Abstract
Study background
DEFB4/103A encoding β-defensin 2 and 3, respectively, inhibit CXCR4-tropic (X4) viruses in vitro. We determined whether DEFB4/103A Copy Number Variation (CNV) influences time-to-X4 and time-to-AIDS outcomes.
Methods
We utilized samples from a previously published Multicenter AIDS Cohort Study (MACS), which provides longitudinal account of viral tropism in relation to the full spectrum of rates of disease progression. Using traditional models for time-to-event analysis, we investigated association between DEFB4/103A CNV and the two outcomes, and interaction between DEFB4/103A CNV and disease progression groups, Fast and Slow.
Results
Time-to-X4 and time-to-AIDS were weakly correlated. There was a stronger relationship between these two outcomes for the fast progressors. DEFB4/103A CNV was associated with time-to-AIDS, but not time-to-X4. The association between higher DEFB4/103A CNV and time-to-AIDS was more pronounced for the slow progressors.
Conclusion
DEFB4/103A CNV was associated with time-to-AIDS in a disease progression group-specific manner in the MACS cohort. Our findings may contribute to enhancing current understanding of how genetic predisposition influences AIDS progression.
Keywords: AIDS progression, β-Defensin, DEFB4, DEFB103A, MACS, X4 emergence
Introduction
The antimicrobial peptides human β-defensin 2 (hBD-2) and -3, encoded by DEFB4 and DEFB103A, respectively, are induced during infections and inflammation, and can reach biologically relevant concentrations in the systemic circulation [1–4]. To date, all evidence suggests that the β-defensin gene cluster on chromosome 8p23.1, carrying DEFB4 and DEFB103A, varies en bloc, with copy numbers ranging from 2 to 12 per diploid genome (PDG) [5,6]. DEFB4 Copy Number Variation (CNV) correlates with its transcript levels [6], and with hBD-2 serum concentrations [2]. Recently, we demonstrated that: (1) CCR5-tropic (R5) and CXCR4-tropic (X4) strains of HIV- 1 induce DEFB4 and DEFB103A transcripts in normal human oral epithelial cells; (2) hBD-2 and -3 inhibit HIV-1 infection by both strains, with greater activity against X4 viruses; and (3) this inhibition is due to direct binding with virions, and through down-modulation of the CXCR4 co-receptor [7]. A later study confirmed the anti-HIV activity of these hBDs [8]. We subsequently discovered that hBD-3 competes with stromal-derived factor 1 (SDF-1), the natural ligand for CXCR4, and acts as an antagonist of CXCR4 on T cells by promoting its internalization without subsequent activation of the cell [9].
Chemokine receptor usage (viral tropism), a complex phenomenon where many aspects are still unresolved, plays a major role in HIV-1 disease pathogenesis. It is generally accepted that R5 viruses predominate at early stages of infection, whereas X4 or R5/X4 (dual- or mixed-tropic) viruses tend to emerge at later stages and their emergence appears to be associated with accelerated progression to disease [10–12]. Viral subtype, drug resistance, and impact of highly active antiretroviral therapy (HAART) may be important factors in influencing viral tropism [13–17]. However, the role of host genetic factors, to our knowledge, is not known. Based on our previous findings [7,9], we hypothesized that there may be an association between DEFB4/103A CNV and time to emergence of X4 viruses (time-to-X4), and thus between DEFB4/103A CNV and time to AIDS. To test our hypothesis, we utilized samples from the previously published Multicenter AIDS Cohort Study (MACS), which provides a longitudinal account of viral tropism in relation to rates of HIV-1 disease progression [18].
Materials and Methods
Study population
De-identified packed white blood cell pellets (n = 50) were obtained from the MACS. These samples were part of the study, conducted on HIV-1 seroconverters (n = 67) for whom emergence of X4 viruses was monitored and used to examine whether X4 emergence before onset of AIDS was related to the rate of AIDS progression [18]. The baseline characteristics of this study population have been previously described [18]. The self-identified racial/ethnic distribution of the 50 subjects was: white, non-Hispanic (White, n = 43), white, Hispanic (Hispanic, n = 4), and black, non-Hispanic (Black, n = 3), which reflects the fact that the MACS cohort is predominantly white (Caucasian). For these subjects, the information regarding their dates of seroconversion, emergence of X4 viruses, defined as the detection of R5/X4 viruses, and AIDS diagnosis was made available for further analysis (Table 1 and Supplemental Figure 1).
