PABP3. To examine whether endogenous ZNF9 interacts with the 5′ UTRs of PABP1, eEF1A, and eEF2 in cytoplasm of DM2 myoblasts, we have performed UV cross-link assay with these RNA probes using cytoplasmic extracts from normal and DM2 myoblasts. As shown in Figure 4C, ZNF9 RNA-binding activity toward PABP1, eEF1A, and eEF2 is reduced in cytoplasm of DM2 myoblasts. These studies indicated that, in addition to RPS17, ZNF9 might regulate several other TOP-containing RNAs, suggesting that the levels of corresponding proteins might be reduced in DM2 myoblasts. Therefore, we have measured the levels of eEF1A and eEF2 in the cytoplasm from normal and DM2 myoblasts, and found that these proteins are decreased in DM2 myoblasts (Fig. 4D). Western blot assay of cytoplasmic proteins from normal and DM2 myoblasts with anti-PABP showed the reduction of the 70 kDa protein migrating in the position of PABP (Fig. 4D). Together, these studies revealed that ZNF9 binds to the 5′ UTRs of key regulators of protein translation, and that the reduction of ZNF9 in DM2 myoblasts leads to the reduction of eEF1A, eEF2, and PABP proteins.
ZNF9 and proteins of translational apparatus are reduced in muscle biopsies derived from patients with DM2
Reduction of ZNF9 and proteins, encoded by TOP-containing mRNAs, in primary myoblasts of DM2 patients suggested that ZNF9 and its targets might be reduced in mature muscle of DM2 patients. To examine this suggestion, we have analyzed ZNF9 RNA-binding activity in cytoplasm from skeletal muscle biopsies derived from normal patients, patients affected with myopathy and neurogenic atrophy, and from patients with DM2. ZNF9 RNA-binding activity was analyzed by UV cross-link assay using the RNA oligos containing the 5′ UTRs of mRNAs encoding RPS17, PABP1, eEF1A, and eEF2. As shown in Figure 5A, ZNF9 RNA-binding activity is reduced in cytoplasm from mature muscle of DM2 patients. In contrast, ZNF9 RNA-binding activity in muscle samples from patients with myopathy or neurogenic atrophy is comparable to that in normal muscle samples (Fig. 5A). Thus, similar to primary myoblasts, ZNF9 RNA-binding activity is diminished in cytoplasm of DM2 muscle. To examine whether the reduction of ZNF9 RNA-binding activity in cytoplasm of DM2 muscle is a result of reduced protein levels of ZNF9, we have measured ZNF9 levels by Western blotting assay. As seen on Figure 5B, the levels of ZNF9 are reduced in cytoplasm of DM2 muscle samples. In agreement with these data, the levels of RPS17 and eEF1A are reduced in cytoplasm of DM2 muscle (Fig. 5B). Similar to myoblasts, anti-PABP (clone 10E10) recognized one protein with molecular weight 70 kDa that is reduced in cytoplasm of DM2 muscle (Fig. 5B). Expression of tubulin, used as control, was not altered in DM2 muscle samples and was comparable to that in normal muscle and in muscle samples from patients with myopathy and neurogenic atrophy. These data show that ZNF9 protein levels are reduced not only in cytoplasm of DM2 primary myoblasts, but also in cytoplasm in mature muscle of DM2 patients.
