Abstract
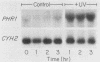
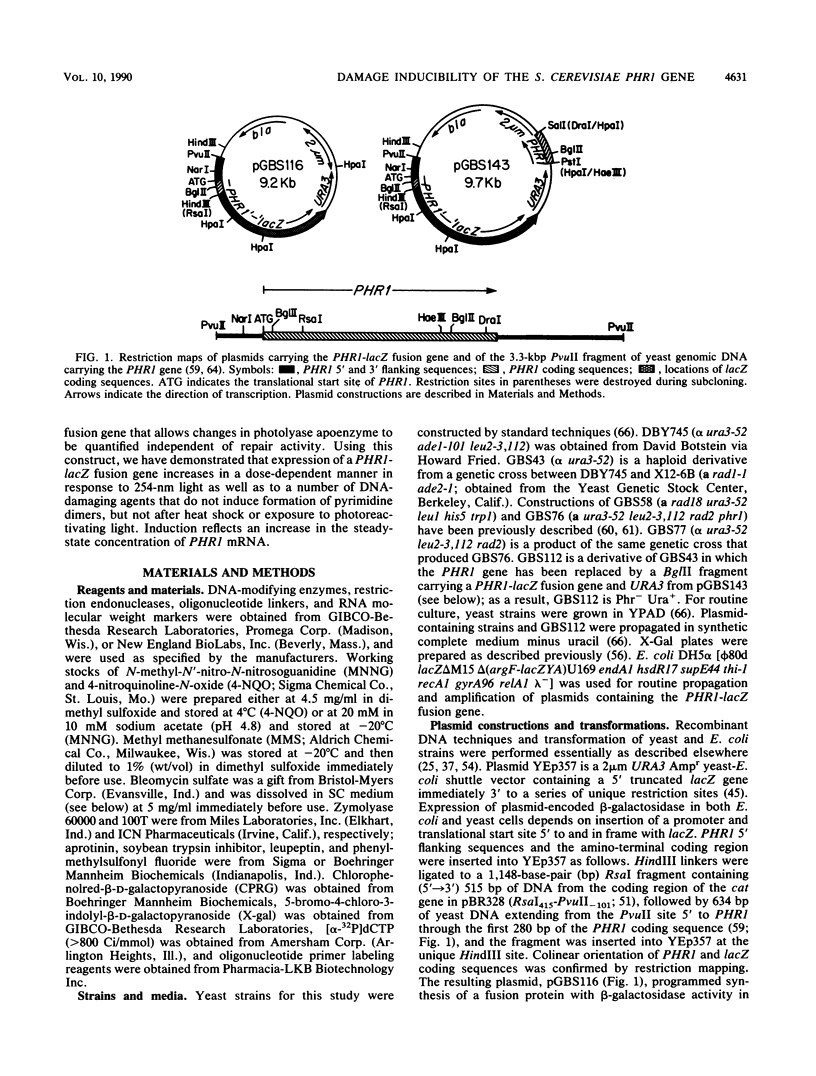
The PHR1 gene of Saccharomyces cerevisiae encodes a photolyase which repairs specifically and exclusively pyrimidine dimers, the most frequent lesions induced in DNA by far-UV radiation. We have asked whether expression of PHR1 is modulated in response to UV-induced DNA damage and to DNA-damaging agents that induce lesions structurally dissimilar to pyrimidine dimers. Using a PHR1-lacZ fusion gene in which expression of beta-galactosidase is regulated by PHR1 5' regulatory elements, we found that exposure of cells to 254-nm light, 4-nitroquinoline-N-oxide, methyl methanesulfonate, and N-methyl-N'-nitro-N-nitrosoguanidine induced synthesis of increased amounts of fusion protein. In contrast to these DNA-damaging agents, neither heat shock nor exposure to photoreactivating light elicited a response. Induction by far-UV radiation was evident both when the fusion gene was carried on a multicopy plasmid and when it replaced the endogenous chromosomal copy of PHR1, and it was accompanied by an increase in the steady-state concentration of PHR1-lacZ mRNA. Northern (RNA) blot analysis of PHR1 mRNA encoded by the chromosomal locus was consistent with either enhanced transcription of PHR1 after DNA damage or stabilization of the transcripts. Neither the intact PHR1 or RAD2 gene was required for induction. Comparison of the region of PHR1 implicated in regulation of its expression with other damage-inducible genes from yeast cells revealed a common conserved sequence that is present in the PHR1, RAD2, and RNR2 genes and is required for damage inducibility of the latter two genes. These sequences may constitute elements of a damage-responsive regulon in S. cerevisiae.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Angel P., Baumann I., Stein B., Delius H., Rahmsdorf H. J., Herrlich P. 12-O-tetradecanoyl-phorbol-13-acetate induction of the human collagenase gene is mediated by an inducible enhancer element located in the 5'-flanking region. Mol Cell Biol. 1987 Jun;7(6):2256–2266. doi: 10.1128/mcb.7.6.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angel P., Pöting A., Mallick U., Rahmsdorf H. J., Schorpp M., Herrlich P. Induction of metallothionein and other mRNA species by carcinogens and tumor promoters in primary human skin fibroblasts. Mol Cell Biol. 1986 May;6(5):1760–1766. doi: 10.1128/mcb.6.5.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell L., Burtscher H., Weiss W. A., Nicolet C. M., Friedberg E. C. Characterization of the RAD10 gene of Saccharomyces cerevisiae and purification of Rad10 protein. Biochemistry. 1990 Mar 27;29(12):3119–3126. doi: 10.1021/bi00464a031. [DOI] [PubMed] [Google Scholar]
- Barker D. G., White J. H., Johnston L. H. The nucleotide sequence of the DNA ligase gene (CDC9) from Saccharomyces cerevisiae: a gene which is cell-cycle regulated and induced in response to DNA damage. Nucleic Acids Res. 1985 Dec 9;13(23):8323–8337. doi: 10.1093/nar/13.23.8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell G. I., DeGennaro L. J., Gelfand D. H., Bishop R. J., Valenzuela P., Rutter W. J. Ribosomal RNA genes of Saccharomyces cerevisiae. I. Physical map of the repeating unit and location of the regions coding for 5 S, 5.8 S, 18 S, and 25 S ribosomal RNAs. J Biol Chem. 1977 Nov 25;252(22):8118–8125. [PubMed] [Google Scholar]
