Abstract
The genetic contribution to the development of bronchopulmonary dysplasia (BPD) in prematurely born infants is substantial, but information related to the specific genes involved is lacking. We conducted a case-control single nucleotide polymorphism (SNP) association study of candidate genes (n=601) or 6,324 SNPs in 1,091 prematurely born infants with gestational age <35 weeks, with or without neonatal lung disease including BPD. BPD was defined as need for oxygen at 28 days. Genotype analysis revealed, after multiple comparisons correction, two significant SNPs, rs3771150 (IL-18RAP) and rs3771171 (IL-18R1), in African Americans (AA) with BPD (vs. AA without BPD; q<0.05). No associations with Caucasian (CA) BPD, AA or CA RDS, or prematurity in either AA or CA, were identified with these SNPs. Respective frequencies were 0.098 and 0.093 without BPD and 0.38 for each SNP in infants with BPD. In the replication set (82 cases; 102 controls), the p-values were 0.012 for rs3771150 and 0.07 for rs3771171. Combining p-values using Fisher's method, overall p-values were 8.31E-07 for rs3771150, and 6.33E-06 for rs3771171. We conclude, IL-18RAP and IL-18R1 SNPs identify AA infants at risk for BPD. These genes may contribute to AA BPD pathogenesis via inflammatory-mediated processes and require further study.
INTRODUCTION
Respiratory distress syndrome (RDS), an acute lung disease occurring in neonates with surfactant deficiency, remains a leading cause of morbidity and mortality in prematurely born infants. The advent of surfactant replacement therapy has dramatically reduced the severity and improved the outcome of RDS, leading to increased survival of neonates born at earlier gestational ages with quite immature lungs. Lung function in survivors is variable with some infants emerging unscathed and others left with long-term pulmonary dysfunction. Collective improvements in obstetric and neonatal care during the past two decades have steadily increased survival of very low birth weight infants (1). This prolonged survival has lead to a gradual rise in the number of infants who develop bronchopulmonary dysplasia (BPD). Each year, between 5,000 and 10,000 prematurely born infants in U.S. are diagnosed with BPD, a form of chronic lung disease characterized by disordered lung growth and associated with respiratory and neurodevelopmental morbidities (2–4). Moreover, surviving prematurely born infants with or without BPD are at increased risk for respiratory disease hospitalizations (5), indicating a long term health disadvantage (6).
Recently, genetic variance has emerged as a significant risk factor for BPD development, accounting for as much as 82% of BPD risk among twins (7–9). Genetic variation in surfactant proteins genes and in SP-B linked microsatellites has been observed as susceptibility markers in BPD (10–13). These genetic variants may differentially affect innate immunity (14), inflammatory processes (15, 16), and/or surfactant-related functions (17, 18). Polymorphisms of MBL, MMP-16, extracellular matrix receptor (dystroglycan), and VEGF factor genes also associate with risk of BPD development among premature infants (19–22). These genetic variations may contribute to infection risk (19) and processes that lead to derangements in lung alveolarization or capillary growth (20, 22, 23). Identifying genetic markers and understanding mechanisms responsible for these variances has the potential to permit early identification of at-risk neonates and allow for experimental interventions in an attempt to decrease the long term pulmonary dysfunction. To date, however, only candidate gene approaches for a small number of polymorphisms have been undertaken.
In the present study, we used high-throughput technology to study SNP associations in a large number of candidate genes (n=601) that spanned all chromosomes with various subgroups of prematurely born infants (n=1,091). We hypothesized that genetic differences in the selected candidate genes identify prematurely born children who are at higher risk for the development of neonatal pulmonary disease.
METHODS
Study group
Following Institutional Review Board approval from the Human Subjects Protection Office at the Pennsylvania State University College of Medicine of a multi-site protocol, DNA samples from prematurely born infants with or without RDS, and/or with or without BPD (n=1,099), were prospectively collected from 1989–2008 and genotyped. Parents of subjects were approached if they had babies born prematurely or with neonatal lung disease (RDS and/or BPD), and informed consent was obtained. Patients treated with surfactant prophylactically were excluded, but patients who received surfactant therapy after diagnosis were included. Prenatal steroid therapy, gestational age (GA), birth weight (BW), the individual’s race admixture estimate, sex, maternal steroid treatment, and surfactant therapy were recorded. The GA and BW of each study group are shown in Table 1. BPD was defined as need for supplemental oxygen therapy at 28 days of life (24). This definition was chosen (as opposed to examining the diagnosis based on supplemental oxygen at 36 postmenstrual weeks) in order to focus on the dichotomous outcome of the diagnosis of BPD, as opposed to studying BPD severity. RDS was diagnosed by the neonatologist based on clinical criteria (grunting, retraction, and flaring), and verified by radiographic analysis (reticulogranular pattern). Eight samples were excluded based on the decision rule for admixture proportions, leaving 922 and 169 samples identified as CA and AA, respectively. Of these, 682 CA and 115 AA had complete data for determination of BPD and were used in the present analysis. The replication study consisted of 82 AA cases and 102 AA controls with mean GA for cases and controls of 26.64 and 30.15 weeks, respectively.
Table 1.
