Abstract
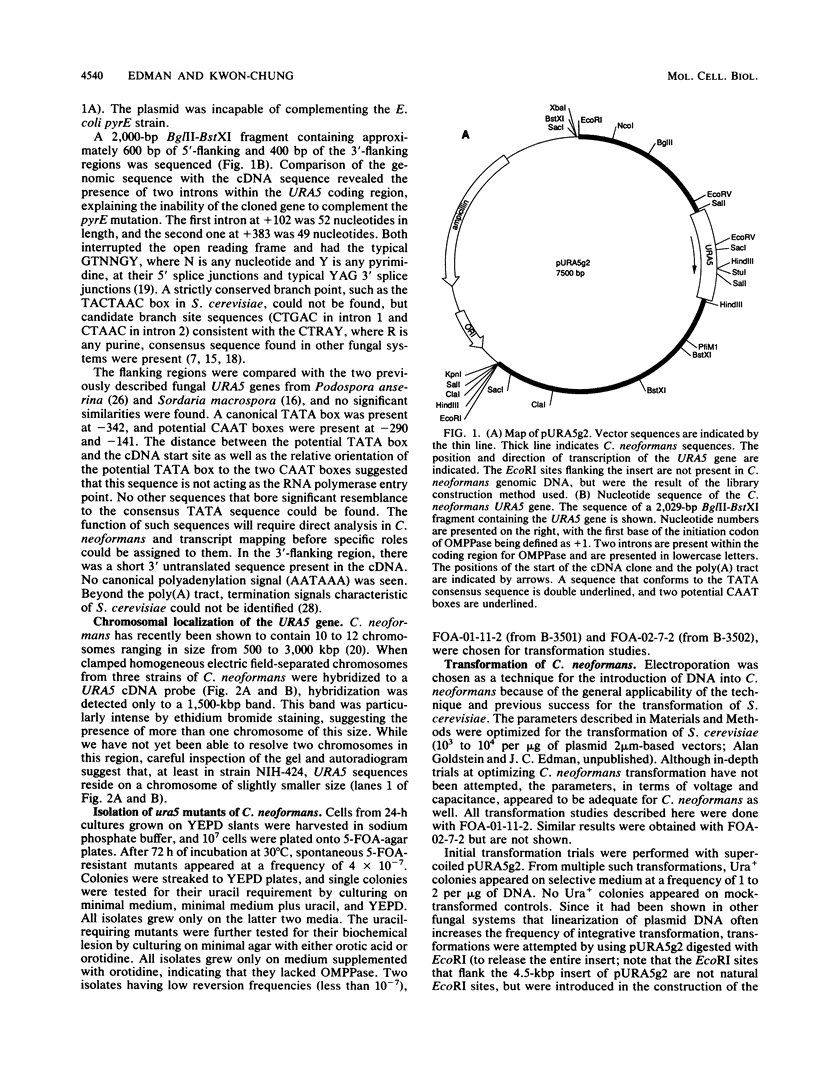
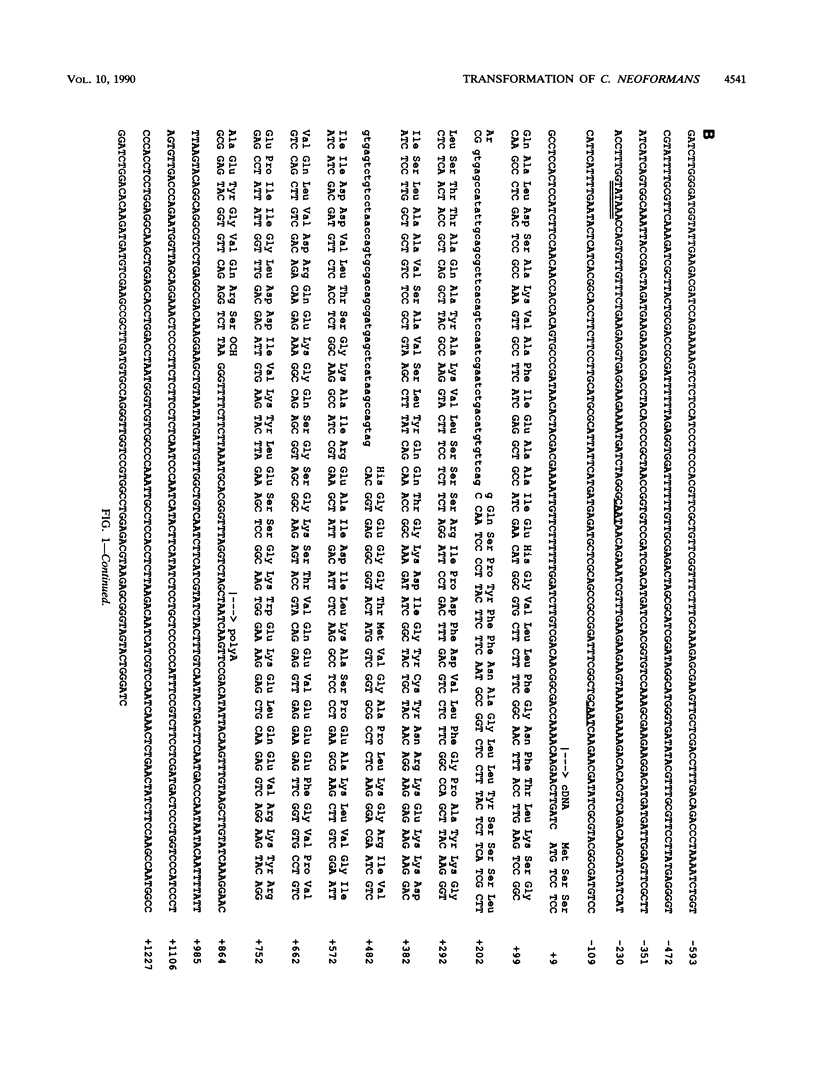
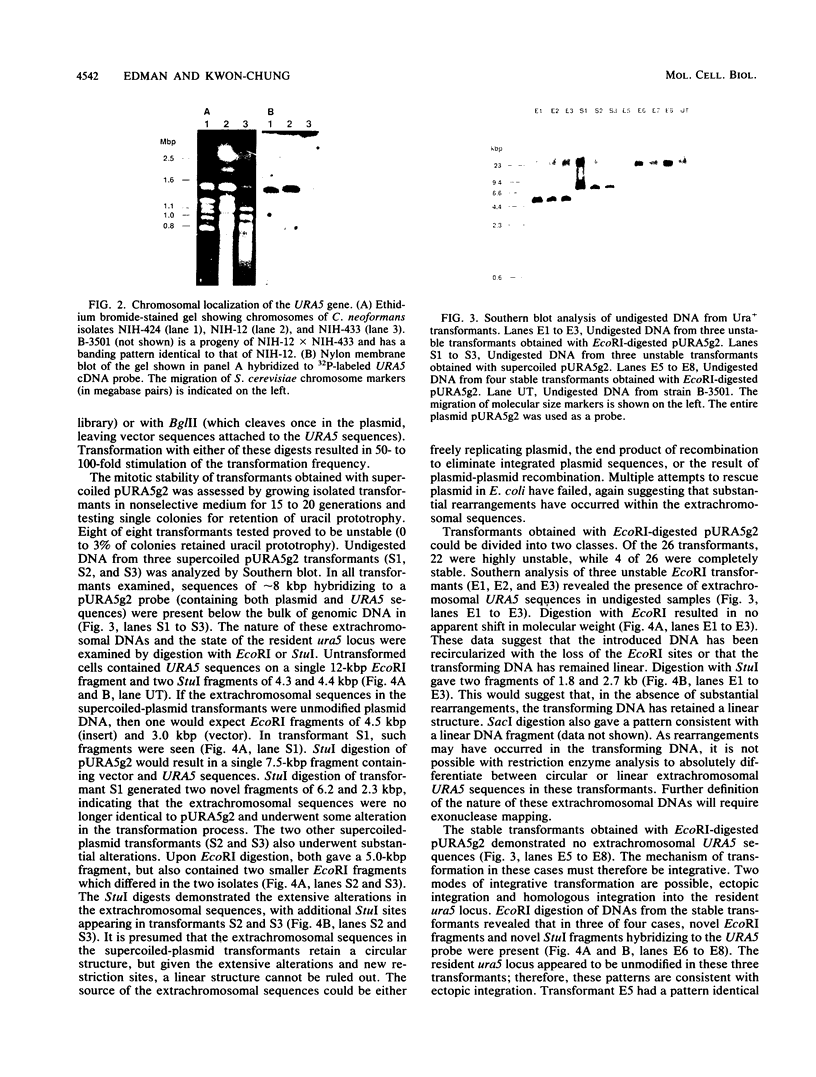
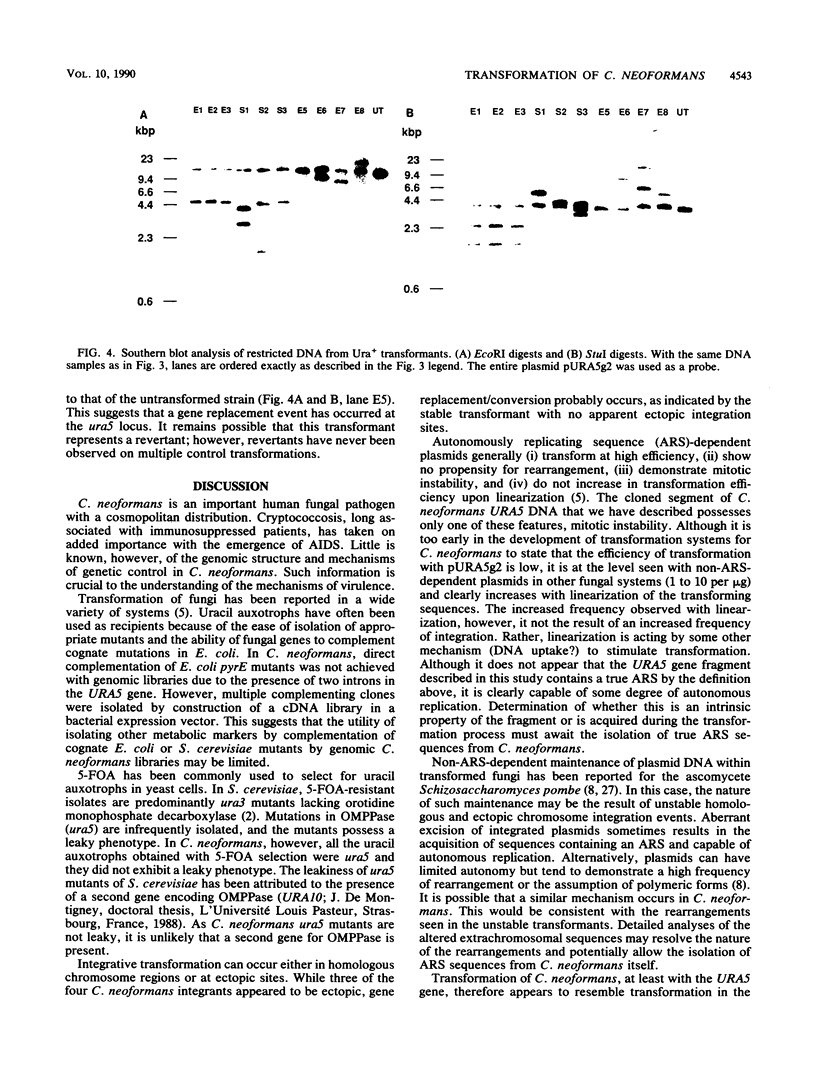
A cDNA encoding Cryptococcus neoformans orotidine monophosphate pyrophosphorylase (OMPPase) has been isolated by complementation of the cognate Escherichia coli pyrE mutant. The cDNA was used as a probe to isolate a genomic DNA fragment encoding the OMPPase gene (URA5). By using electroporation for the introduction of plasmid DNA containing the URA5 gene, C. neoformans ura5 mutants could be transformed at low efficiency. Ura+ transformants obtained with supercoiled plasmids containing the URA5 gene showed marked mitotic instability and contained extrachromosomal URA5 sequences, suggesting limited ability to replicate within C. neoformans. Transformants obtained with linear DNA were of two classes: stable transformants with integrated URA5 sequences, and unstable transformants with extrachromosomal URA5 sequences.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Banks G. R., Taylor S. Y. Cloning of the PYR3 gene of Ustilago maydis and its use in DNA transformation. Mol Cell Biol. 1988 Dec;8(12):5417–5424. doi: 10.1128/mcb.8.12.5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., LaCroute F., Fink G. R. A positive selection for mutants lacking orotidine-5'-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197(2):345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Edman J. C., Kovacs J. A., Masur H., Santi D. V., Elwood H. J., Sogin M. L. Ribosomal RNA sequence shows Pneumocystis carinii to be a member of the fungi. Nature. 1988 Aug 11;334(6182):519–522. doi: 10.1038/334519a0. [DOI] [PubMed] [Google Scholar]