- Cole G. M., Mortimer R. K. Failure to induce a DNA repair gene, RAD54, in Saccharomyces cerevisiae does not affect DNA repair or recombination phenotypes. Mol Cell Biol. 1989 Aug;9(8):3314–3322. doi: 10.1128/mcb.9.8.3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole G. M., Schild D., Lovett S. T., Mortimer R. K. Regulation of RAD54- and RAD52-lacZ gene fusions in Saccharomyces cerevisiae in response to DNA damage. Mol Cell Biol. 1987 Mar;7(3):1078–1084. doi: 10.1128/mcb.7.3.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couto L. B., Friedberg E. C. Nucleotide sequence of the wild-type RAD4 gene of Saccharomyces cerevisiae and characterization of mutant rad4 alleles. J Bacteriol. 1989 Apr;171(4):1862–1869. doi: 10.1128/jb.171.4.1862-1869.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DasGupta U. B., Summers W. C. Ultraviolet reactivation of herpes simplex virus is mutagenic and inducible in mammlian cells. Proc Natl Acad Sci U S A. 1978 May;75(5):2378–2381. doi: 10.1073/pnas.75.5.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge S. J., Davis R. W. Identification and isolation of the gene encoding the small subunit of ribonucleotide reductase from Saccharomyces cerevisiae: DNA damage-inducible gene required for mitotic viability. Mol Cell Biol. 1987 Aug;7(8):2783–2793. doi: 10.1128/mcb.7.8.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge S. J., Davis R. W. Identification of the DNA damage-responsive element of RNR2 and evidence that four distinct cellular factors bind it. Mol Cell Biol. 1989 Dec;9(12):5373–5386. doi: 10.1128/mcb.9.12.5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D., Ozkaynak E., Varshavsky A. The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell. 1987 Mar 27;48(6):1035–1046. doi: 10.1016/0092-8674(87)90711-2. [DOI] [PubMed] [Google Scholar]
- Fleer R., Nicolet C. M., Pure G. A., Friedberg E. C. RAD4 gene of Saccharomyces cerevisiae: molecular cloning and partial characterization of a gene that is inactivated in Escherichia coli. Mol Cell Biol. 1987 Mar;7(3):1180–1192. doi: 10.1128/mcb.7.3.1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornace A. J., Jr, Alamo I., Jr, Hollander M. C. DNA damage-inducible transcripts in mammalian cells. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8800–8804. doi: 10.1073/pnas.85.23.8800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornace A. J., Jr, Nebert D. W., Hollander M. C., Luethy J. D., Papathanasiou M., Fargnoli J., Holbrook N. J. Mammalian genes coordinately regulated by growth arrest signals and DNA-damaging agents. Mol Cell Biol. 1989 Oct;9(10):4196–4203. doi: 10.1128/mcb.9.10.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg E. C. Deoxyribonucleic acid repair in the yeast Saccharomyces cerevisiae. Microbiol Rev. 1988 Mar;52(1):70–102. doi: 10.1128/mr.52.1.70-102.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui A., Hieda K., Matsudaira Y. Light-flash analysis of the photoenzymic repair process in yeast cells. I. Determination of the number of photoreactivating enzyme molecules. Mutat Res. 1978 Sep;51(3):435–439. doi: 10.1016/0027-5107(78)90133-1. [DOI] [PubMed] [Google Scholar]
- Fukui A., Laskowski W. Light-illumination effects on the cellular concentration of photolyase molecules in yeast. Radiat Environ Biophys. 1985;24(4):251–258. doi: 10.1007/BF01210932. [DOI] [PubMed] [Google Scholar]
- Fukui A., Laskowski W. Modifying factors of the cellular concentration of photolyase molecules in Saccharomyces cerevisiae cells--I. Effects of temperature and light. Photochem Photobiol. 1984 May;39(5):613–617. doi: 10.1111/j.1751-1097.1984.tb03899.x. [DOI] [PubMed] [Google Scholar]
- Fukui A., Laskowski W. Modifying factors of the cellular concentration of photolyase molecules in Saccharomyces cerevisiae--II. Effects of preillumination with light flashes. Photochem Photobiol. 1984 Aug;40(2):227–230. doi: 10.1111/j.1751-1097.1984.tb04580.x. [DOI] [PubMed] [Google Scholar]