Table 1.
Racial/ethnic distribution and clinical information of study subjects.
| Race/ethnicity | X4 emergence | AIDS | |||||
|---|---|---|---|---|---|---|---|
| n | TTX | n | TTA | ||||
| n | + | − | Years* | + | − | Years* | |
| White/non-Hispanic | 43 | 29 | 14 | [0.99, 9.93] | 43 | 0 | [0.99, 14.33] |
| White/Hispanic | 4 | 2 | 2 | [0.95, 1.45] | 4 | 0 | [0.95, 8.18] |
| Black/non-Hispanic | 3 | 1 | 2 | 3.68 | 2 | 1 | [3.68, 6.08] |
| Total | 50 | 32 | 18 | [0.95, 9.93] | 49 | 1 | [0.95, 14.33] |
+ = event present, − = event absent
TTX = Time-to-X4
TTA = Time-to-AIDS
range, in subjects with events
Disease progression groups
Previously, the rate of AIDS progression was defined in terms of the interval (t) between seroconversion and onset of an AIDS-defining event (CD4+ T cell count of <200 cells/µL on two consecutive visits and/ or an AIDS-defining illness) [18]. AIDS progression was considered Rapid (t ≤ 5 years), Moderate (5 < t ≤ 9 years), Slow (9 < t ≤ 11 years), and Very Slow (t > 11 years) [18]. To increase statistical power, the disease progression groups were merged by similarities of CD4+ and CD3+ T cell count profiles and HIV tropism patterns (Figure 1 of [18]). We thus identified a “Fast” group (t ≤ 9 years, n = 36) after merging the Rapid and Moderate groups, and a “Slow” group (t > 9 years, n = 14) after merging the Slow and Very Slow groups.
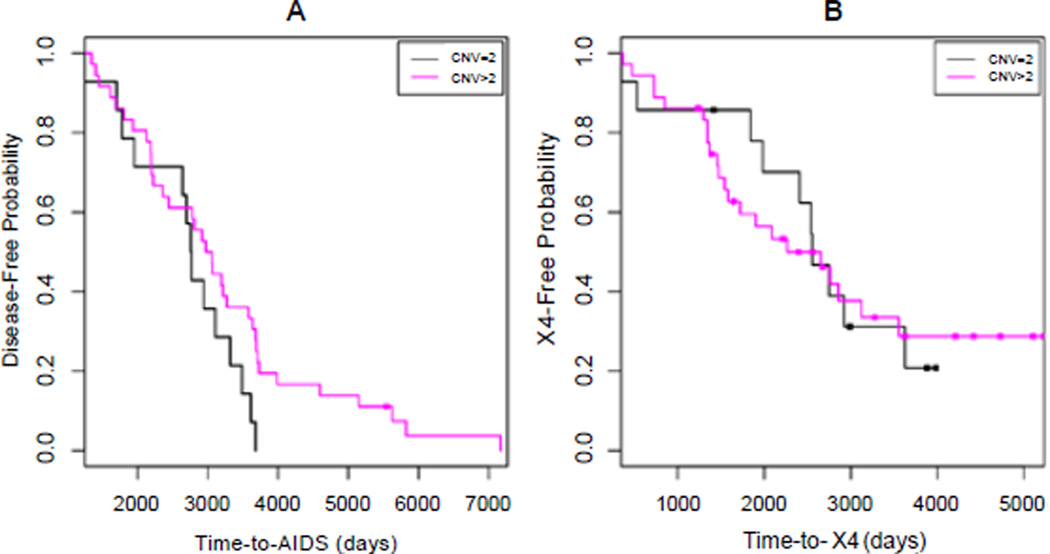
Figure 1.