Rate of translation is reduced in DM2 myoblasts
The reduction of the proteins which are important for the general translation suggested that the rate of protein biosynthesis might be reduced in DM2 muscle cells. To test this suggestion, we have grown cultured myoblasts derived from normal and DM2 patients in the Met-free medium supplemented with 35S-Met. In 2 h after addition of 35S-Met, cytoplasmic proteins were collected and analyzed by gel electrophoresis after exposure to x-ray film. As seen in Figure 6A, the amounts of newly synthesized radioactive proteins after 2 h labeling are lower compared with those in normal myoblasts. Calculations of the 35S incorporation in proteins showed ∼2-fold reduction of the protein synthesis in myoblasts from DM2 patients (Fig. 6A, bottom). These studies clearly indicated that the rate of global protein synthesis is decreased in DM2 myoblasts. There are several factors that may inhibit protein synthesis in DM2 cytoplasm. One of these factors is the reduction of protein translation by ZNF9 through the decrease of proteins of translational machinery. To determine the role of the reduction of ZNF9 RNA-binding activity in the inhibition of protein synthesis, DM2 myoblasts were transfected with ZNF9-expressing plasmid and with an empty vector. As can be seen in Figure 6B, the levels of ZNF9 are corrected by the ectopic expression of ZNF9 from the translated plasmid. The amounts of proteins synthesized in DM2 myoblasts with corrected levels of ZNF9 are identical to those in cells transfected with an empty vector. As shown in Figure 6B (bottom), ectopic expression of ZNF9 increases the amounts of newly synthesized proteins in DM2 myoblasts to the levels observed in normal cells. To obtain additional evidence for the role of ZNF9 in the reduction of the rate of total translation, we have inhibited expression of ZNF9 with siRNA in normal human myoblasts, and found that this inhibition also reduces the amounts of newly synthesized proteins (Fig. 6B). These data suggest that the decline of ZNF9 in cytoplasm plays a critical role in the reduction of the rate of total protein synthesis in DM2 myoblasts.
Figure 6.
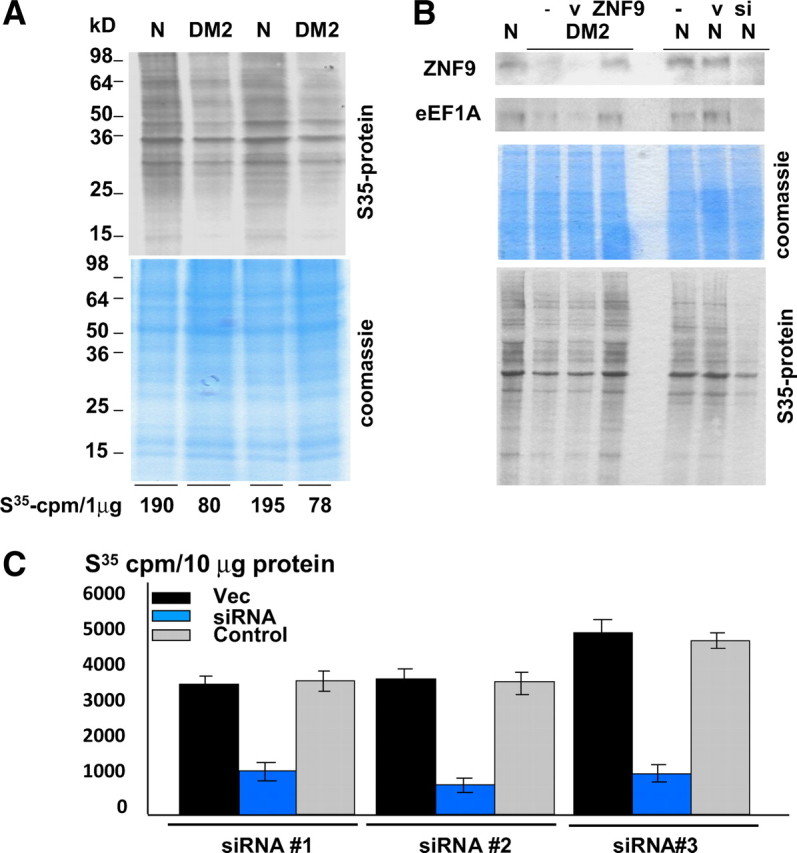
The reduction of the rate of protein synthesis in DM2 myoblasts. A, Comparison of the total amounts of radioactive proteins in the cytoplasms of normal (N) and DM2 myoblasts after 2 h growth in the Met-free medium supplemented with 35S-Met. Data with two normal and two DM2 myoblast cell lines are shown. The membrane was stained with Coomassie blue. 35S-labeled proteins were precipitated with TCA and counted on scintillation counter. Amount of 35S radioactivity (cpm) per microgram of protein is shown below. B, Correction of ZNF9 in DM2 myoblasts corrects the rate of protein translation. Top, Western blot of the cytoplasmic protein extracts isolated from normal and DM2 myoblasts transfected with ZNF9-expressing plasmid, empty vector (GFP) to visualize transfected cells or siRNA to ZNF9 (shown on the top). The membrane was sequentially incubated with antibodies to ZNF9 and eEF1A. Middle, The membrane was stained with Coomassie blue. Bottom, The electrophoretic analysis of the 35S-labeled proteins from cytoplasm of normal and DM2 myoblasts transfected with empty vector (v) and with plasmid expressing ZNF9 or siRNA (si) to ZNF9 is shown. C, Inhibition of ZNF9 by three additional siRNAs reduces incorporation of 35S-methionine into protein. Myoblasts from normal patients were transfected with three siRNAs to ZNF9 (blue), empty vector (Vec; black), and control siRNA (gray) and cells were grown with 35S-methionine as described above. Protein extracts were isolated in 2 h and the incorporation of 35S-methionine was calculated per 10 μg of protein.
