Abstract
MicroRNAs are endogenous posttranscriptional modulators that negatively control the expression of their target genes and play an important role in the development and progression of many malignancies, including colorectal carcinoma. In particular, expression of microRNA-21 (miR-21) is greatly increased in chemotherapy-resistant (CR) colon cancer cells that are enriched in undifferentiated cancer stem/stem-like cells (CSCs/CSLCs). We hypothesize that miR-21 plays a critical role in regulating differentiation of CR colon cancer cells. Indeed, we observed that downregulation of miR-21 in CR colon cancer cells (HCT-116 or HT-29) by antisense miR-21 induced differentiation, as evidenced by marked increases in cytokeratin-20 (CK-20) expression and alkaline phosphatase activity. These changes were accompanied by a significant reduction in the expression of colon CSC/CSLC marker CD44, colonosphere formation, and T-cell factor/lymphoid enhancer factor (TCF/LEF) activity but increased the expression of proapoptotic programmed cell death 4 gene. Induction of differentiation greatly increased sensitivity of CR colon cancer cells to the growth inhibitory properties of all three regimens tested: 5-fluorouracil + oxaliplatin (FUOX), difluorinated curcumin (CDF), and the combination of CDF and FUOX. However, the magnitude of inhibition of growth by either CDF (75%) alone or CDF + FUOX (80%) was much higher than that observed with only FUOX (40%). Growth inhibition by CDF and CDF + FUOX in differentiating CR colon cancer cells was associated with a 98% to 99% reduction in the expression of CD44 and epidermal growth factor receptor (EGFR). However, down-regulation of CK-20 in CR colon cancer cells produced no significant change in cellular growth in the absence or presence of FUOX, when compared with the corresponding controls. The current observation suggests that CDF and CDF + FUOX are highly effective in inhibiting growth and reducing colon CSCs/CSLCs in anti-miR-21-induced differentiating CR colon cancer cells and supports our contention that differentiation enhances susceptibility of CR cancer cells to conventional and nonconventional therapeutic regimen.
Introduction
Colorectal cancer is one of the most common cancers and the second leading cause of cancer-related deaths in the United States with about 140,000 newly diagnosed cases per year. Although current treatments involving a combination of surgical resection, radiation, and chemotherapy have increased the patient's five-year survival, nearly 50% of patients with colorectal carcinoma show recurrence of the disease with about 50,000 deaths per year [1]. Therefore, an improved therapeutic strategy specifically targeting the recurrent disease is desperately needed.
Carcinoma recurrence in part is due to the fact that conventional chemotherapy only targets the rapidly dividing cells that form bulk of the tumor. While chemotherapy can shrink the size of the tumor, the tumor initiating or cancer stem/stem-like cells (CSCs/CSLCs) that are resistant to chemotherapy are thought to be the leading cause of cancer recurrence [2]. Moreover, continued use of conventional chemotherapeutics is associated with added toxicities, some of which are even fatal.
CSCs/CSLCs are defined as self-renewing population of undifferentiated cells within a tumor that are primarily responsible for populating the bulk of the tumor. They have been shown to be resistant to radiation and chemotherapy and are thought to be responsible for tumor initiation, progression, metastasis, and relapse [3]. CSCs/CSLCs isolated from different solid tumors, including the colon, are usually identified by specific surface epitopes. Colon CSCs/CSLCs have been shown to express CD44, CD166, CD133, and epithelial-specific antigen surface markers [4]. We have observed that in humans, colon CSCs/CSLCs are present not only in premalignant adenomas but also in normal appearing colonic mucosa and that the population of CSCs/CSLCs increases with advancing age, suggesting that they may be partly responsible for the age-related rise in colorectal cancer [5].
Like normal stem cells, CSCs/CSLCs grow slowly and are more likely to survive chemotherapy than other cells [6]. Hence, the proportion of CSCs/CSLCs in the tumor increases after conventional chemotherapy. We have recently reported that exposure of colon cancer HCT-116 or HT-29 cells to the combination of 5-fluorouracil (5-FU) and oxaliplatin (Ox), the backbone of colorectal cancer conventional chemotherapy, inhibited their growth and led to enrichment of CSC/CSLC phenotype where Wnt/β-catenin signaling played a critical role in regulating their growth and maintenance [7,8]. More interestingly, the levels of microRNA-21 (miR-21), an oncomiR, were found to be greatly increased in CSCs/CSLCs-enriched chemotherapy-resistant (CR) colon cancer HCT-116 and HT-29 cells, indicating that miR-21 may play a role in regulating the growth of CSCs/CSLCs [9].