Kaplan-Meier plots of aggregated DEFB4/103A copy number effect on time-to-AIDS (A) and time-to-X4 (B).
Determination of CNV
Genomic DNAs were extracted from 200 µl of each of the pellets using the QIAamp DNA Blood Mini Kit (Qiagen). For the determination of DEFB4 and DEFB103A copy numbers, the real-time quantitative PCR assay was used as described [19]. Reference genes TBP (TATA-Box Binding Protein, GenBank accession #AL031259) and DEFB1 (hBD-1, GenBank accession #NT_023736), specific primer sets producing only one specific product of ~150 bp at 54°C annealing temperature, reaction mix, and conditions were used as described [19], and the Bio-Rad CFX96™ system (Bio-Rad Laboratories) was used for the PCR analysis. Each sample was run in triplicate. Data were analyzed by the comparative Ct method, and the copy numbers were calculated as described [19]. Recently, we assessed the reliability and validity of this assay using 151 multi-population samples, which included 46 well-characterized samples from five diverse populations from the Coriell Cell Repositories [20].
Statistical analysis
All 50 subjects were assumed independent from each other. Missing DEFB4/103A copy numbers (n = 5) were determined by simple-imputation results from the expectation-maximization algorithm, assuming missing data at random [21]. The missing CNVs were imputed by using other genotype data (CCR5 -2459G>A, CCR5 32-bp deletion, CCR2 190G>A and CXCL12 801G>A [unpublished]), as well as disease progression groups and race. Initial exploratory data analyses to compare trends between the two time-to-event outcomes, and to investigate a trend in a given time-to-event as a function of CNVs were carried out with a standard non-parametric trend test requiring minimal assumptions (1-sided Cox-Stuart test) [22]. For measuring the monotonic association between the two right-censored time to events, a non-parametric partial correlation coefficient (partial Kendall’s τ) was used [23].
We used the right-censoring modeling framework of risk or survival analysis to model either time-to-X4 or time-to-AIDS outcome, for which the event was either X4 emergence or AIDS diagnosis, each of which was observed or not during each patient’s follow-up time (Table 1,Supplemental Figure 1). We assumed non-informative (or random) censoring and non-competing risks for the two outcomes (e.g., Supplemental Figure 2). We used the non-parametric Kaplan- Meier estimator [24] to model the relationship of the Disease-Free (DF) or X4-Free (XF) probability as a function of the right-censored time-to-event outcome of interest. We report from Kaplan-Meier models the time-to-event at median DF or XF probability and log-rank test results adjusting for race/ethnicity. We also used Cox proportional hazards modeling (Cox model) [25] adjusting for race/ethnicity.
For all statistical analyses, differences were considered significant at the α = 0.05 level. All statistical modeling, computations, and plotting were performed in the R language and environment for statistical computing (http://www.r-project.org/). We used our own code in addition to the following R packages: “survival” for Kaplan-Meier and Cox regression modeling and “NADA” for correlation analysis in the presence of censoring (http://cran.r-project.org/).
Results
Distribution of CNV
The DEFB4/103A copy numbers ranged from 2 to 5 (median = 3). The distribution of copy numbers was: 2 (n = 13), 3 (n = 11), 4 (n = 17) and 5 (n = 4). The copy numbers could not be determined for five samples due to low genomic DNA concentrations (<50 ng). After imputation, the distribution of copy numbers was: 2 (n = 14), 3 (n = 13), 4 (n = 19), and 5 (n = 4). To increase statistical power, we aggregated DEFB4/103A CNV into either two categories: a “minimal” (CNV = 2, n = 14) and a “higher” (CNV > 2, n = 36), or three categories: below the median (CNV < 3 (CNV = 2), n = 14), median (CNV = 3, n = 13), and above the median (CNV > 3, n = 23).
Relationship between time-to-event outcomes
Although a positive slope was observed between time-to-X4 and time-to-AIDS (Cox-Stuart P = 0.007), these two outcomes were weakly correlated (τ = 0.284). When we performed this analysis using the Fast and Slow progression groups, we observed a stronger relationship between the two outcomes in the Fast group (Supplemental Figure 2A) than in the Slow group (Supplemental Figure 2B).