Comparisons of BW and GA between cases and controls in the different pairs of study groups
| Study group | n | BW | GA | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean | Min | Max | t-test p-value | Mean | Min | Max | t-test p-value | |||
| 1 | AA_BPD_Cas | 17 | 954.53 | 400 | 2184 | 1.34E-08 | 26.71 | 24.2 | 32.5 | 1.49E-09 |
| 2 | AA_BPD_Con | 98 | 1835.69 | 577 | 3416 | 32.49 | 25.0 | 39.4 | ||
| 3 | AA_RDS_Cas | 85 | 1223.12 | 400 | 3090 | 3.69E-12 | 28.85 | 24.2 | 37.0 | 4.18E-14 |
| 4 | AA_RDS_Con | 84 | 1920.62 | 666 | 3416 | 32.94 | 24.5 | 39.4 | ||
| 5 | CA_BPD_Cas | 153 | 1081.51 | 400 | 2378 | 2.2E-16 | 27.67 | 24.0 | 33.2 | 2.2E-16 |
| 6 | CA_BPD_Con | 529 | 1854.53 | 410 | 3930 | 32.13 | 24.0 | 40.4 | ||
| 7 | CA_RDS_Cas | 487 | 1532.65 | 400 | 4035 | 8.47E-16 | 29.99 | 22.0 | 40.4 | 2.2E-16 |
| 8 | CA_RDS_Con | 434 | 1910.30 | 620 | 4220 | 32.79 | 24.0 | 40.4 | ||
BPD_Cas: BPD cases; BPD_Con: BPD control=subjects without BPD; RDS_Cas: RDS cases; RDS_Con: RDS control=subjects without RDS. Comparisons of BW and GA between cases and controls in all pairs of study groups were statistically significant (p<0.001). The study group comparisons included study group 1 vs. 2; 3 vs. 4; 5 vs. 6; and 7 vs. 8
DNA preparation and genotyping
Genomic DNA was extracted with phenol/chloroform and quantified by Nanodrop. Samples with low DNA concentration (n=30) underwent whole genome amplification (WGA) by the WGA-REPLI-g kit (Qiagen, Velencia, CA). A fraction of the samples with adequate DNA concentration (n=61) were genotyped using genomic and WGA DNA. Of the initial 1,267 genotyped samples, only 32 failed, and of these only 5 were from WGA DNA. The Illumina high-throughput platform was used in two independent runs, by genotyping 57 samples prepared by different individuals and different protocols and concentrations ranging from 21–883 ng/µl. All but two of the test samples had gene call (GC) scores >0.99, a quality metric that indicates the reliability of the genotype called (maximum GC = 1.0).
Selection of targeted genes and SNPs
Candidate genes were chosen from expression profiles of animal models of neonatal lung disease and lung inflammation (25, 26), from NIH-sponsored Programs for Genomic Applications related to human lung disease development (http://innateimmunity.net, www.hopkins-genomics.org, http://pga.mbt.washington.edu), and by fulfilling at least one of the following criteria: a) involvement in innate immune response, inflammation, and tissue repair, and b) biologic plausibility in relation to lung disease development. Thus the candidate genes include genes of cytokines, inflammation, growth factors, antioxidants, cell adhesion receptors and proteins, apoptosis-associated proteins, cytoskeletal and mobility proteins, ion channels and transport proteins, receptors, proteases, transcription factors and DNA binding proteins, modulator-effectors-edema and water channels, coagulation and fibrinolysis, extracellular cell signaling and communication, protein turnover, and other miscellaneous genes. A current website that may include the information of the earlier website which we used in this study is http://public.nhlbi.nih.gov/GeneticsGenomics/home/.
Both non-synonymous and tagSNPs were selected from HapMap (www.hapmap.org) and studied for each target gene plus 5kb upstream and downstream of each target gene sequence, based on the quality metric (GC score ≥ 0.8, validation class ≥ 2). A tagging algorithm was then run using the selected SNPs with a minor allele frequency (MAF) of 0.2 for CA or MAF=0.4 for AA, and all the selected SNPs were assayed in genomic DNA samples from both CA and AA groups. The final list (n=6926) of SNPs selected included: a) Tag SNPs (n=4974). Of these 3789 were CEU and 1195 YRI; b) Non-synonymous SNPs (n=1392); and c) European stratification SNPs (n=560) (27). Of the 6926 SNPs, 450 (6.5%) failed primer manufacturing and 152 (2.3%) failed genotyping, and these were excluded.
Addressing population stratification
Our initial sample is a mixture of AA and CA. We used the method implemented in STRUCTURE (version 2.2) (28) on Ancestry Informative Markers (AIMS) designed to distinguish between CA and AA, and on AIMS by Seldin et al. (27) to distinguish between Northern and Southern European descent. The replication set consisted of all parent-reported race of AA.
Association testing and group comparison
We used linear trend test as implemented in PLINK (version 1.05) (29–31) in discovery and replication sets. The data sets analyzed included AA and CA groups using the admixture proportions identified by STRUCTURE as covariates. The latter association was tested using logistic regression. Welch’s t-test (32) was used to test for the difference in mean GA and mean BW between cases and controls.
Correction for multiple testing
To correct for multiple testing, we used the q-value method v1.1 developed by Storey and Tibshirani (33), and applied it to group’s set of p-values. Instead of controlling the probability of one or more false positives in a family of tests (the family-wise error rate), the q-value controls the expected proportion of false positives among all rejected hypotheses (the false discovery rate – FDR) (34). Q-value takes a given set of p values and, estimates the minimum FDR that is incurred when calling a particular test significant (the q-value of the test). An FDR of 0.05 is used as the significance level.
Replication study
We genotyped a second, distinct set of samples (n=194) derived from AA cases (44 males; 38 females) and controls (52 males; 50 females). Genomic DNA was amplified with the Taqman® universal PCR master mix and SNP genotyping assays (c__27514233_10 for rs3771171, and c__25808669_10 for rs3771150) (Applied Biosystems). Results were monitored by the ABI PRISM® 7900 sequence detection system (Applied Biosystems), and analyzed by allelic discrimination. No correction for multiple testing was performed for these two SNPs.
Computing combined p-values for discovery set and replication set
We used Fisher's method as implemented in PVALUES (35) to combine p-values across discovery and replication sets.
Linkage disequilibrium (LD) analysis
We used Haploview (36) to examine LD relationships for rs3771171 and rs3771150 in YRI. The SNP input information was obtained from HapMap, and the Yoruban population was chosen because of its closeness to AA. The Hedricks multiallelic association measure D’ was calculated for all pairs of SNPs by the confidence interval method (37).
Methods for data management are available online (Methods Supplement).
RESULTS
Population stratification
The initial study group of CA and AA, was stratified according to ethnicity using genetic markers. The decision rule was used, where individuals with admixture proportion values between 0.45–0.55 were excluded. Those with admixture proportion below 0.45 were placed into one group and anyone above 0.55 into the other group. Using this method we identified 922 CA and 169 AA, and 8 subjects were excluded (out of 1099).
The admixture testing for CA to distinguish between Northern and Southern European decent placed 897 in one group and 25 in another. Association testing was performed with all CA together and with stratified data using the covariates. Because one of the CA groups was small (n=25), no differences were observed with or without this admixture covariates. Therefore, we report data without the admixture adjustment.