- Edman U., Edman J. C., Lundgren B., Santi D. V. Isolation and expression of the Pneumocystis carinii thymidylate synthase gene. Proc Natl Acad Sci U S A. 1989 Sep;86(17):6503–6507. doi: 10.1073/pnas.86.17.6503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincham J. R. Transformation in fungi. Microbiol Rev. 1989 Mar;53(1):148–170. doi: 10.1128/mr.53.1.148-170.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler U., Hoffman B. J. A simple and very efficient method for generating cDNA libraries. Gene. 1983 Nov;25(2-3):263–269. doi: 10.1016/0378-1119(83)90230-5. [DOI] [PubMed] [Google Scholar]
- Heyer W. D., Sipiczki M., Kohli J. Replicating plasmids in Schizosaccharomyces pombe: improvement of symmetric segregation by a new genetic element. Mol Cell Biol. 1986 Jan;6(1):80–89. doi: 10.1128/mcb.6.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronstad J. W., Wang J., Covert S. F., Holden D. W., McKnight G. L., Leong S. A. Isolation of metabolic genes and demonstration of gene disruption in the phytopathogenic fungus Ustilago maydis. Gene. 1989 Jun 30;79(1):97–106. doi: 10.1016/0378-1119(89)90095-4. [DOI] [PubMed] [Google Scholar]
- Kwon-Chung K. J. A new genus, filobasidiella, the perfect state of Cryptococcus neoformans. Mycologia. 1975 Nov-Dec;67(6):1197–1200. [PubMed] [Google Scholar]