MicroRNAs (miRNAs) comprise a broad class of small (19–22 nucleotide) endogenous RNAs that negatively control the expression of the target genes by cleaving mRNA or through translation repression [10]. It is estimated that miRNAs can control the expression of approximately 30% of all proteins in humans and regulate various cellular processes. Many miRNAs are found to be aberrantly expressed in several pathologic conditions, including cancer, leading to the identification of “miRNA signature” characteristics of certain tumors [11]. Tumor-specific miRNA expression profiles are also functionally relevant because many miRNAs act as tumor suppressors or as oncogenes (oncomiRs) [12].
miR-21 has been found to be overexpressed in most epithelial cancers including colorectal cancers. Knockdown of miR-21 in cancer cells impairs growth, induces apoptosis, and reduces migration and invasion of cancer cells [13]. Forced expression of miR-21 leads to increased T-cell factor/lymphoid enhancer factor (TCF/LEF) activity, augmentation of the expression of c-Myc and cyclin D accompanied by increased sphere forming ability in vitro, and tumor formation in severe combined immunodeficiency (SCID) mice [9].
The target genes of miR-21 were determined to be tumor-suppressive genes such as PTEN, programmed cell death 4 gene (PDCD4), and transforming growth factor βR2 [14]. miR-21 is believed to play a pivotal role in the progression of many malignancies. Therefore, it is not surprising that miR-21 is involved in diverse biologic processes, including cell differentiation, proliferation, and apoptosis, presumably by modulating target proteins [15]. The primary objective of the current investigation was to examine whether down-regulation of miR-21 in CSC/CSLC-enriched CR colon cancer cells would induce differentiation. Herein, we report that down-regulation of miR-21 by anti-miR-21 in CR colon cancer cells induces differentiation and increases their susceptibility to conventional or/and nonconventional therapeutic regimens.
Materials and Methods
Cell Lines and Generation of CR Cells
Human colon cancer HCT-116 and HT-29 cells were obtained from the American Type Culture Collection (ATCC, Rockville, MD). They were maintained in Dulbecco's modified Eagle's medium (4.5 g/l D-glucose) supplemented with 10% FBS (Invitrogen, Grand Island, NY) and 1% antibiotic/antimycotic in tissue culture flasks in a humidified incubator at 37°C in an atmosphere of 95% air and 5% carbon dioxide. The medium was changed two times a week, and cells were passaged using 0.05% trypsin/EDTA (Invitrogen).
FOLFOX/FUOX (5-FU + Oxaliplatin)-resistant (CR) cell lines were generated in our laboratory as described previously [8,16], Briefly, HCT-116 or HT-29 cells were incubated with a clinically relevant dose of FUOX (25 µM 5-FU and 0.625 µM oxaliplatin) for 72 hours. The medium was removed and the adherent cells, which survived the FUOX insult, were cultured in Dulbecco's modified Eagle's medium containing 10% FBS without the drugs for 3 to 4 days. The addremove FUOX cycle was repeated 12 times. The surviving cells were then passaged and exposed to higher doses of combination of 5-FU and Ox (100 µM 5-FU + 2.5 µM oxaliplatin) for approximately 2 to 3 weeks. Finally, the CR cells were maintained in normal culture medium containing FUOX (50 µM 5-FU + 1.25 µM oxaliplatin). The medium was changed two times a week and the cells were passaged using trypsin/EDTA.
Anti-miR-21 and Cytokeratin-20 siRNA Transfection
For transfection of siRNA into the parental and CR colon cells, Oligofectamine reagent (Invitrogen Corp, Carlsbad, CA) and serum-free Opti-MEM (Invitrogen Corp) medium was used to prepare transfection complexes according to the manufacturer's instructions. Briefly, single-cell suspension was plated onto six-well tissue culture plates with normal growth medium overnight to achieve 25% to 30% confluence. Next day, the medium was removed, washed twice with serum-free Opti-MEM (Invitrogen Corp) medium before adding the complexes containing nontargeted, anti-miR-21 or cytokeratin-20 (CK-20) siRNA (Integrated DNA Technologies Inc, Coralville, IA). After 2 days of transfection, the cells were collected and analyzed for mRNA, protein expression, or cellular growth assay.