Association testing
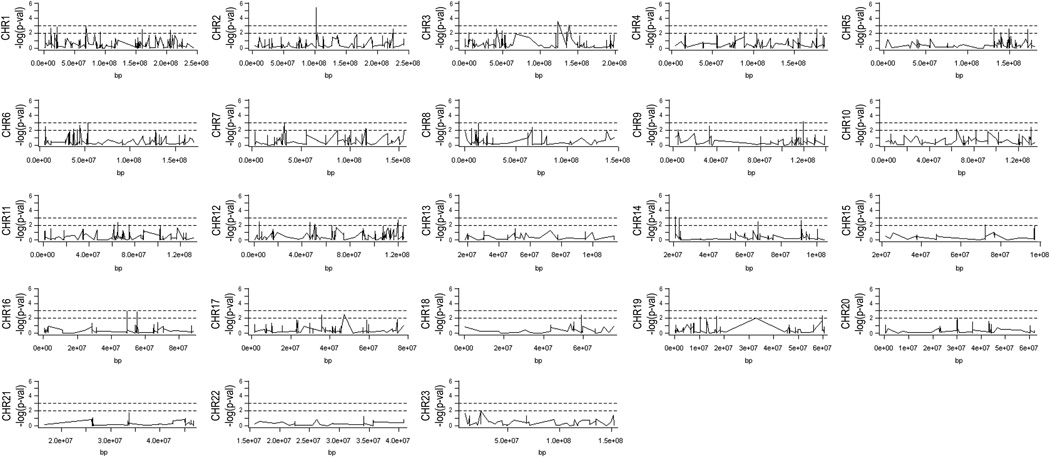
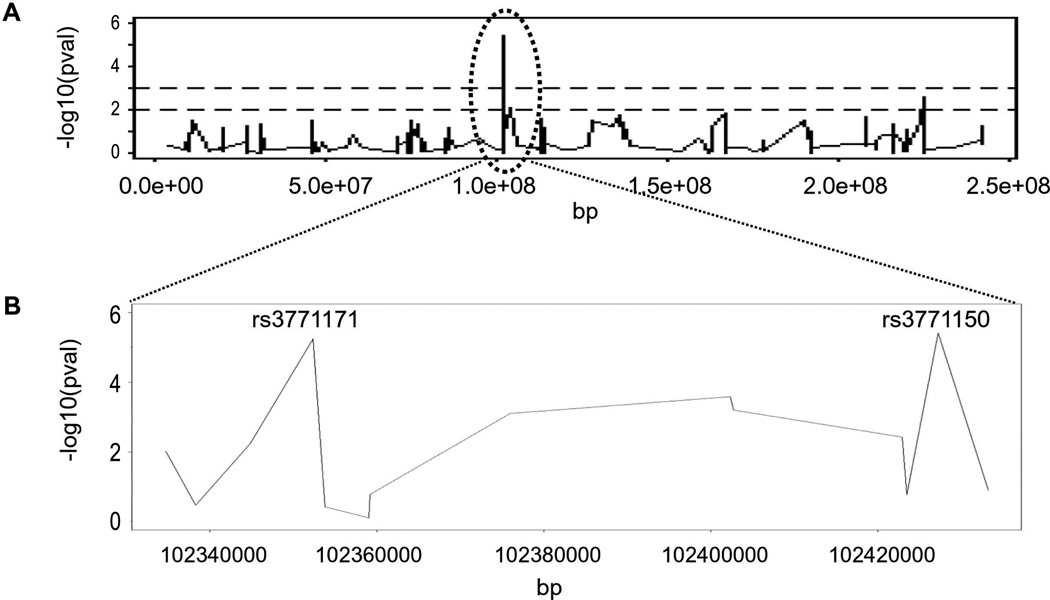
Association test analysis revealed significant findings after correcting for multiple testing only for the AA with or without BPD. A locus on chromosome 2 contained SNPs with a significantly higher frequency in AA with BPD (AA_BPD). No significant SNPs after correction for multiple testing were observed in other chromosomes (Figure 1). Upon enlargement of the significant chromosome 2 locus, two SNPs, rs3771150 and rs3771171 (approximately 80kb apart) were identified with uncorrected p-values of 3.87E-06 and 5.75E-06, respectively (Figure 2). After correction for multiple testing (33), these SNPs were significant at the 0.05 FDR level.
Figure 1.
Distribution of SNPs along chromosomes (CHR) 1–23. Vertical axis indicates the negative log10 p-value. The dotted lines represent thresholds (nominal levels) of p=0.01 (at 2) and p=0.001 (at 3). Horizontal axis indicates the CHR position in bp.
Figure 2.
A: Distribution of SNPs along chromosome 2. Dotted lines represent thresholds (nominal levels) of negative log10 p-values: p=0.01 (at 2) and p=0.001 (at 3). The circle depicts region containing the two significant SNPs, the enlargement of which is shown in B.