Consistent with this observation, Kaplan-Meier and Cox model analyses showed a much stronger significant difference between the Fast and Slow groups in time-to-AIDS (Fast, 6.46 years; Slow, 10.14 years; log-rank P = 9.91E-12; Cox P < 1E-12; Supplemental Figure 2C), compared with time-to-X4 (Fast, 4.71 years; Slow, 8.00 years; log-rank P = 0.124; Cox P = 0.046; Supplemental Figure 2D). This was also evident among the four disease progression groups (see Methods) (time-to- AIDS (log-rank P < 1E-12; Cox P = 2.83E-8), time-to-X4 (log-rank P = 0.004; Cox P = 0.031)).
Time-to-event analysis of DEFB4/103A CNV
A trend analysis revealed a stronger relationship between DEFB4/103A CNV and time-to-AIDS (Cox-Stuart P = 0.084; Supplemental Figure 3A) than between DEFB4/103A CNV and timeto- X4 (Cox-Stuart P = 0.412; Supplemental Figure 3B).
In line with the above trend analysis, both Kaplan-Meier and Cox model analyses detected a trend towards significant difference between DEFB4/103A CNV categories in time-to-AIDS (CNV = 2, 7.55 years; CNV > 2, 8.13 years; log-rank P = 6.2E-4; Cox P = 0.113; Figure 1A), but not in time-to-X4 (CNV = 2, 6.96 years; CNV > 2, 5.21 years; log-rank P = 0.83; Cox P = 0.947; Figure 1B). We observed no difference in the results obtained by using either two or three categories of DEFB4/103A CNV (data not shown).
Interaction effect of DEFB4/103A CNV with AIDS progression groups
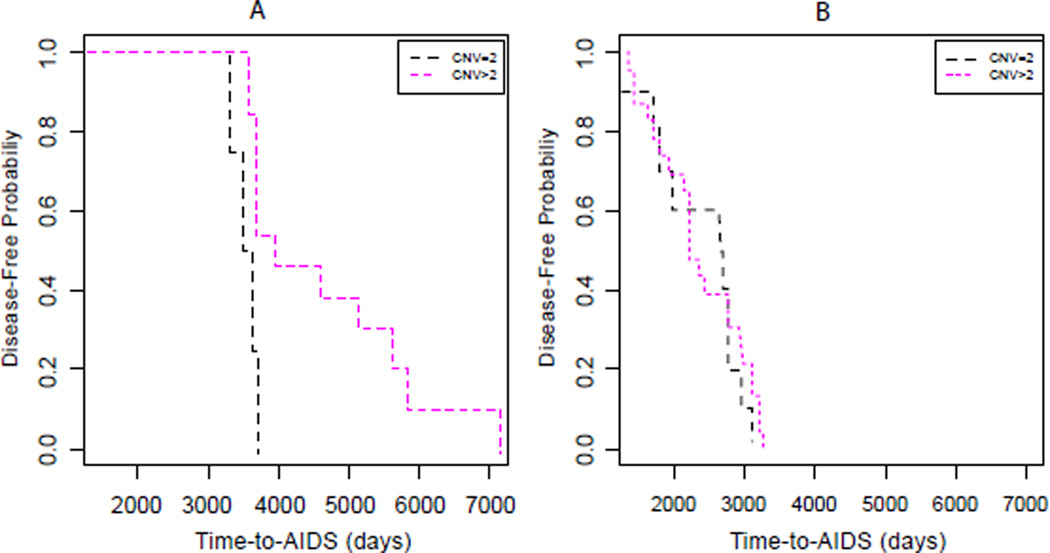
Whereas a trend towards significant difference in time-to-AIDS was observed between DEFB4/103A CNV categories (Figure 1A), a stronger significant difference was detected by both Kaplan-Meier and Cox model analyses, after fixing the AIDS progression groups. In the Slow group, time-to-AIDS appeared to be significantly slower when the DEFB4/103A CNV > 2 (CNV = 2, 9.54 years; CNV > 2, 10.92 years; log-rank P = 9.6E-4; Cox P = 0.005; Figure 2A), in contrast with the Fast group, where no significant difference in time-to-AIDS was observed between the CNV categories (CNV = 2, 7.22 years; CNV > 2, 6.09 years; log-rank P = 0.127; Cox P = 0.888; Figure 2B). We observed no difference in the results obtained by using either two or three categories of DEFB4/103A CNV (data not shown). Because a significant difference in time-to-AIDS between the CNV categories was observed in the slow group, but not in the Fast group, it strongly implies an interaction effect between DEFB4/103A CNV and disease progression.