Determination of Cellular Growth
The growth of CR colon cancer HCT-116 cells in response to FUOX (100 µM 5-FU + 2.5 µM oxaliplatin), difluorinated curcumin (CDF; 2.0 µM) and FUOX + CDF was assessed by 3-(4,5-dimethylthiazol- 2yl)-2, 5-diphenyltetrazolium bromide (MTT) assay as described previously [17].
Western Blot Analysis
Western blot analysis was performed essentially according to our standard protocol [18,19]. Briefly, the cells were solubilized in lysis buffer and the protein concentration was determined by the Bio-Rad Protein Assay Kit (Bio-Rad, Hercules, CA). After electrophoresis, proteins were transferred electrophoretically onto the supported polyvinylidene difluoride membrane (Millipore Corp, Bedford, MA) and incubated for 1 hour at room temperature with blocking buffer, TBS-T [20 mM Tris (pH 7.6), 100 µM NaCl, 0.1% Tween-20], and 5% nonfat dry milk with gentle agitation. After washing the membranes with TBS-T, they were incubated overnight with primary antibodies in TBS-T buffer containing 5% milk at 4°C. The membranes were washed with TBS-T, subsequently incubated with appropriate secondary antibodies in TBS-T/5% milk for 1 to 2 hours at room temperature. The membranes were washed again with TBS-T, and the protein bands were visualized by enhanced chemiluminescence detection system (Amersham, Piscataway, NJ). The membranes were stripped (2x for 15 minutes at 55°C) in stripping buffer containing 100 mM 2-mercaptoethanol, 2% sodium dodecyl sulfate, and 62.5 mM Tris-HCl (pH 6.7) and reprobed for β-actin. All Western blots were performed at least three times for each experiment.
Isolation of RNA and Quantitative Polymerase Chain Reaction Analysis
Total RNA was extracted from different cells using RNA-STAT solution (Tel Test, Friendswood, TX) according to the manufacturer's instruction. The total RNA was treated with DNase I and purified with phenol-chloroform. RNA concentration was measured spectrophotometrically at an OD of 260 nm.
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was performed using the GeneAmp RNA PCR Kit (Applied Biosystems, Foster City, CA). Five microliters of cDNA products were amplified with SYBR Green Quantitative PCR Master Mix (Applied Biosystems). PCR primers were used as follows: pri-miR-21, forward: 5′-tgagactgatgttgactgttgaa-3′ and reverse: 5′-tgtcagacagcccatcgac-3′; CK-20, forward: 5′-tgaagagctgcgaagtcaga-3′ and reverse: 5′-gaagtcctcagcagccagtt-3′; CD44, forward: 5′-aaggtggagcaaacacaacc-3′, reverse: 5′-actgcaatgcaaactgcaag-3′; epidermal growth factor receptor (EGFR), forward: 5′-cagcgctaccttgtcattca-3′, reverse: 5′-cgtcgtccatgtcttcttca-3′; β-actin, forward: 5′-cccagcacaatgaagatcaa-3′ and reverse 5′-acatctgctggaaggtggac- 3′. Reactions were carried out in Applied Biosystems 7500 Real-Time PCR System; the conditions for PCR running were given as follows: for activating the DNA polymerase, hot start was performed for 10 minutes at 95°C and then cycling at 95°C for 15 seconds and 60°C for 1 minute for a total of 40 cycles.
Quantitation of miR-21
TaqMan microRNA assays were used to quantitate miR-21 in different colon cancer cells according to the manufacturer's instruction (Applied Biosystems). Briefly, cDNA synthesis was carried out with the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems). The miRNA RT-PCR primers for miR-21 and endogenous control RNU6B were purchased from Applied Biosystems. Real-time qRTPCR analysis was carried out using Applied Biosystems 7500 Real-Time PCR System. The PCR mix containing TaqMan 2x Universal PCR Master Mix were processed as follows: 95°C for 10 minutes and then 95°C for 15 seconds and 60°C for 60 seconds for up to 40 cycles. Signal was collected at the endpoint of every cycle. The gene expression ΔCT values of miRNAs from each sample were calculated by normalizing with internal control RNU6B, and relative quantitation values were plotted.