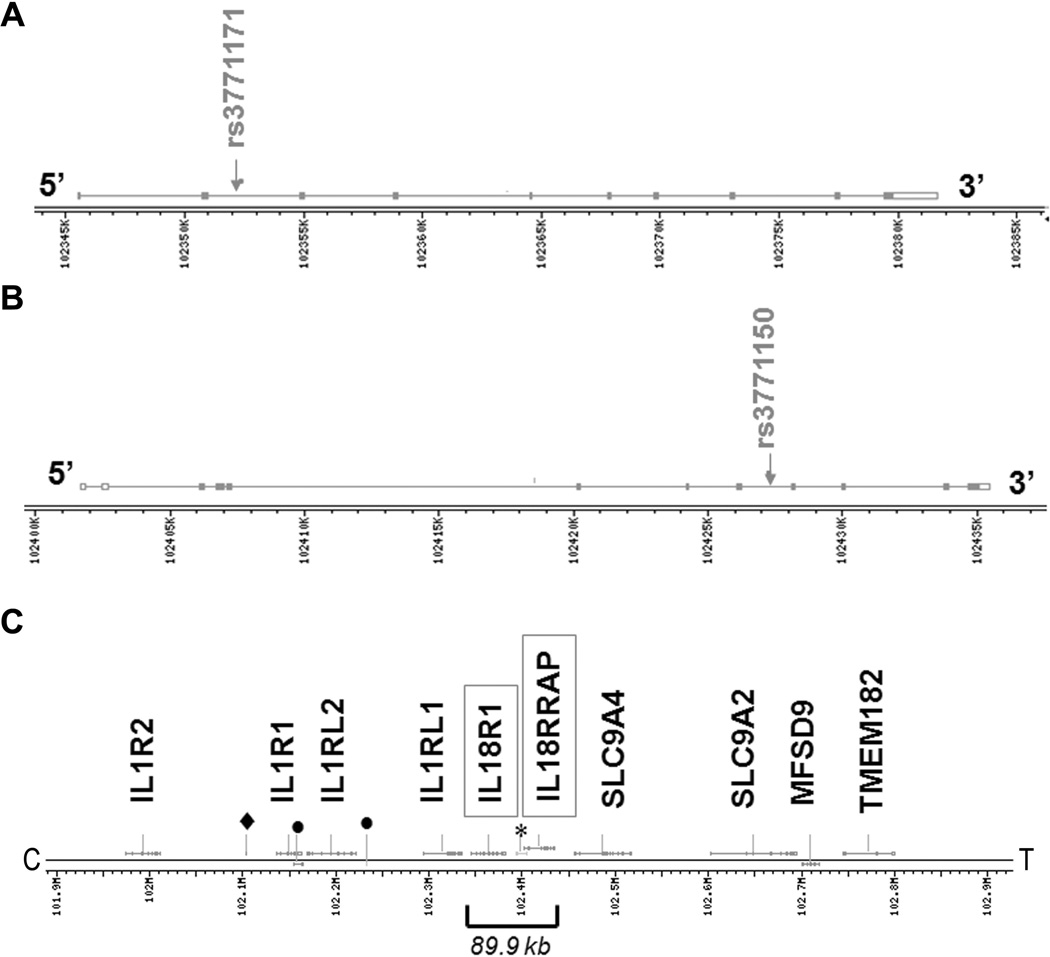
Complete data for determination of BPD were available for 115 AA infants (out of 169) and of these, 17 were identified with BPD (6 females and 11 males, and of these, 13 had been diagnosed with RDS) and 98 with no BPD (52 females, 45 males, 1 missing, and of these, 35 diagnosed with RDS). Of the 17 AA_BPD, all but 5 had at least one copy of the risk allele (one individual had no data for marker rs3771171). Marker rs3771150 has alleles C and T, where T is the minor allele with frequency 0.1213 based on the entire AA sample (n=169). MAF in affected and unaffected AA samples are 0.3824 and 0.0979, respectively (Table 2). The odds ratio for this marker is 5.7 with 95% confidence interval (2.5, 13.2), suggesting that AA individuals with the T allele are almost 6 times as likely to develop BPD as individuals with the C allele. Marker rs3771171 has alleles A and G, where G is the minor allele with frequency 0.1161. MAF in affected and unaffected AA samples are 0.375 and 0.0928, respectively (Table 2). The odds ratio for this marker is 5.9 with 95% confidence interval (2.4, 13.9), suggesting that AA individuals with the G allele are almost 6 times as likely to develop BPD as individuals with the A allele. Both SNPs, rs3771150 and rs3771171, are located within intron sequences of the interleukin 18 receptor accessory protein (IL-18RAP) and interleukin 18 receptor 1 (IL-18R1) genes, respectively (Figure 3A), and both genes are located in a region where other interleukin genes are found (Figure 3B). Moreover, SNPs genotyped in the present study that are 50kb upstream of one of the significant markers and 50kb downstream of the other are shown in Table 3. All the SNPs upstream of rs3771171 with the exception of rs974389, rs13015714, and rs3755276 reside within the IL-1RL1 gene, which mediates IL-33 signaling, and belongs to the same family as the IL-18R1 and IL-18RAP (38). These receptors share common signaling pathways, including that mediated via MyD88. The remaining SNPs (with the exception of rs11886793) reside within introns of either the IL-18R1 or the IL-18RAP gene.
Table 2.
Frequency of Significant Alleles in Discovery and Replication Sets
| SNP | CHR position |
Alleles | MAF in Discovery Set | p-value | MAF in Replication Set |
p-value | ||
|---|---|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | |||||
| rs3771150 | 102427283 | T/C | 0.3824 | 0.0979 | 3.87E-06 | 0.1375 | 0.0495 | 0.012 |
| rs3771171 | 102352382 | G/A | 0.375 | 0.0928 | 5.75E-06 | 0.1429 | 0.06566 | 0.07 |
Minor allele is underlined. CHR: chromosome. MAF: minor allele frequency.
Figure 3.
A–B: Location of each significant SNP within its respective gene (panel A: IL-18R1, panel B: IL-18RAP). Solid boxes: translated exons; open boxes: untranslated exons. C: Chromosome location of IL-18RAP and IL-18R1, and adjacent genes *similar to NAD(P) dependent steroid dehydrogenase-like protein; ● hypothetical protein; ♦ similar to AHPA9419.
Table 3.