To examine whether the normalization of protein synthesis in DM2 myoblasts transfected with ZNF9 occurs because of correction of proteins, encoded by the TOP-containing mRNAs, we have measured the levels of eEF1A in DM2 myoblasts transfected with ZNF9 and grown in the medium with 35S-Met. As shown in Figure 6B, the levels of eEF1A are low in untransfected DM2 myoblasts or in DM2 myoblasts transfected with an empty vector. However, expression of ZNF9 in DM2 myoblasts increases the levels of eEF1A. In agreement with the increase of eEF1A, the amounts of newly synthesized proteins are also increased in DM2 myoblasts, whereas the inhibition of ZNF9 by siRNA in normal myoblasts reduces eEF1A leading to the reduction of the rate of protein synthesis. To verify that the siRNA to ZNF9 has the specific effect on the rate of protein translation through the inhibition of ZNF9, we have examined the rate of protein synthesis in normal myoblasts transfected with three additional siRNAs to ZNF9 (see Materials and Methods). As can be seen in Figure 6C, all three siRNAs to ZNF9 reduced the rate of protein synthesis in normal myoblasts, whereas control siRNA did not change the rate of proteins synthesis. These data are consistent with the hypothesis that the reduction of ZNF9 activity in DM2 leads to the reduction of the rate of translation in DM2.
Discussion
DM1 and DM2 diseases are caused by an expansion of noncoding RNA repeats. The DM2 expansion is located within ZNF9 gene, which encodes a protein with predicted RNA binding activity. Although previous studies showed that RNA CCUG repeats disrupt splicing in DM2 cells through reduction of MBNL1 and/or increase of CUGBP1 (Savcur et al., 2004), several additional mechanisms might be involved in the development of DM2 pathology. In this study, we present evidence that DM2 muscle cells are characterized by the reduced rate of global translation. These data provide an additional mechanism by which expansion of CCUG repeats might cause pathological alterations in DM2 patients.