TCF/LEF Dual-Luciferase Assay
The activation of transcription factor TCF/LEF was evaluated by using Cignal TCF/LEF Reporter Assay Kit (SA Biosciences, Frederick, MD) as described previously [20]. Briefly, the cells were grown to 25% to 30% confluence and co-transfected with TCF/LEF reporter constructs along with either nontargeted (control), anti-miR-21, or β-catenin siRNAs (Integrated DNA Technologies Inc) using SureFECT transfection reagent (SA Biosciences) according to the manufacturer's instructions. After transfection, the TCF/LEF activity reporter assays were performed 48 hours post-transfection using a Dual-Luciferase Assay Kit (Promega Biosciences, San Luis Obispo, CA) following the instructions outlined by the manufacturer.
Statistical Analysis
Unless otherwise stated, data are expressed as means ± SEM. Where applicable, the results were analyzed using analysis of variance followed by Fisher protected least significant difference or Scheffe test. P < .05 was designated as the level of significance.
Results
Down-regulation of miR-21 Induces Differentiation of CR Colon Cancer Cells
miR-21 has been recently referred to as an “oncomiR” (an miRNA with oncogenic properties) [21]. This miRNA, which is normally upregulated in many solid tumors including colon tumors [22], promotes cell transformation by targeting the PDCD4 [13] through posttranscriptional down-regulation [22]. We have reported that CR colon cancer HCT-116 and HT-29 cells exhibit enrichment of CSCs/CSLCs and elevated levels of mature miR-21 and that miR-21 induces stemness in colon cancer cells [9]. These properties of miR-21 prompted us to examine whether down-regulation of miR-21 in CR colon cancer cells would induce differentiation of CSCs.
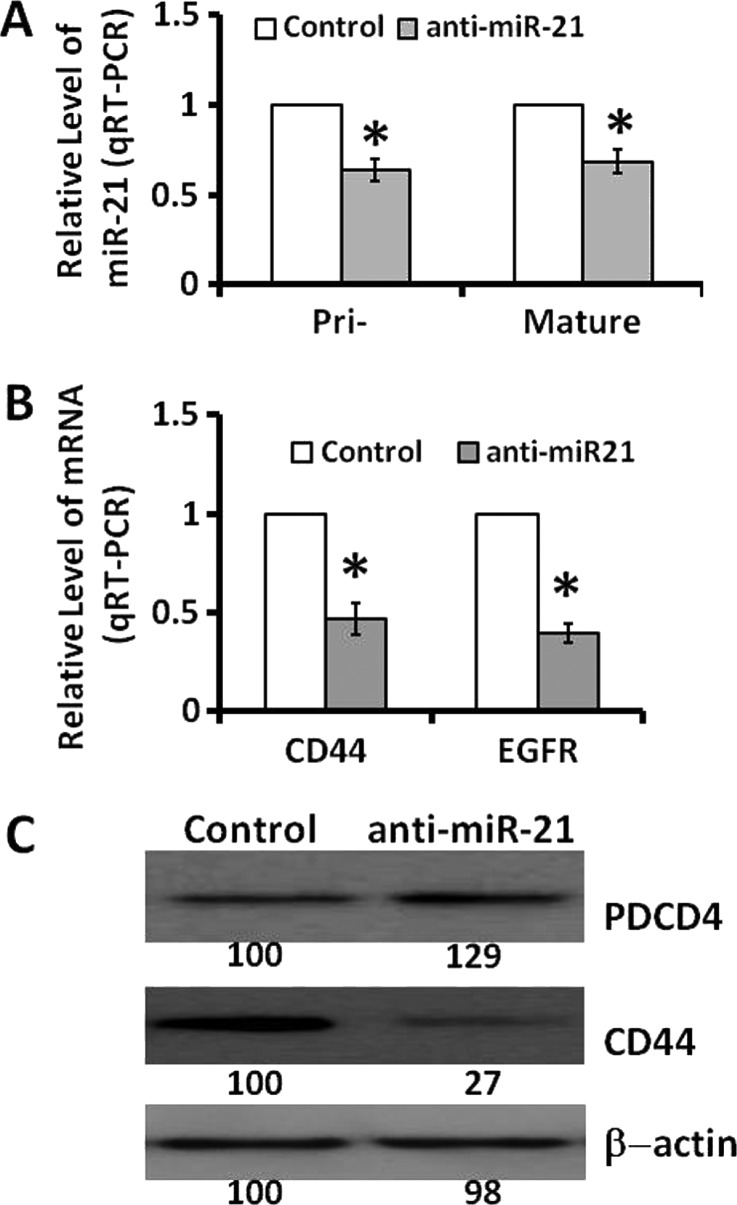
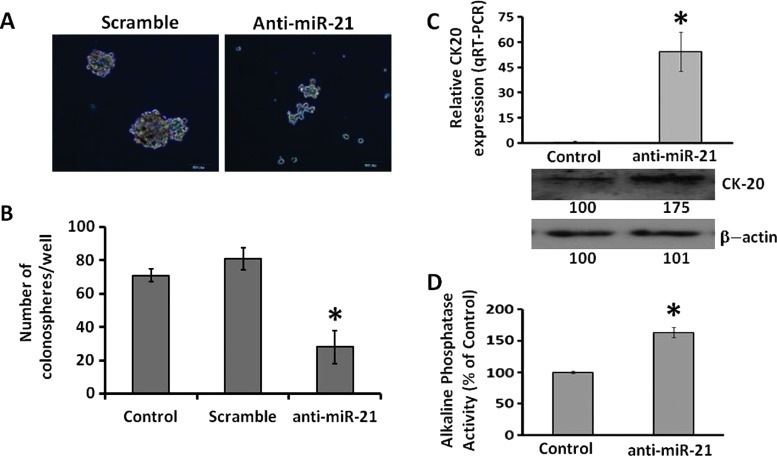