SNPs †present upstream and downstream of the two significant SNPs (rs3771150 and rs3771171)
| CHR | SNP | CHR position | Alleles | Frequency in Cases | Frequency in Controls | p-value |
|---|---|---|---|---|---|---|
| 2 | rs974389 | 102303413 | C/T | 0.3235 | 0.3557 | 0.7116 |
| 2 | rs12712141 | 102319499 | C/T | 0.5294 | 0.4375 | 0.2933 |
| 2 | rs1041973 | 102321900 | A/C | 0.2812 | 0.5106 | 0.02247 |
| 2 | rs1420101 | 102324148 | A/G | 0.5882 | 0.3814 | 0.02044 |
| 2 | rs12712142 | 102327016 | A/C | 0.5588 | 0.3866 | 0.04901 |
| 2 | rs1921622 | 102332499 | A/G | 0.5588 | 0.2526 | 0.000525 |
| 2 | rs10208293 | 102332742 | G/A | 0.6176 | 0.4794 | 0.1404 |
| 2 | rs4988955 | 102334360 | A/G | 0.5 | 0.2835 | 0.009945 |
| 2 | rs4988956 | 102334439 | G/A | 0.5 | 0.2835 | 0.009945 |
| 2 | rs10192036 | 102334643 | C/A | 0.5 | 0.2835 | 0.009945 |
| 2 | rs10204137 | 102334644 | A/G | 0.5 | 0.2835 | 0.009945 |
| 2 | rs10192157 | 102334788 | C/T | 0.5 | 0.2835 | 0.009945 |
| 2 | rs10206753 | 102334794 | T/C | 0.5 | 0.2835 | 0.009945 |
| 2 | rs13015714 | 102338297 | G/T | 0.05882 | 0.1094 | 0.3399 |
| 2 | rs3755276 | 102344891 | G/A | 0.5882 | 0.3402 | 0.005714 |
| 2 | §rs3771171 | 102352382 | G/A | 0.375 | 0.09278 | 5.75E-06 |
| 2 | rs7579737 | 102353793 | G/A | 0.2353 | 0.3053 | 0.3804 |
| 2 | rs2270297 | 102359107 | A/G | 0.1176 | 0.1031 | 0.786 |
| 2 | rs7558013 | 102359238 | T/G | 0.1471 | 0.2577 | 0.1678 |
| 2 | rs4851004 | 102375969 | C/T | 0.5625 | 0.2553 | 0.000791 |
| 2 | rs2293225 | 102402321 | T/C | 0.2647 | 0.07216 | 0.000263 |
| 2 | rs6743516 | 102402767 | A/G | 0.5588 | 0.2577 | 0.000635 |
| 2 | rs13021177 | 102422925 | A/G | 0.6176 | 0.3802 | 0.003749 |
| 2 | rs17027173 | 102423475 | A/G | 0.1471 | 0.2552 | 0.1681 |
| 2 | §rs3771150 | 102427283 | T/C | 0.3824 | 0.09794 | 3.87E-06 |
| 2 | rs11465730 | 102433290 | A/G | 0.3235 | 0.4639 | 0.126 |
| 2 | rs11886793 | 102438652 | G/T | 0.2647 | 0.3802 | 0.1891 |
These are within 50kb upstream and downstream the two significant SNPs. Their chromosome (CHR) position, frequency in cases and controls, and p-values are shown;
The two significant SNPs are shown in bold; minor alleles are underlined.
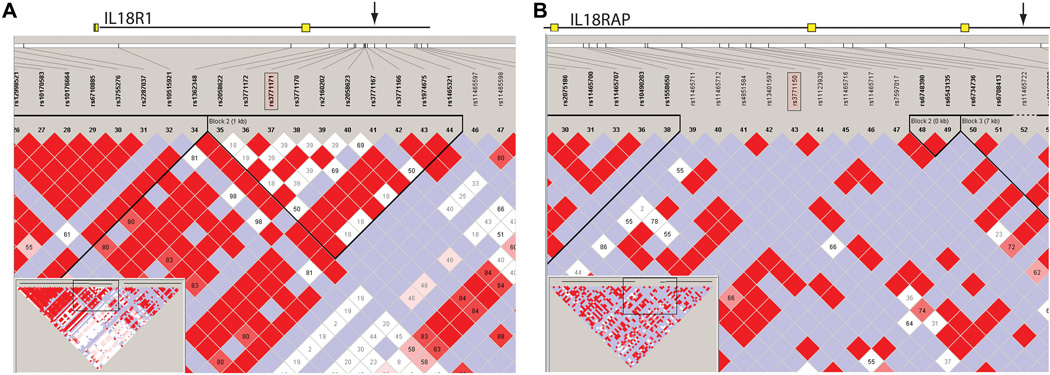
To determine whether these SNPs were located in regions of LD, we performed a haplotypic analysis. Figure 4 displays the haplotype block structure for IL-18R1 and IL-18RAP, as depicted by Haploview (36). As seen in Figure 4, rs3771171 is located in a region of strong LD with other regions of the IL-18R1 gene, raising the possibility that this SNP, rather than being of pathogenic significance itself, is a marker for another polymorphism that confers altered risk. In contrast, rs3771150 is not in a region of high LD.
Figure 4.
Haplotypic analysis (LD plots) of IL-18R1 and IL-18RAP genes obtained with Haploview and HapMap data from YRI, showing the region surrounding the SNPs of interest (pink rectangles).
GA and BW in AA_BPD and controls
Significant differences (p<0.001) in GA and BW were observed between cases and controls. To determine whether the significant SNP frequency differences described reflect GA or BW differences between cases and controls, we performed comparisons of groups of prematurely born infants with BPD or RDS from AA or CA descent (Table 1). Regardless of race, BPD and RDS cases were identified with significantly lower BW and GA compared to their corresponding controls (Table 1). However, frequency comparisons of the two aforementioned significant SNPs between cases and controls in each set depicted (Table 1), showed significant differences only for the AA_BPD group.
Replication Study
The replication p-values were 0.012 for rs3771150 and 0.07 for rs3771171 (Table 2). Power and minimum sample size calculations were performed by methods implemented in PAWE (39, 40). For 80% power and 0.05 significance level, under a genetic model free method, the minimal sample sizes are 139 cases and 184 controls for rs3771150, and 180 cases and 238 controls for rs3771171, respectively. The replication study did not reach the minimal required number for 80% power. However, the combined p-values for both studies remained significant for both SNPs: 8.31E-07 (rs3771150) and 6.33E-06 (rs3771171).