- Kwon-Chung K. J., Polacheck I., Popkin T. J. Melanin-lacking mutants of Cryptococcus neoformans and their virulence for mice. J Bacteriol. 1982 Jun;150(3):1414–1421. doi: 10.1128/jb.150.3.1414-1421.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Chung K. J., Rhodes J. C. Encapsulation and melanin formation as indicators of virulence in Cryptococcus neoformans. Infect Immun. 1986 Jan;51(1):218–223. doi: 10.1128/iai.51.1.218-223.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langford C. J., Gallwitz D. Evidence for an intron-contained sequence required for the splicing of yeast RNA polymerase II transcripts. Cell. 1983 Jun;33(2):519–527. doi: 10.1016/0092-8674(83)90433-6. [DOI] [PubMed] [Google Scholar]
- Le Chevanton L., Leblon G. The ura5 gene of the ascomycete Sordaria macrospora: molecular cloning, characterization and expression in Escherichia coli. Gene. 1989 Apr 15;77(1):39–49. doi: 10.1016/0378-1119(89)90357-0. [DOI] [PubMed] [Google Scholar]
- Mertins P., Gallwitz D. Nuclear pre-mRNA splicing in the fission yeast Schizosaccharomyces pombe strictly requires an intron-contained, conserved sequence element. EMBO J. 1987 Jun;6(6):1757–1763. doi: 10.1002/j.1460-2075.1987.tb02428.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfect J. R., Magee B. B., Magee P. T. Separation of chromosomes of Cryptococcus neoformans by pulsed field gel electrophoresis. Infect Immun. 1989 Sep;57(9):2624–2627. doi: 10.1128/iai.57.9.2624-2627.