Figure 2.
Kaplan-Meier plots of aggregated DEFB4/103A copy number effect, after fixing the AIDS progression groups Slow (A) and Fast (B).
In an alternative approach, we compared time-to-AIDS between AIDS progression groups, after fixing the DEFB4/103A CNV categories. We observed a larger difference between the Fast and Slow groups when the DEFB4/103A CNV > 2 (log-rank P = 6.14E-8), compared with the DEFB4/103A CNV = 2 (log-rank P = 4.03E-4). Collectively, these results suggest that the protective effect of higher DEFB4/103A copy number (>2) strongly depends on whether the patients are slow or fast progressors.
Discussion
By utilizing samples previously analyzed for viral tropism in relation to rates of HIV-1 disease progression [18], we observed a weak correlation between time-to-X4 and time-to-AIDS. However, there was a stronger relationship between these two outcomes in the Fast progression group than in the Slow progression group, which is consistent with previous findings [10–12,14,18]. Although the exact biological mechanisms that regulate viral tropism in vivo remain elusive, it has been clear that emergence of the X4 phenotype heralds a rapid clinical deterioration, suggesting that co-receptor usage strongly influences disease progression [10–12]. A literature analysis of 498 patients, who were in late-stage disease, found high frequencies (60–77%) of CXCR4 tropism for five major HIV-1 subtypes [14]. Similarly, in the sample set used here, the frequency of X4 viruses and the likelihood of X4 virus emergence were significantly higher among patients with more rapid disease progression than among those with slower disease progression [18]. In the present study, our direct analysis to measure a relationship between emergence of X4 viruses and progression to AIDS suggests that these two outcomes are distinct, and that their relationship strongly depends on whether the patients are fast or slow progressors. Time-to-AIDS showed a much larger difference between fast and slow progressors than did time-to-X4, further supporting that these two outcomes reflect different levels of biological complexity [12,18].
In line with our first observation, we observed that higher DEFB4/103A copy number was associated with a delayed onset of AIDS, but not with delayed X4 virus emergence. Furthermore, the effect of DEFB4/103A CNV on AIDS progression was more pronounced when we considered the interaction between DEFB4/103A CNV categories and AIDS progression groups: copy number >2 PDG was significantly associated with slower time-to-AIDS in the Slow group, but not in the Fast group. A considerable advancement has been made in understanding the ways that slow progressors can control HIV-1 infection and AIDS progression naturally. It is understood that the slow progression phenotype is an outcome of interplay among viral, host genetic, and immunologic components [26]. Recent studies have shown that higher copy numbers of certain host genes, such as chemokine (CCL3L and CCL4L, ligands for CCR5) [27–30] and immune receptor family (KIR) [31], exert a protective effect against HIV infection and AIDS progression. In addition, CNV of β-defensin genes may also play an important role in susceptibility to HIV infection [32,33]. In Brazilian HIV-positive children, median copy number of DEFB104, which encodes hBD-4 and is located in the same chromosomal region as DEFB4 and DEFB103A, was lower compared with HIV-exposed uninfected children and healthy controls, suggesting that DEFB104 may be involved in protection from vertical transmission of HIV [33]. On the other hand, in Ethiopian and Tanzanian patients, higher β-defensin CNV was associated with increased HIV viral load prior to HAART, and poor immune reconstitution following initiation of HAART, suggesting that higher β-defensin copy number may be a risk factor [32]. Here we show that higher DEFB4/103A copy number is an additional genetic factor associated with slower progression to AIDS. Since the MACS cohort is predominantly Caucasian, our finding, together with the results of recent studies [32,33], indicates that the association between β-defensin CNV and HIV infection/disease progression may be population-specific.