- Hinnen A., Hicks J. B., Fink G. R. Transformation of yeast. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1929–1933. doi: 10.1073/pnas.75.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hittelman W. N., Rao P. N. Bleomycin-induced damage in prematurely condensed chromosomes and its relationship to cell cycle progression in CHO cells. Cancer Res. 1974 Dec;34(12):3433–3439. [PubMed] [Google Scholar]
- Holliday R. Further evidence for an inducible recombination repair system in Ustilago maydis. Mutat Res. 1975 Jul;29(1):149–153. doi: 10.1016/0027-5107(75)90029-9. [DOI] [PubMed] [Google Scholar]
- Hurd H. K., Roberts J. W. Upstream regulatory sequences of the yeast RNR2 gene include a repression sequence and an activation site that binds the RAP1 protein. Mol Cell Biol. 1989 Dec;9(12):5359–5372. doi: 10.1128/mcb.9.12.5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch S., McGrath J. P., Varshavsky A. The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Nature. 1987 Sep 10;329(6135):131–134. doi: 10.1038/329131a0. [DOI] [PubMed] [Google Scholar]
- Johnson A. L., Barker D. G., Johnston L. H. Induction of yeast DNA ligase genes in exponential and stationary phase cultures in response to DNA damaging agents. Curr Genet. 1986;11(2):107–112. doi: 10.1007/BF00378201. [DOI] [PubMed] [Google Scholar]
- Johnson J. L., Hamm-Alvarez S., Payne G., Sancar G. B., Rajagopalan K. V., Sancar A. Identification of the second chromophore of Escherichia coli and yeast DNA photolyases as 5,10-methenyltetrahydrofolate. Proc Natl Acad Sci U S A. 1988 Apr;85(7):2046–2050. doi: 10.1073/pnas.85.7.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston L. H., White J. H., Johnson A. L., Lucchini G., Plevani P. The yeast DNA polymerase I transcript is regulated in both the mitotic cell cycle and in meiosis and is also induced after DNA damage. Nucleic Acids Res. 1987 Jul 10;15(13):5017–5030. doi: 10.1093/nar/15.13.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larimer F. W., Perry J. R., Hardigree A. A. The REV1 gene of Saccharomyces cerevisiae: isolation, sequence, and functional analysis. J Bacteriol. 1989 Jan;171(1):230–237. doi: 10.1128/jb.171.1.230-237.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. G., Yarranton G. T. Inducible DNA repair in Ustilago maydis. Mol Gen Genet. 1982;185(2):245–250. doi: 10.1007/BF00330793. [DOI] [PubMed] [Google Scholar]
- Madura K., Prakash S. Nucleotide sequence, transcript mapping, and regulation of the RAD2 gene of Saccharomyces cerevisiae. J Bacteriol. 1986 Jun;166(3):914–923. doi: 10.1128/jb.166.3.914-923.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClanahan T., McEntee K. DNA damage and heat shock dually regulate genes in Saccharomyces cerevisiae. Mol Cell Biol. 1986 Jan;6(1):90–96. doi: 10.1128/mcb.6.1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClanahan T., McEntee K. Specific transcripts are elevated in Saccharomyces cerevisiae in response to DNA damage. Mol Cell Biol. 1984 Nov;4(11):2356–2363. doi: 10.1128/mcb.4.11.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh E. M., Haynes R. H. Sequence and expression of the dCMP deaminase gene (DCD1) of Saccharomyces cerevisiae. Mol Cell Biol. 1986 May;6(5):1711–1721. doi: 10.1128/mcb.6.5.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchel R. E., Morrison D. P. Heat-shock induction of ultraviolet light resistance in Saccharomyces cerevisiae. Radiat Res. 1983 Oct;96(1):95–99. [PubMed] [Google Scholar]
- Mitchel R. E., Morrison D. P. Is DNA damage the signal for induction of thermal resistance? induction by radiation in yeast. Radiat Res. 1984 Aug;99(2):383–393. [PubMed] [Google Scholar]