Down-regulation of miR-21 through transfection of anti-miR-21 in CR HCT-116 cells caused about 40% reduction in pri-miR-21 and mature miR-21 levels (Figure 1A) and elevated the levels of PDCD4 (Figure 1C), one of the targets of miR-21, and produced approximately 50% reduction in the expression of EGFR and CD44 (Figure 1B), the latter being one of the markers of colon CSCs/CSLCs. Additionally, down-regulation of miR-21 in CR HCT-116 cells was found to lower the ability of these cells to form colonospheres (Figure 2, A and B), one of the primary properties of colon CSCs/CSLCs. These changes were associated with marked increases in both mRNA and protein expression of CK-20 [23] and a 50% increase in alkaline phosphatase activity (Figure 2, C and D), indicating induction of differentiation of CR HCT-116 cells following down-regulation of miR-21.
Figure 1.
Down-regulation of miR-21 in CR colon cancer HCT-116 cells through transfection of anti-miR-21 (A) lowers the levels of both pri-miR-21 and mature miR-21, (B) decreases the expression of CD44 and EGFR, and (C) stimulates the expression of PDCD4 but reduces CD44. In this and all subsequent studies, the control cells were transfected with scrambled miRNAs. The data represent means ± SD of three independent experiments.
Figure 2.
Down-regulation of miR-21 in CR HCT-116 cells by anti-miR-21 (A) decreases the ability of cells to form spheroids as shown by the representative photographs (magnification, x10) taken after 7 days of transfection with either anti-miR-21 or scrambled miRNA (control), (B) the number of spheres formed 7 days following transfection of anti-miR-21 or scrambled miRNA; nontransfected CR-HCT-116 cells were used as controls. Values are means ± SD. P < . 05, compared with scramble or untreated controls. (C) qRT-PCR (upper panel) showing up-regulation of CK-20 mRNA in CR HCT-116 cells 48 hours following transfection of anti-miR-21, when compared with corresponding scramble control (*P < .001). Western blot (lower panel) showing increased expression of CK-20 in CR HCT-116 cells following transfection of anti-miR-21 when compared with the corresponding control cells; β-actin was used as a loading control. (D) Induction of alkaline phosphatase activity in CR HCT-116 cells 48 hours following transfection of anti-miR-21 (n = 3). Values are means ± SD. *P < .05, compared with scramble transfected control.