Higher β-defensin CNV correlates with higher peptide concentration in serum [2]. The anti-HIV activity of β-defensins [7,8], together with down-modulation of CXCR4 [7], and antagonism and internalization of CXCR4 by hBD-3 [9] may be responsible for the observed association between DEFB4/103A CNV and AIDS progression. Testing this plausible mechanistic model requires further studies.
We acknowledge that our study has some limitations. We combined the three racial/ethnic groups, due to very low numbers of Hispanic and Black subjects, and thus were not able to perform a comparative analysis of DEFB4/103A CNV vis-à-vis AIDS progression in various racial/ethnic groups. The observed association between DEFB4/103A CNV and AIDS progression is based on either two or three categories of DEFB4/103A CNV. A more in depth analysis using distinct copy numbers, and their diplotypes [34], may provide a better resolution of DEFB4/103A CNV vis-à-vis AIDS progression. Addressing these limitations and reproducing our findings will need longitudinal analysis of viral tropism in larger HIV/AIDS cohorts. For such a study, the cost, in addition to accessibility and performance, of assay for the determination of viral tropism may pose a challenge [35–37].
In conclusion, we observed that higher DEFB4/103A CNV was associated with a significant delay in time-to-AIDS in individuals with slow disease progression phenotype. Further exploration to unravel the possible mechanism(s) underlying this association may enhance our understanding of genetic predisposition to AIDS progression.
Supplementary Material
Acknowledgments
This study was supported by a grant from the National Institute of Dental and Craniofacial Research at the National Institutes of Health (1P01DE019759, A.W.; Project 4, R.J.J.). J-E.D. was supported by this grant as a Statistician (Core B).
We are thankful to Ms. Janet Schollenberger, Senior Project Coordinator, CAMACS, for providing us the samples as well as data related to the samples to perform the present study. We thank Dr. Vinay Cheruvu for his initial efforts on this project, and Dr. John Tilton, Dave McNamara, Dr. Carolyn Myers, and Tenisha Phipps for their critical comments on the manuscript.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- 1.Ishimoto H, Mukae H, Date Y, Shimbara T, Mondal MS, et al. Identification of hBD-3 in respiratory tract and serum: the increase in pneumonia. Eur Respir J. 2006;27:253–260. doi: 10.1183/09031936.06.00105904. [DOI] [PubMed] [Google Scholar]
- 2.Jansen PA, Rodijk-Olthuis D, Hollox EJ, Kamsteeg M, Tjabringa GS, et al. Beta-defensin-2 protein is a serum biomarker for disease activity in psoriasis and reaches biologically relevant concentrations in lesional skin. PLoS One. 2009;4:e4725. doi: 10.1371/journal.pone.0004725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lippross S, Klueter T, Steubesand N, Oestern S, Mentlein R, et al. Multiple trauma induces serum production of host defence peptides. Injury. 2012;43:137–142. doi: 10.1016/j.injury.2011.03.044. [DOI] [PubMed] [Google Scholar]
- 4.Vordenbäumen S, Fischer-Betz R, Timm D, Sander O, Chehab G, et al. Elevated levels of human beta-defensin 2 and human neutrophil peptides in systemic lupus erythematosus. Lupus. 2010;19:1648–1653. doi: 10.1177/0961203310377089. [DOI] [PubMed] [Google Scholar]
- 5.Groth M, Szafranski K, Taudien S, Huse K, Mueller O, et al. Highresolution mapping of the 8p23.1 beta-defensin cluster reveals strictly concordant copy number variation of all genes. Hum Mutat. 2008;29:1247–1254. doi: 10.1002/humu.20751. [DOI] [PubMed] [Google Scholar]