Available information for the role of ZNF9 in DM2 is contradictory. Margolis et al. (2006) have shown no alterations in the expression of total ZNF9 protein and ZNF9 mRNA in human DM2 myoblasts using a polyclonal antibody generated to the middle portion of the ZNF9 protein. Another study of immortalized lymphoblastoid cultures from DM2 patients also did not find significant changes in expression of ZNF9 using C-terminal ZNF9 antibody (Botta et al., 2006). Splicing of the ZNF9 gene is also not changed by DM2 mutation (Botta et al., 2006; Margolis et al., 2006). Despite these observations, DM2-like phenotype in the ZNF9 knock-out mice has clearly demonstrated that alterations in ZNF9 expression are involved in DM2 disease (Chen et al., 2007). Therefore, we have performed a careful analysis of ZNF9 in DM2 cells using different approaches and newly generated antibodies to ZNF9. In agreement with data for ZNF9 knock-out mice, our data show that protein levels of ZNF9 are reduced in cytoplasm of both DM2 primary myoblasts and DM2 mature muscle. Moreover, we found that this reduction is a key event in DM2, which leads to the decline in the rate of protein synthesis. The reduction of ZNF9 levels and its RNA-binding activity in cytoplasm of DM2 myoblasts is shown by three independent approaches: by Western blotting, by immunostaining, and by UV cross-link assay. In addition to cultured cells, we found the reduction of ZNF9 protein levels and its RNA-binding activity in cytoplasm of muscle biopsies from DM2 patients. We present evidence that ZNF9 binds to the 5′ UTR of several TOP mRNAs, including mRNAs encoding ribosomal protein RPS17, PABP1, and two elongation factors, eEF1A and eEF2. Our data suggest that, under normal conditions, ZNF9 supports translation of the TOP-containing mRNAs. This suggestion is based on the experiments which show that a direct inhibition of ZNF9 by siRNAs in normal cells leads to the reduction of RPS17 and proteins encoded by TOP mRNAs to which ZNF9 binds (Figs. 1, 6). The reduction of these proteins correlates with the reduction of the rate of protein synthesis (Fig. 6). Our data demonstrate that the reduction of ZNF9 in the cytoplasm of DM2 myoblasts is caused by the expansion of CCUG RNA repeats. Ectopic expression of the pure mutant CCUG repeats reduces protein levels of ZNF9 and the levels of targets of ZNF9 in cytoplasm of normal myoblasts. The precise mechanism by which CCUG RNA reduces ZNF9 in cytoplasm remains to be investigated.
The main result of this work is that DM2 myoblasts have a reduced rate of global protein synthesis (Fig. 6). Although our data show a critical role of ZNF9, other factors might also contribute to the reduction of the rate of protein synthesis in DM2. As we have previously showed, CUGBP1 regulates translation of proteins in DM1 myoblasts (Timchenko et al., 2001, 2004; Salisbury et al., 2008). Depending on the phosphorylation status of CUGBP1, it may have positive or negative effects on translation. It is possible that in DM2, CUGBP1 might reduce translation or increase degradation of mRNAs, encoding proteins of translational apparatus. We have recently shown that CCUG repeats interact with CUGBP1-eIF2 complex and with proteasome. Therefore, the rate of translation in DM2 muscle cells may be also reduced as a result of binding of the CUGBP1-eIF2 complexes to CCUG repeats leading to the sequestration of the eIF2 factor or as a result of dysfunction of the proteasome (Salisbury et al., 2009). Since DM2 is late-onset disease, it would be important to determine whether the inhibition of the rate of protein synthesis in DM2 muscle is worsening with age. It is possible that the negative effect of CCUG expansion on protein synthesis is increased with age because of accumulation of additional age-dependent modifiers. In this study, we have examined the rate of protein synthesis in DM2 myoblasts. It would be important to test whether the rate of protein synthesis is reduced in other tissues, affected in DM2, such as cardiac muscle, eye, and brain.
Footnotes
This work was supported by National Institutes of Health Grants AR052791 and NS063298 (to L.T.) and by grants from the Deutsche Gesellschaft fur Muskelkranke (B.S.). B.S. is member of the German network on muscular dystrophies (MD-NET, 01GM0601). We thank Muscle Tissue Culture Collection (MTCC) for providing the samples. MTCC is a partner of the EuroBioBank Network established in 2001 thanks to European Commission funding (01/2003-03/2006), www.eurobiobank.org. We thank Bob Chen and Priya Weerasinghe for excellent help with experiments and Bjarne Udd and Ralf Krahe for discussion of data.