- Morrison A., Christensen R. B., Alley J., Beck A. K., Bernstine E. G., Lemontt J. F., Lawrence C. W. REV3, a Saccharomyces cerevisiae gene whose function is required for induced mutagenesis, is predicted to encode a nonessential DNA polymerase. J Bacteriol. 1989 Oct;171(10):5659–5667. doi: 10.1128/jb.171.10.5659-5667.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers A. M., Tzagoloff A., Kinney D. M., Lusty C. J. Yeast shuttle and integrative vectors with multiple cloning sites suitable for construction of lacZ fusions. Gene. 1986;45(3):299–310. doi: 10.1016/0378-1119(86)90028-4. [DOI] [PubMed] [Google Scholar]
- Nicolet C. M., Chenevert J. M., Friedberg E. C. The RAD2 gene of Saccharomyces cerevisiae: nucleotide sequence and transcript mapping. Gene. 1985;36(3):225–234. doi: 10.1016/0378-1119(85)90177-5. [DOI] [PubMed] [Google Scholar]
- Parker R. C., Watson R. M., Vinograd J. Mapping of closed circular DNAs by cleavage with restriction endonucleases and calibration by agarose gel electrophoresis. Proc Natl Acad Sci U S A. 1977 Mar;74(3):851–855. doi: 10.1073/pnas.74.3.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perozzi G., Prakash S. RAD7 gene of Saccharomyces cerevisiae: transcripts, nucleotide sequence analysis, and functional relationship between the RAD7 and RAD23 gene products. Mol Cell Biol. 1986 May;6(5):1497–1507. doi: 10.1128/mcb.6.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson T. A., Prakash L., Prakash S., Osley M. A., Reed S. I. Regulation of CDC9, the Saccharomyces cerevisiae gene that encodes DNA ligase. Mol Cell Biol. 1985 Jan;5(1):226–235. doi: 10.1128/mcb.5.1.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzagalli A., Valsasnini P., Plevani P., Lucchini G. DNA polymerase I gene of Saccharomyces cerevisiae: nucleotide sequence, mapping of a temperature-sensitive mutation, and protein homology with other DNA polymerases. Proc Natl Acad Sci U S A. 1988 Jun;85(11):3772–3776. doi: 10.1073/pnas.85.11.3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentki P., Karch F., Iida S., Meyer J. The plasmid cloning vector pBR325 contains a 482 base-pair-long inverted duplication. Gene. 1981 Sep;14(4):289–299. doi: 10.1016/0378-1119(81)90161-x. [DOI] [PubMed] [Google Scholar]
- Reynolds P., Prakash L., Dumais D., Perozzi G., Prakash S. Nucleotide sequence of the RAD10 gene of Saccharomyces cerevisiae. EMBO J. 1985 Dec 16;4(13A):3549–3552. doi: 10.1002/j.1460-2075.1985.tb04115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson G. W., Nicolet C. M., Kalainov D., Friedberg E. C. A yeast excision-repair gene is inducible by DNA damaging agents. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1842–1846. doi: 10.1073/pnas.83.6.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Ruby S. W., Szostak J. W., Murray A. W. Cloning regulated yeast genes from a pool of lacZ fusions. Methods Enzymol. 1983;101:253–269. doi: 10.1016/0076-6879(83)01019-8. [DOI] [PubMed] [Google Scholar]
- Ruby S. W., Szostak J. W. Specific Saccharomyces cerevisiae genes are expressed in response to DNA-damaging agents. Mol Cell Biol. 1985 Jan;5(1):75–84. doi: 10.1128/mcb.5.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A., Sancar G. B. DNA repair enzymes. Annu Rev Biochem. 1988;57:29–67. doi: 10.1146/annurev.bi.57.070188.000333. [DOI] [PubMed] [Google Scholar]
- Sancar G. B. Expression of a Saccharomyces cerevisiae photolyase gene in Escherichia coli. J Bacteriol. 1985 Feb;161(2):769–771. doi: 10.1128/jb.161.2.769-771.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar G. B. Sequence of the Saccharomyces cerevisiae PHR1 gene and homology of the PHR1 photolyase to E. coli photolyase. Nucleic Acids Res. 1985 Nov 25;13(22):8231–8246. doi: 10.1093/nar/13.22.8231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar G. B., Smith F. W. Construction of plasmids which lead to overproduction of yeast PHR1 photolyase in Saccharomyces cerevisiae and Escherichia coli. Gene. 1988 Apr 15;64(1):87–96. doi: 10.1016/0378-1119(88)90483-0. [DOI] [PubMed] [Google Scholar]