Wnt/β-Catenin Signaling Pathway Plays a Role in Regulating miR-21-Mediated Differentiation of CR Colon Cancer Cells
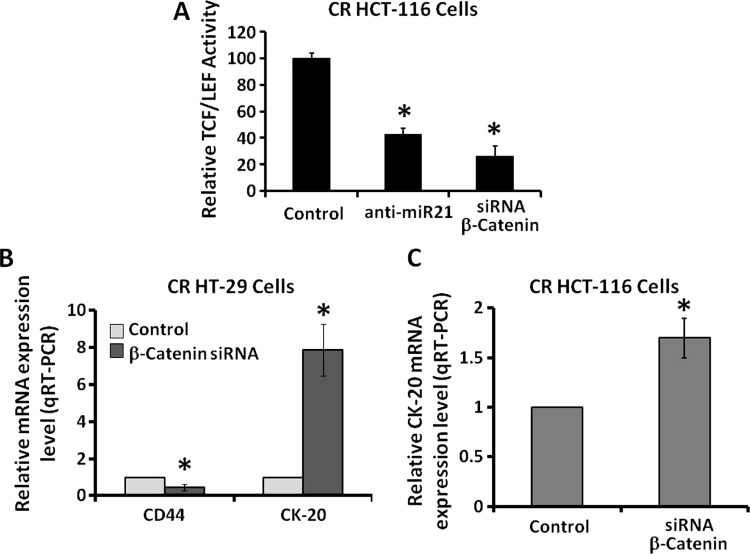
The next set of experiments was undertaken to examine the potential mechanisms of miR-21-mediated differentiation in CR colon cancer cells. The Wnt/β-catenin signaling pathway that regulates β-catenin/TCF-driven gene transcription is known to be activated in many malignancies, including colorectal cancer [24]. This signaling pathway also plays a critical role in regulating proliferation of colon CSCs/CSLCs [20].We have reported that Wnt/β-catenin signaling plays a pivotal role in regulating the miR-21 induction of stemness of colon cancer cells [9]. Although down-regulation of miR-21 is found to be associated with a reduction in β-catenin levels in CR colon cancer cells [9], it is not known whether down-regulation of either miR-21 or β-catenin would also lead to attenuation of transcriptional activity of TCF/LEF. Our results demonstrate that down-regulation of either miR-21 by anti-miR-21 or β-catenin by the corresponding siRNA in CR colon cancer HCT-116 or HT-29 cells caused a 50% to 60% reduction in transcriptional activity of TCF/LEF (Figure 3A), accompanied by a significant reduction in CD44 expression (Figure 3B) and a marked increase in CK20 expression (Figure 3, B and C), suggesting induction of differentiation. Taken together, the results suggest a role for Wnt/β-catenin signaling pathway in regulating miR-21-mediated differentiation of CR colon cancer cells.
Figure 3.
Down-regulation of either miR-21 or β-catenin in CR colon cancer cells (HCT-116 or HT-29) leads to attenuation of transcriptional activation of TCF/LEF. (A) Relative transcriptional activities of TCF/LEF in CR colon cancer HCT-116 cells after down-regulation of either miR-21 by anti-miR-21 or β-catenin by the corresponding siRNA, when compared with the corresponding controls that were similarly treated with scramble miRNA or siRNA. *P < .001, compared with the corresponding scramble controls. (B and C) qRT-PCR showing down-regulation of β-catenin in CR colon cancer HT-29 or HCT-116 cells by the corresponding siRNA leads to reduction in CD44 expression and increase in CK20 expression in CR colon cancer HT-29 cells (B) and HCT-116 cells (C), compared with the corresponding controls. The data represent means ± SD of three independent experiments, *P < .001.