References
- Aslanidis C, Jansen G, Amemiya C, Shutler G, Mahadevan M, Tsilfidis C, Chen C, Alleman J, Wormskamp NG, Vooijs, Buxton J, Johnson K, Smeets HJM, Lennon GG, Carrano AV, Korneluk RG, Wieringa B, de Jong PJ. Cloning of the essential myotonic dystrophy region and mapping of the putative defect. Nature. 1992;355:548–551. doi: 10.1038/355548a0. [DOI] [PubMed] [Google Scholar]
- Botta A, Caldarola S, Vallo L, Bonifazi E, Fruci D, Gullotta F, Massa R, Novelli G, Loreni F. Effect of the [CCTG]n repeat expansion on ZNF9 expression in myotonic dystrophy II (DM2) Biochem Biophys Acta. 2006;1762:329–334. doi: 10.1016/j.bbadis.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Chen W, Wang Y, Abe Y, Cheney L, Udd B, Li YP. Haploinsufficiency for Znf9 in Znf9+/− mice is associated with multiorgan abnormalities resembling myotonic dystrophy. J Mol Biol. 2007;368:8–17. doi: 10.1016/j.jmb.2007.01.088. [DOI] [PubMed] [Google Scholar]
- Day JW, Ricker K, Jacobsen JF, Rasmussen LJ, Dick KA, Kress W, Schneider C, Koch MC, Beilman GJ, Harrison AR, Dalton JC, Ranum LP. Myotonic dystrophy type 2. Neurology. 2003;60:657–664. doi: 10.1212/01.wnl.0000054481.84978.f9. [DOI] [PubMed] [Google Scholar]
- Edwards JG, Bahl JJ, Flink I, Milavetz J, Goldman S, Morkin E. A repressor region in the human beta-myosin heavy chain gene that has a partial position dependency. Biochem Biophys Res Commun. 1992;189:504–510. doi: 10.1016/0006-291x(92)91586-f. [DOI] [PubMed] [Google Scholar]
- Flink IL, Morkin E. Alternatively processed isoforms of cellular nucleic acid-binding protein interact with a suppressor region of the human beta-myosin heavy chain gene. J Biol Chem. 1995;270:6959–6965. doi: 10.1074/jbc.270.12.6959. [DOI] [PubMed] [Google Scholar]
- Flink IL, Blitz I, Morkin E. Characterization of cellular nucleic binding protein from Xenopus laevis: expression in all three germ layers during development. Dev Dyn. 1998;211:123–130. doi: 10.1002/(SICI)1097-0177(199802)211:2<123::AID-AJA1>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Gerbasi VR, Link AJ. The myotonic dystrophy 2 protein ZNF9 is part of an ITAF complex that promotes cap-independent translation. Mol Cell Proteomics. 2007;6:1049–1058. doi: 10.1074/mcp.M600384-MCP200. [DOI] [PubMed] [Google Scholar]
- Liquori CL, Ricker K, Moseley ML, Jacobsen JF, Kress W, Naylor SL, Day JW, Ranum LP. Myotonic dystrophy 2 is caused by CCTG expansion in intron 1 of ZNF9. Science. 2001;293:864–867. doi: 10.1126/science.1062125. [DOI] [PubMed] [Google Scholar]
- Margolis JM, Schoser BG, Moseley ML, Day JW, Ranum LPW. DM2 intronic expansions: evidence for CCUG accumulation without flanking sequence or effects on ZNF9 mRNA processing or protein expression. Hum Mol Genet. 2006;15:1808–1815. doi: 10.1093/hmg/ddl103. [DOI] [PubMed] [Google Scholar]
- Meyuhas O. Synthesis of the translational apparatus is regulated at the translational level. Eur J Biochem. 2000;267:6321–6330. doi: 10.1046/j.1432-1327.2000.01719.x. [DOI] [PubMed] [Google Scholar]
- Michelotti EF, Tomonaga T, Krutzsch H, Levens D. Cellular nucleic acid binding protein regulates the CT element of the human c-myc protooncogene. J Biol Chem. 1995;270:9494–9499. doi: 10.1074/jbc.270.16.9494. [DOI] [PubMed] [Google Scholar]
- Ordway JM, Detloff PJ. In vitro synthesis and cloning of long CAG repeats. Biotechniques. 1996;21:609–610. 612. doi: 10.2144/96214bm08. [DOI] [PubMed] [Google Scholar]