DISCUSSION
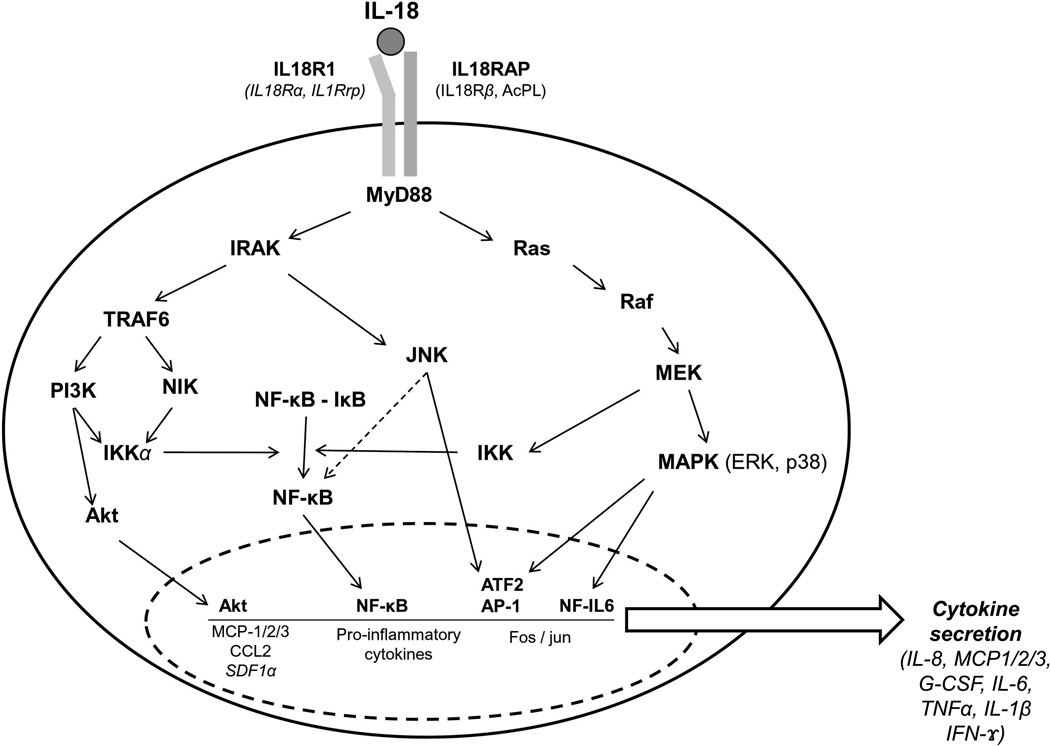
BPD is characterized by arrested lung development whereby alveolar septation and pulmonary vascularization are impaired culminating in a reduction in alveolar number and abnormal pulmonary microvasculature (41, 42). Both prenatal and postnatal factors, including infection and the requirement of mechanical ventilation, stimulate inflammatory processes that contribute to the BPD phenotype. Studies of genetic factor contributions have identified associations (10–13, 19, 20, 22, 23) of polymorphisms with BPD in genes involved in processes known to be deranged in BPD. Given the potential complexity of mechanisms and number of genes that may contribute to BPD, we used a high-throughput approach to study genetic associations of a large number of candidate genes and their impact on BPD development The analysis revealed two SNPs corresponding to IL-18R1 and IL-18RAP that were associated with BPD development in AA infants. The products of these genes are necessary to effectively mediate interleukin-18 (IL-18) signal transduction and subsequent activation of NF-κB and MAPK8 (JNK) pathways in response to IL-18 (Figure 5). There is limited information in the literature about association of IL-18R1, IL-18RAP, or SNPs present in these two genes with disease. Thus, to the best of our knowledge, this is the first study that describes an association of rs3771150 and rs3771171 with disease in AA.
Figure 5.
IL-18-induced signaling via IL-18RAP and IL-18R1 results in cytokine secretion, a number of which (IL-8, MCP-1/2/3, G-CSF and IL-6) have been associated with BPD.
Although the role of these genes in BPD has not been previously investigated at the genetic level, we postulate that altered expression of several pro-inflammatory cytokines in response to IL-18 contributes to BPD. Regulation of cytokine gene expression and secretion is critical in the inflammatory response, and a variety of cytokines have been reported to be stimulated by IL-18 (43). Several of the IL-18-stimulated proinflammatory cytokines in Figure 5 have been found to be increased in BPD (43–45). However, further studies are needed to assess the plausibility of the scenario presented in Figure 5.
The IL-18R1 gene intron 2 SNP rs3771171 is found at similar frequencies between our AA subjects and the YRI population in HapMap. Interestingly, ancestral allele frequency is somewhat lower in other populations. The SNP in IL-18RAP, however, is present in markedly different frequencies in different populations. The ancestral allele is found in 92% of the YRI and our AA populations, but in other populations (Papua New Guinea) is as low as 27%. The Yoruban population is of interest as it is the HapMap population most closely related to AA. Thus, the frequencies of these alleles in AA group, although similar to the “parent” population, can differ in other populations, underscoring the importance of ethnicity considerations in genetic study associations.
The incidence of BPD development is indirectly correlated with GA and BW (24). Although the incidence of BPD was somewhat higher in the CA group, this trend was not significant (CA: 22.4%; AA: 14.8%, p=0.103, z-test). However, because BPD and control groups differed significantly in both BW and GA, it was necessary to ascertain whether the two significant SNPs were markers of prematurity or BPD. Comparison of four sets of groups of prematurely born CA or AA infants with lung disease (RDS or BPD) versus their corresponding controls revealed significant BW and GA differences in each set (case, control) (Table 1). However, the frequency of each of the two SNPs did not differ between cases and controls except for the AA_BPD group. Therefore, it is unlikely that the two SNPs associated with risk in AA_BPD are markers of prematurity.
As with many genetic epidemiology studies using the candidate gene approach, replication with a distinct study population is necessary. However, when studying a more rare disease (such as BPD) in a single race (such as AA), collection of replication samples may take years. However, we were able to form a new collaboration to attempt to replicate the results. Because the absolute (but not the relative) differences in MAF of the two studied alleles differ between the discovery and the replication studies, it is possible that unrecognized confounding factors exist between the two study groups. Moreover, by limiting our SNPs selection and not performing a genome-wide SNP analysis, we may have missed SNPs important for BPD. Nonetheless, these results should be tested by different investigators before being used to guide targeted interventions that may carry some risk. Furthermore, changes in the clinical definition of BPD over the period of sample collection, lack of knowledge of oxygen saturation limits or other management strategies, could potentially limit the application of these findings. However, the nearly significant association in the replication group indicates that changes in clinical management may not be a limitation factor.
While this manuscript was in preparation, a report was published of association studies between IL-18 SNPs and BPD in CA and between IL-18 SNPs and prematurity in CA (46). The authors found no correlation between IL-18 SNPs and BPD or prematurity in CA, an observation similar to our findings with the two IL-18 receptor molecules (Figure 5). These findings indicate that IL-18, and the cell surface molecules with which it interacts (IL-18R1, IL-18RAP), do not contribute to the genetics of either BPD or prematurity in CA. Within the AA population, however, the SNPs described in this report may contribute to the pathogenesis of BPD and warrants further investigation.
In summary, we identified two SNPs associated with BPD risk in AA. The involvement of IL-18R1 and IL-18RAP further supports a role of inflammation in BPD pathogenesis.
Supplementary Material
Acknowledgements
The authors thank Rosanne Pogash and Mark Hansen for valuable input, Anamika Singh for help with DNA extraction, Julie Graham and Gina Marcucci for typing, and the Pennsylvania State University College of Medicine Macromolecular core facility for Real Time PCR.
Supported by NIH HL-34788 [J.F.], R01HL71113 and R01HL87166 [R.M.V.] grants.