- 6.Hollox EJ, Armour JA, Barber JC. Extensive normal copy number variation of a beta-defensin antimicrobial-gene cluster. Am J Hum Genet. 2003;73:591–600. doi: 10.1086/378157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quiñones-Mateu ME, Lederman MM, Feng Z, Chakraborty B, Weber J, et al. Human epithelial beta-defensins 2 and 3 inhibit HIV-1 replication. AIDS. 2003;17:F39–F48. doi: 10.1097/00002030-200311070-00001. [DOI] [PubMed] [Google Scholar]
- 8.Sun L, Finnegan CM, Kish-Catalone T, Blumenthal R, Garzino-Demo P, et al. Human beta-defensins suppress human immunodeficiency virus infection: potential role in mucosal protection. J Virol. 2005;79:14318–14329. doi: 10.1128/JVI.79.22.14318-14329.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng Z, Dubyak GR, Lederman MM, Weinberg A. Cutting edge: human beta defensin 3--a novel antagonist of the HIV-1 coreceptor CXCR4. J Immunol. 2006;177:782–786. doi: 10.4049/jimmunol.177.2.782. [DOI] [PubMed] [Google Scholar]
- 10.Alkhatib G. The biology of CCR5 and CXCR4. Curr Opin HIV AIDS. 2009;4:96–103. doi: 10.1097/COH.0b013e328324bbec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cavarelli M, Scarlatti G. Phenotype variation in human immunodeficiency virus type 1 transmission and disease progression. Dis Markers. 2009;27:121–136. doi: 10.3233/DMA-2009-0656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mosier DE. How HIV changes its tropism: evolution and adaptation? Curr Opin HIV AIDS. 2009;4:125–130. doi: 10.1097/COH.0b013e3283223d61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Briz V, Poveda E, del Mar González M, Martín-Carbonero L, González-González R, et al. Impact of antiretroviral therapy on viral tropism in HIV-infected patients followed longitudinally for over 5 years. J Antimicrob Chemother. 2008;61:405–410. doi: 10.1093/jac/dkm469. [DOI] [PubMed] [Google Scholar]
- 14.Esbjörnsson J, Månsson F, Martínez-Arias W, Vincic E, Biague AJ, et al. Frequent CXCR4 tropism of HIV-1 subtype A and CRF02_AG during late-stage disease--indication of an evolving epidemic in West Africa. Retrovirology. 2010;7:23. doi: 10.1186/1742-4690-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green TN, Archary M, Gordon ML, Padayachi N, Lie Y, et al. Drug resistance and coreceptor usage in HIV type 1 subtype C-infected children initiating or failing highly active antiretroviral therapy in South Africa. AIDS Res Hum Retroviruses. 2012;28:324–332. doi: 10.1089/aid.2011.0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin NH, Smeaton LM, Giguel F, Novitsky V, Moyo S, et al. Prevalence and clinical associations of CXCR4-using HIV-1 among treatment-naive subtype C-infected women in Botswana. J Acquir Immune Defic Syndr. 2011;57:46–50. doi: 10.1097/QAI.0b013e318214fe27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saracino A, Monno L, Cibelli DC, Punzi G, Brindicci G, et al. Co-receptor switch during HAART is independent of virological success. J Med Virol. 2009;81:2036–2044. doi: 10.1002/jmv.21598. [DOI] [PubMed] [Google Scholar]
- 18.Shepherd JC, Jacobson LP, Qiao W, Jamieson BD, Phair JP, et al. Emergence and persistence of CXCR4-tropic HIV-1 in a population of men from the multicenter AIDS cohort study. J Infect Dis. 2008;198:1104–1112. doi: 10.1086/591623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Linzmeier RM, Ganz T. Human defensin gene copy number polymorphisms: comprehensive analysis of independent variation in alpha- and beta-defensin regions at 8p22-p23. Genomics. 2005;86:423–430. doi: 10.1016/j.ygeno.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 20.Mehlotra RK, Zimmerman PA, Weinberg A, Jurevic RJ. Variation in human β-defensin genes: new insights from a multi-population study. Int J Immunogenet. 2012 doi: 10.1111/iji.12021. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dempster AP, Laird NM, Rubin DB. Maximum likelihood from incomplete data via the EM algorithm. J R Stat Soc Series B Stat Methodol. 1977;39:1–38. [Google Scholar]