Down-regulation of miR-21 Increases the Cell Responsiveness to Conventional and Nonconventional Therapeutics
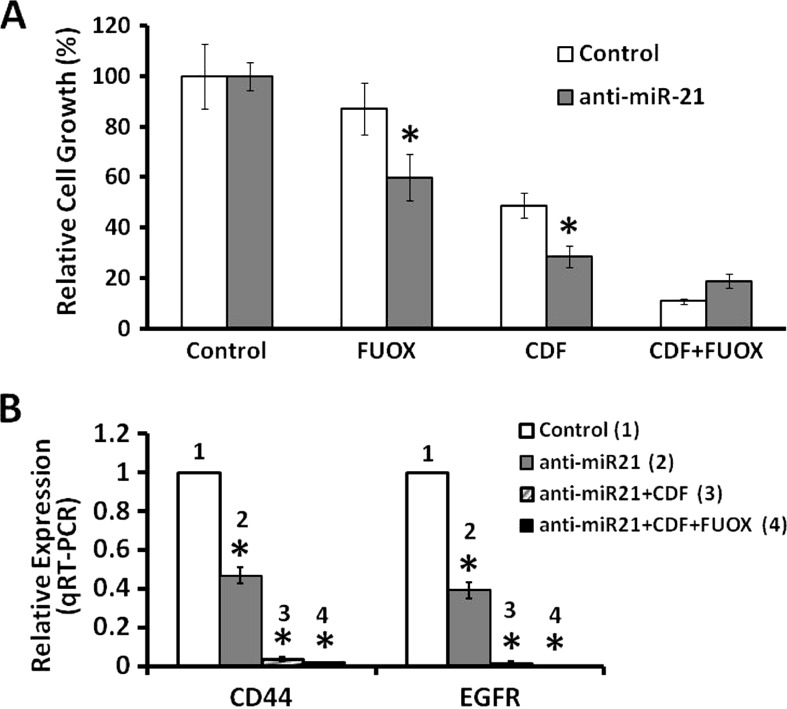
CR colon cancer cells that are highly enriched in CSCs/CSLCs are also poorly differentiated. To further determine whether induction of differentiation would increase susceptibility to different therapeutics, CR HCT-116 cells transfected with either the scrambled anti-miR (controls) or anti-miR-21 were subsequently incubated for 48 hours with FUOX (100 µM 5-FU + 2.5 µM oxaliplatin), CDF (2.0 µM), or the combination of CDF + FUOX. Indeed, we observed that induction of differentiation of CR colon cancer cells increased their sensitivity to the growth inhibitory properties of all three regimens tested. However, the magnitude of inhibition of growth by either CDF (75%) or CDF + FUOX (80%) was much higher than that observed with FUOX alone (40%), suggesting that CDF and the combination of CDF and FUOX are more effective than FUOX in inhibiting the growth of differentiating CR colon cancer cells (Figure 4A). This inference is further supported by the observation that CDF and the combination of CDF and FUOX produced an incredible 98% to 99% reduction in the expression of CD44 and EGFR in the differentiating CR HCT-116 cells (Figure 4B). In contrast, FUOX by itself caused only a 60% reduction in CD44 and EGFR expression (Figure 4B). Our current observation that CDF and CDF + FUOX were highly effective in inhibiting growth and reducing the expression of CD44 and EGFR in anti-miR-21-induced differentiating CR HCT-116 cells supports our contention that differentiating cells become susceptible to conventional as well as nonconventional therapeutic regimens.
Figure 4.
Down-regulation of miR-21 increases the CR colon cancer cell susceptibility to conventional therapeutic FUOX, nonconventional therapeutic CDF, and the combination of FUOX plus CDF. (A) MTT assay showing growth inhibition of anti-miR-21 or scramble miRNA-transfected CR HCT-116 cells in response to FUOX (100 µM 5-FU + 2.5 µM oxaliplatin), CDF (2.0 µM), or the combination of CDF+FUOX compared to the untreated cells (control). The cells were exposed to the therapeutic agents for 48 hours. Each value represents mean ± SD of six observations; *P < .001. (B) qRT-PCR showing anti-miR-21.transfected CR HCT-116 cells; expression of CD44 and EGFR is greatly inhibited in response to FUOX, CDF, or CDF + FUOX. Scramble miRNA-transfected CR cells were used as control; *P < .001. The CR cells were maintained in FUOX (50 µM 5-FU + 1.25 µM oxaliplatin) throughout as stated above in Materials and Methods section.