Abbreviations
- AA
African American
- AA_BPD
AA with BPD
- BPD
Bronchopulmonary Dysplasia
- BW
Birth weight
- CA
Caucasian
- FDR
False discovery rate
- GC
Gene call
- IL-18R1
Interleukin-18 Receptor 1
- IL-18RAP
Interleukin-18 receptor accessory protein
- LD
Linkage Disequilibrium
- MAF
Minor allele frequency
- SNP
Single nucleotide polymorphism
REFERENCES
- 1.Horbar JD, Badger GJ, Carpenter JH, Fanaroff AA, Kilpatrick S, LaCorte M, Phibbs R, Soll RF. Trends in mortality and morbidity for very low birth weight infants, 1991–1999. Pediatrics. 2002;110:143–151. doi: 10.1542/peds.110.1.143. [DOI] [PubMed] [Google Scholar]
- 2.Baraldi E, Filippone M. Chronic lung disease after premature birth. N Engl J Med. 2007;357:1946–1955. doi: 10.1056/NEJMra067279. [DOI] [PubMed] [Google Scholar]
- 3.Short EJ, Kirchner HL, Asaad GR, Fulton SE, Lewis BA, Klein N, Eisengart S, Baley J, Kercsmar C, Min MO, Singer LT. Developmental sequelae in preterm infants having a diagnosis of bronchopulmonary dysplasia: analysis using a severity-based classification system. Arch Pediatr Adolesc Med. 2007;161:1082–1087. doi: 10.1001/archpedi.161.11.1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vrijlandt EJ, Boezen HM, Gerritsen J, Stremmelaar EF, Duiverman EJ. Respiratory health in prematurely born preschool children with and without bronchopulmonary dysplasia. J Pediatr. 2007;150:256–261. doi: 10.1016/j.jpeds.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 5.Walter EC, Ehlenbach WJ, Hotchkin DL, Chien JW, Koepsell TD. Low birth weight and respiratory disease in adulthood: a population-based case-control study. Am J Respir Crit Care Med. 2009;180:176–180. doi: 10.1164/rccm.200901-0046OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greenough A. Does low birth weight confer a lifelong respiratory disadvantage? Am J Respir Crit Care Med. 2009;180:107–108. doi: 10.1164/rccm.200904-0643ED. [DOI] [PubMed] [Google Scholar]
- 7.Hallman M, Marttila R, Pertile R, Ojaniemi M, Haataja R. Genes and environment in common neonatal lung disease. Neonatology. 2007;91:298–302. doi: 10.1159/000101345. [DOI] [PubMed] [Google Scholar]
- 8.Lavoie PM, Pham C, Jang KL. Heritability of bronchopulmonary dysplasia, defined according to the consensus statement of the National Institutes of Health. Pediatrics. 2008;122:479–485. doi: 10.1542/peds.2007-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meng H, Gruen JR. Genetic approaches to complications of prematurity. Front Biosci. 2007;12:2344–2351. doi: 10.2741/2236. [DOI] [PubMed] [Google Scholar]
- 10.Pavlovic J, Papagaroufalis C, Xanthou M, Liu W, Fan R, Thomas NJ, Apostolidou I, Papathoma E, Megaloyianni E, DiAngelo S, Floros J. Genetic variants of surfactant proteins A, B, C, and D in bronchopulmonary dysplasia. Dis Markers. 2006;22:277–291. doi: 10.1155/2006/817805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weber B, Borkhardt A, Stoll-Becker S, Reiss I, Gortner L. Polymorphisms of surfactant protein A genes and the risk of bronchopulmonary dysplasia in preterm infants. Turk J Pediatr. 2000;42:181–185. [PubMed] [Google Scholar]
- 12.Rova M, Haataja R, Marttila R, Ollikainen V, Tammela O, Hallman M. Data mining and multiparameter analysis of lung surfactant protein genes in bronchopulmonary dysplasia. Hum Mol Genet. 2004;13:1095–1104. doi: 10.1093/hmg/ddh132. [DOI] [PubMed] [Google Scholar]
- 13.Makri V, Hospes B, Stoll-Becker S, Borkhardt A, Gortner L. Polymorphisms of surfactant protein B encoding gene: modifiers of the course of neonatal respiratory distress syndrome? Eur J Pediatr. 2002;161:604–608. doi: 10.1007/s00431-002-1046-1. [DOI] [PubMed] [Google Scholar]
- 14.Mikerov A, Umstead T, Gan X, Huang W, Guo X, Wang G, Phelps D, Floros J. Impact of ozone exposure on the phagocytic activity of human surfactant protein A (SP-A) and SP-A variants. Am J Physiol Lung Cell Mol Physiol. 2008;294:L121–L130. doi: 10.1152/ajplung.00288.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang G, Umstead TM, Phelps DS, Al-Mondhiry H, Floros J. The effect of ozone exposure on the ability of human surfactant protein a variants to stimulate cytokine production. Environ Health Perspect. 2002;110:79–84. doi: 10.1289/ehp.0211079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang W, Wang G, Phelps DS, Al-Mondhiry H, Floros J. Human SP-A genetic variants and bleomycin-induced cytokine production by THP-1 cells: effect of ozone-induced SP-A oxidation. Am J Physiol Lung Cell Mol Physiol. 2004;286:L546–L553. doi: 10.1152/ajplung.00267.2003. [DOI] [PubMed] [Google Scholar]
- 17.Wang G, Taneva S, Keough KM, Floros J. Differential effects of human SP-A1 and SP-A2 variants on phospholipid monolayers containing surfactant protein B. Biochim Biophys Acta. 2007;1768:2060–2069. doi: 10.1016/j.bbamem.2007.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang G, Bates-Kenney SR, Tao JQ, Phelps DS, Floros J. Differences in biochemical properties and in biological function between human SP-A1 and SP-A2 variants, and the impact of ozone-induced oxidation. Biochemistry. 2004;43:4227–4239. doi: 10.1021/bi036023i. [DOI] [PubMed] [Google Scholar]
- 19.Hilgendorff A, Heidinger K, Pfeiffer A, Bohnert A, Konig IR, Ziegler A, Merz C, Frey G, Chakraborty T, Gortner L, Bein G. Association of polymorphisms in the mannose-binding lectin gene and pulmonary morbidity in preterm infants. Genes Immun. 2007;8:671–677. doi: 10.1038/sj.gene.6364432. [DOI] [PubMed] [Google Scholar]