- Sancar G. B., Smith F. W., Heelis P. F. Purification of the yeast PHR1 photolyase from an Escherichia coli overproducing strain and characterization of the intrinsic chromophores of the enzyme. J Biol Chem. 1987 Nov 15;262(32):15457–15465. [PubMed] [Google Scholar]
- Sancar G. B., Smith F. W. Interactions between yeast photolyase and nucleotide excision repair proteins in Saccharomyces cerevisiae and Escherichia coli. Mol Cell Biol. 1989 Nov;9(11):4767–4776. doi: 10.1128/mcb.9.11.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarasin A., Benoit A. Induction of an error-prone mode of DNA repair in UV-irradiated monkey kidney cells. Mutat Res. 1980 Mar;70(1):71–81. doi: 10.1016/0027-5107(80)90059-7. [DOI] [PubMed] [Google Scholar]
- Schild D., Johnston J., Chang C., Mortimer R. K. Cloning and mapping of Saccharomyces cerevisiae photoreactivation gene PHR1. Mol Cell Biol. 1984 Sep;4(9):1864–1870. doi: 10.1128/mcb.4.9.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira S. K., Chou J., Richaud F. V., Casadaban M. J. New versatile plasmid vectors for expression of hybrid proteins coded by a cloned gene fused to lacZ gene sequences encoding an enzymatically active carboxy-terminal portion of beta-galactosidase. Gene. 1983 Nov;25(1):71–82. doi: 10.1016/0378-1119(83)90169-5. [DOI] [PubMed] [Google Scholar]
- Siede W., Robinson G. W., Kalainov D., Malley T., Friedberg E. C. Regulation of the RAD2 gene of Saccharomyces cerevisiae. Mol Microbiol. 1989 Dec;3(12):1697–1707. doi: 10.1111/j.1365-2958.1989.tb00155.x. [DOI] [PubMed] [Google Scholar]
- Teo I., Sedgwick B., Demple B., Li B., Lindahl T. Induction of resistance to alkylating agents in E. coli: the ada+ gene product serves both as a regulatory protein and as an enzyme for repair of mutagenic damage. EMBO J. 1984 Sep;3(9):2151–2157. doi: 10.1002/j.1460-2075.1984.tb02105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobey R. A. Different drugs arrest cells at a number of distinct stages in G2. Nature. 1975 Mar 20;254(5497):245–247. doi: 10.1038/254245a0. [DOI] [PubMed] [Google Scholar]
- Torczynski R., Bollon A. P., Fuke M. The complete nucleotide sequence of the rat 18S ribosomal RNA gene and comparison with the respective yeast and frog genes. Nucleic Acids Res. 1983 Jul 25;11(14):4879–4890. doi: 10.1093/nar/11.14.4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treger J. M., Heichman K. A., McEntee K. Expression of the yeast UB14 gene increases in response to DNA-damaging agents and in meiosis. Mol Cell Biol. 1988 Mar;8(3):1132–1136. doi: 10.1128/mcb.8.3.1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G. C. Inducible DNA repair systems. Annu Rev Biochem. 1985;54:425–457. doi: 10.1146/annurev.bi.54.070185.002233. [DOI] [PubMed] [Google Scholar]
- White J. H., Barker D. G., Nurse P., Johnston L. H. Periodic transcription as a means of regulating gene expression during the cell cycle: contrasting modes of expression of DNA ligase genes in budding and fission yeast. EMBO J. 1986 Jul;5(7):1705–1709. doi: 10.1002/j.1460-2075.1986.tb04414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasui A., Laskowski W. Determination of the number of photoreactivating enzyme molecules per haploid Saccharomyces cells. Int J Radiat Biol Relat Stud Phys Chem Med. 1975 Dec;28(6):511–518. doi: 10.1080/09553007514551371. [DOI] [PubMed] [Google Scholar]