- 22.Cox DR, Stuart A. Some quick tests for trend and dispersion. Biometrika. 1955;42:80–95. [Google Scholar]
- 23.Akritas MG, Murphy SA, LaValley MP. The Theil-Sen estimator with doubly censored data and applications to astronomy. J Am Statist Assoc. 1995;90:170–177. [Google Scholar]
- 24.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Statist Assoc. 1958;53:457–481. [Google Scholar]
- 25.Cox DR. Regression models and life-tables. J R Stat Soc Series B Stat Methodol. 1972;34:187–220. [Google Scholar]
- 26.Poropatich K, Sullivan DJ., Jr Human immunodeficiency virus type 1 longterm non-progressors: the viral, genetic and immunological basis for disease non-progression. J Gen Virol. 2011;92:247–268. doi: 10.1099/vir.0.027102-0. [DOI] [PubMed] [Google Scholar]
- 27.Gonzalez E, Kulkarni H, Bolivar H, Mangano A, Sanchez R, et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 28.Kuhn L, Schramm DB, Donninger S, Meddows-Taylor S, Coovadia AH, et al. African infants’ CCL3 gene copies influence perinatal HIV transmission in the absence of maternal nevirapine. AIDS. 2007;21:1753–1761. doi: 10.1097/QAD.0b013e3282ba553a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu S, Yao L, Ding D, Zhu H. CCL3L1 copy number variation and susceptibility to HIV-1 infection: a meta-analysis. PLoS One. 2010;5:e15778. doi: 10.1371/journal.pone.0015778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shostakovich-Koretskaya L, Catano G, Chykarenko ZA, He W, Gornalusse G, et al. Combinatorial content of CCL3L and CCL4L gene copy numbers influence HIV-AIDS susceptibility in Ukrainian children. AIDS. 2009;23:679–688. doi: 10.1097/QAD.0b013e3283270b3f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pelak K, Need AC, Fellay J, Shianna KV, Feng S, et al. Copy number variation of KIR genes influences HIV-1 control. PLoS Biol. 2011;9:e1001208. doi: 10.1371/journal.pbio.1001208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hardwick RJ, Amogne W, Mugusi S, Yimer G, Ngaimisi E, et al. β-defensin genomic copy number is associated with HIV load and immune reconstitution in sub-saharan Africans. J Infect Dis. 2012;206:1012–1019. doi: 10.1093/infdis/jis448. [DOI] [PubMed] [Google Scholar]
- 33.Milanese M, Segat L, Arraes LC, Garzino-Demo A, Crovella S. Copy number variation of defensin genes and HIV infection in Brazilian children. J Acquir Immune Defic Syndr. 2009;50:331–333. doi: 10.1097/QAI.0b013e3181945f39. [DOI] [PubMed] [Google Scholar]
- 34.Hollox EJ. Copy number variation of beta-defensins and relevance to disease. Cytogenet Genome Res. 2008;123:148–155. doi: 10.1159/000184702. [DOI] [PubMed] [Google Scholar]
- 35.Raymond S, Delobel P, Izopet J. Phenotyping methods for determining HIV tropism and applications in clinical settings. Curr Opin HIV AIDS. 2012;7:463–469. doi: 10.1097/COH.0b013e328356f6d7. [DOI] [PubMed] [Google Scholar]
- 36.Trabaud MA, Icard V, Scholtes C, Perpoint T, Koffi J, et al. Discordance in HIV-1 co-receptor use prediction by different genotypic algorithms and phenotype assay: intermediate profile in relation to concordant predictions. J Med Virol. 2012;84:402–413. doi: 10.1002/jmv.23209. [DOI] [PubMed] [Google Scholar]
- 37.Vandekerckhove L, Verhofstede C, Demecheleer E, De Wit S, Florence E, et al. Comparison of phenotypic and genotypic tropism determination in tripleclass-experienced HIV patients eligible for maraviroc treatment. J Antimicrob Chemother. 2011;66:265–272. doi: 10.1093/jac/dkq458. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.