- 20.Hadchouel A, Decobert F, Franco-Montoya ML, Halphen I, Jarreau PH, Boucherat O, Martin E, Benachi A, Amselem S, Bourbon J, Danan C, Delacourt C. Matrix metalloproteinase gene polymorphisms and bronchopulmonary dysplasia: identification of MMP16 as a new player in lung development. PLoS One. 2008;3:e3188. doi: 10.1371/journal.pone.0003188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abman SH. Impaired vascular endothelial growth factor signaling in the pathogenesis of neonatal pulmonary vascular disease. Adv Exp Med Biol. 2010;661:323–335. doi: 10.1007/978-1-60761-500-2_21. [DOI] [PubMed] [Google Scholar]
- 22.Concolino P, Capoluongo E, Santonocito C, Vento G, Tana M, Romagnoli C, Zuppi C, Ameglio F, Brancaccio A, Sciandra F. Genetic analysis of the dystroglycan gene in bronchopulmonary dysplasia affected premature newborns. Clin Chim Acta. 2007;378:164–167. doi: 10.1016/j.cca.2006.11.016. [DOI] [PubMed] [Google Scholar]
- 23.Kwinta P, Bik-Multanowski M, Mitkowska Z, Tomasik T, Legutko M, Pietrzyk JJ. Genetic risk factors of bronchopulmonary dysplasia. Pediatr Res. 2008;64:682–688. doi: 10.1203/PDR.0b013e318184edeb. [DOI] [PubMed] [Google Scholar]
- 24.Jobe A. The new BPD: an arrest of lung development. Pediatr Int. 1999:641–643. doi: 10.1203/00006450-199912000-00007. [DOI] [PubMed] [Google Scholar]
- 25.Banerjee SK, Young HW, Volmer JB, Blackburn MR. Gene expression profiling in inflammatory airway disease associated with elevated adenosine. Am J Physiol Lung Cell Mol Physiol. 2002;282:L169–L182. doi: 10.1152/ajplung.00243.2001. [DOI] [PubMed] [Google Scholar]
- 26.Wagenaar GT, ter Horst SA, van Gastelen MA, Leijser LM, Mauad T, van der Velden PA, de Heer E, Hiemstra PS, Poorthuis BJ, Walther FJ. Gene expression profile and histopathology of experimental bronchopulmonary dysplasia induced by prolonged oxidative stress. Free Radic Biol Med. 2004;36:782–801. doi: 10.1016/j.freeradbiomed.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 27.Seldin MF, Shigeta R, Villoslada P, Selmi C, Tuomilehto J, Silva G, Belmont JW, Klareskog L, Gregersen PK. European population substructure: clustering of northern and southern populations. PLoS Genet. 2006;2:e143. doi: 10.1371/journal.pgen.0020143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pritchard JK, Rosenberg NA. Use of unlinked genetic markers to detect population stratification in association studies. Am J Hum Genet. 1999;65:220–228. doi: 10.1086/302449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cochran WG. Some methods for strengthening the common chi-squared tests. Biometrics. 1954;10:417–451. [Google Scholar]
- 30.Armitage P. Tests for linear trends in proportions and frequencies. Biometrics. 1955;11:375–386. [Google Scholar]
- 31.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Welch BL. The generalisation of student's problems when several different population variances are involved. Biometrika. 1947;34:28–35. doi: 10.1093/biomet/34.1-2.28. [DOI] [PubMed] [Google Scholar]
- 33.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 35.Ott J. Analysis of Human Genetic Linkage. Baltimore: Johns Hopkins University Press; 1999. [Google Scholar]
- 36.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 37.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 38.Kakkar R, Lee RT. The IL-33/ST2 pathway: therapeutic target and novel biomarker. Nat Rev Drug Discov. 2008;7:827–840. doi: 10.1038/nrd2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gordon D, Finch SJ, Nothnagel M, Ott J. Power and sample size calculations for case-control genetic association tests when errors are present: application to single nucleotide polymorphisms. Hum Hered. 2002;54:22–33. doi: 10.1159/000066696. [DOI] [PubMed] [Google Scholar]
- 40.Gordon D, Levenstien MA, Finch SJ, Ott J. Errors and linkage disequilibrium interact multiplicatively when computing sample sizes for genetic case-control association studies. Pac Symp Biocomput. 2003:490–501. doi: 10.1142/9789812776303_0046. [DOI] [PubMed] [Google Scholar]
- 41.Coalson JJ. Pathology of new bronchopulmonary dysplasia. Semin Neonatol. 2003;8:73–81. doi: 10.1016/s1084-2756(02)00193-8. [DOI] [PubMed] [Google Scholar]
- 42.Bhandari A, Bhandari V. Pitfalls, problems, and progress in bronchopulmonary dysplasia. Pediatrics. 2009;123:1562–1573. doi: 10.1542/peds.2008-1962. [DOI] [PubMed] [Google Scholar]
- 43.Tsuji-Takayama K, Aizawa Y, Okamoto I, Kojima H, Koide K, Takeuchi M, Ikegami H, Ohta T, Kurimoto M. Interleukin-18 induces interferon-gamma production through NF-kappaB and NFAT activation in murine T helper type 1 cells. Cell Immunol. 1999;196:41–50. doi: 10.1006/cimm.1999.1542. [DOI] [PubMed] [Google Scholar]
- 44.Bose CL, Dammann CE, Laughon MM. Bronchopulmonary dysplasia and inflammatory biomarkers in the premature neonate. Arch Dis Child Fetal Neonatal Ed. 2008;93:F455–F461. doi: 10.1136/adc.2007.121327. [DOI] [PubMed] [Google Scholar]
- 45.Paananen R, Husa AK, Vuolteenaho R, Herva R, Kaukola T, Hallman M. Blood cytokines during the perinatal period in very preterm infants: relationship of inflammatory response and bronchopulmonary dysplasia. J Pediatr. 2009;154:39.e33–43.e33. doi: 10.1016/j.jpeds.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 46.Krueger M, Heinzmann A, Mailaparambil B, Hartel C, Gopel W. Polymorphisms of interleukin 18 in the genetics of preterm birth and bronchopulmonary dysplasia. Arch Dis Child Fetal Neonatal Ed. 2010 doi: 10.1136/adc.2009.174862. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.