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen P., Bonekamp F., Jensen K. F. Structure of the Escherichia coli pyrE operon and control of pyrE expression by a UTP modulated intercistronic attentuation. EMBO J. 1984 Aug;3(8):1783–1790. doi: 10.1002/j.1460-2075.1984.tb02046.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes J. C., Kwon-Chung K. J. Production and regeneration of protoplasts from Cryptococcus. Sabouraudia. 1985 Feb;23(1):77–80. [PubMed] [Google Scholar]
- Rhodes J. C., Polacheck I., Kwon-Chung K. J. Phenoloxidase activity and virulence in isogenic strains of Cryptococcus neoformans. Infect Immun. 1982 Jun;36(3):1175–1184. doi: 10.1128/iai.36.3.1175-1184.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short J. M., Fernandez J. M., Sorge J. A., Huse W. D. Lambda ZAP: a bacteriophage lambda expression vector with in vivo excision properties. Nucleic Acids Res. 1988 Aug 11;16(15):7583–7600. doi: 10.1093/nar/16.15.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcq B., Bégueret J. The ura5 gene of the filamentous fungus Podospora anserina: nucleotide sequence and expression in transformed strains. Gene. 1987;53(2-3):201–209. doi: 10.1016/0378-1119(87)90008-4. [DOI] [PubMed] [Google Scholar]
- Wright A. P., Maundrell K., Shall S. Transformation of Schizosaccharomyces pombe by non-homologous, unstable integration of plasmids in the genome. Curr Genet. 1986;10(7):503–508. doi: 10.1007/BF00447383. [DOI] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]