- Pellizzoni L, Lotti F, Maras B, Pierandrei-Amaldi P. Cellular nucleic acid binding protein binds a conserved region of the 5′ UTR of Xenopus laevis ribosomal protein mRNAs. J Mol Biol. 1997;267:264–275. doi: 10.1006/jmbi.1996.0888. [DOI] [PubMed] [Google Scholar]
- Rajavashisth TB, Taylor AK, Andalibi A, Svenson KL, Lusis AJ. Identification of a zinc finger protein that binds to the sterol regulatory element. Science. 1989;245:640–643. doi: 10.1126/science.2562787. [DOI] [PubMed] [Google Scholar]
- Ranum LP, Day JW. Myotonic dystrophy: RNA pathogenesis comes into focus. Am J Hum Genet. 2004;74:793–804. doi: 10.1086/383590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury E, Sakai K, Schoser B, Huichalaf C, Schneider-Gold C, Nguyen H, Wang GL, Albrecht JH, Timchenko LT. Ectopic expression of cyclin D3 corrects differentiation of DM1 myoblasts through activation of RNA CUG-binding protein, CUGBP1. Exp Cell Res. 2008;314:2266–2278. doi: 10.1016/j.yexcr.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury E, Schoser B, Schneider-Gold C, Wang GL, Huichalaf C, Jin B, Sirito M, Sarkar P, Krahe R, Timchenko NA, Timchenko LT. Expression of RNA CCUG repeats dysregulates translation and degradation of proteins in DM2 patients. Am J Pathol. 2009 doi: 10.2353/ajpath.2009.090047. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savcur RS, Philips AV, Cooper TA, Dalton JC, Moseley ML, Ranum LP, Day JW. Insulin receptor splicing alteration in myotonic dystrophy 2. Am J Hum Genet. 2004;74:1309–1313. doi: 10.1086/421528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schara U, Schoser BG. Myotonic dystrophies type 1 and 2—a summary of current aspects. Semin Pediatr Neurol. 2006;13:71–79. doi: 10.1016/j.spen.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Schoser BG, Kress W, Walter MC, Halliger-Keller B, Lochmüller H, Ricker K. Homozygosity for CCTG mutation in myotonic dystrophy type 2. Brain. 2004;127:1868–1877. doi: 10.1093/brain/awh210. [DOI] [PubMed] [Google Scholar]
- Timchenko LT. Myotonic dystrophy: the role of RNA CUG repeats. Am J Hum Genet. 1999;64:360–364. doi: 10.1086/302268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timchenko LT, Miller JW, Timchenko NA, DeVore DR, Datar KV, Lin L, Roberts R, Caskey CT, Swanson MS. Identification of a (CUG)n triplet repeat binding protein and its expression in myotonic dystrophy. Nucleic Acids Res. 1996;24:4407–4414. doi: 10.1093/nar/24.22.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timchenko NA, Iakova P, Cai ZJ, Smith JR, Timchenko LT. Molecular basis for impaired muscle differentiation in myotonic dystrophy. Mol Cell Biol. 2001;21:6927–6938. doi: 10.1128/MCB.21.20.6927-6938.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timchenko NA, Patel R, Iakova P, Cai ZJ, Quan L, Timchenko LT. Overexpression of CUG triplet repeat-binding protein, CUGBP1, in mice inhibits myogenesis. J Biol Chem. 2004;279:13129–13139. doi: 10.1074/jbc.M312923200. [DOI] [PubMed] [Google Scholar]
- Warden CH, Krisans SK, Purcell-Huynh D, Leete LM, Daluiski A, Diep A, Taylor BA, Lusis AJ. Mouse cellular nucleic acid binding proteins: a highly conserved family identified by genetic mapping and sequencing. Genomics. 1994;24:14–19. doi: 10.1006/geno.1994.1576. [DOI] [PubMed] [Google Scholar]
- Yasuda J, Mashiyama S, Makino R, Ohyama S, Sekiya T, Hayashi K. Cloning and characterization of rat cellular nucleic acid binding protein (CNBP) cDNA. DNA Res. 1995;2:45–49. doi: 10.1093/dnares/2.1.45. [DOI] [PubMed] [Google Scholar]