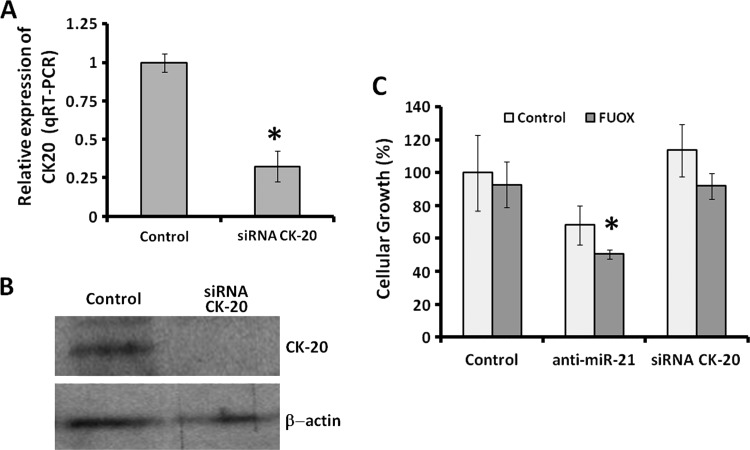
Down-regulation of CK-20 in CR Colon Cancer Cells Causes No Apparent Change in Proliferation
Although knocking down miR-21 or β-catenin induces differentiation, as evidenced by marked increases in CK-20 expression and alkaline phosphatase activity and also increases susceptibility of CR colon cancer cells to therapeutics, it is unclear if these changes are causally related to growth inhibition or epiphenomena of loss of CSC/CSLC characteristics that accompany down-regulation of miR-21. To address this issue, we downregulated CK-20 in CR HT-29 cells using the corresponding siRNA. The controls were transfected with scrambled siRNA. Following transfection, the cells were incubated in the absence or presence of FUOX for 48 hours. Transfection of CK-20-siRNA in CR HT-29 cells produced a marked reduction in CK-20 expression (Figure 5, A and B). The protein and mRNA levels of CK-20 were substantially reduced (70–95% reduction), as determined by qRT-PCR and Western blot analyses, when compared with the corresponding controls (Figure 5, A and B). However, down-regulation of CK-20 in CR HT-29 cells produced no significant change in cellular growth in the absence or presence of FUOX, when compared with the corresponding controls (Figure 5C). In contrast, down-regulation of miR-21 by antisense miR-21 that resulted in up-regulation of CK-20 significantly decreased cellular growth in the absence (basal) or presence of FUOX (Figure 5C). The results suggest that CK-20 acts as a marker of differentiation and does not play any pivotal role in regulating the growth of CR colon cancer cells.
Figure 5.
Down-regulation of CK-20 in CR colon cancer HT-29 cells causes no apparent change in proliferation and susceptibility to FUOX. (A) Real-time qRT-PCR showing down-regulation of CK-20 48 hours following transfection with CK-20 siRNA. Data represent means ± SD of three independent experiments; *P < .001, compared to the control. (B) Western blot analysis of CK-20 following downregulation of CK-20. (C) MTT assay showing the CR HT-29 cell proliferation and its susceptibility to FUOX after transfection with CK-20 siRNA, anti-miR-21, and the scramble transfected (control). Data represent means ± SD of three independent experiments.
Discussion
Although chemotherapeutic regimens consisting of 5-FU and other anticancer drugs, such as FOLFIRI or FOLFOX, are used as a first line of standard chemotherapy for patients with advanced colorectal cancer, virtually all the responses are incomplete and emergence of resistance, with subsequent recurrence of the cancer, is universal. Currently, effectiveness of conventional cancer therapeutics is usually evaluated by the reduction in tumor mass, resulting from elimination/killing of differentiated or undifferentiated cells that form the bulk of the tumor. However, the population of CSCs/CSLCs remains untouched and may even be enriched resulting in relapse of the disease. Earlier, we reported that exposure of colon cancer HCT-116 or HT-29 cells to FOLFOX that inhibited their growth led to enrichment of CSC/CSLC phenotype as evidenced by significantly increased proportion of CD44-, CD166-, and/or CD133-positive cells [8] accompanied by increased colonosphere forming ability in vitro and tumor formation in SCID mice [9].
It has been well documented that dysregulation of Wnt/β-catenin signaling plays a pivotal role in the regulation of colonic stem cells, which has also been implicated in colon carcinogenesis [25]. Our earlier studies have also shown that Wnt/β-catenin signaling pathway plays a pivotal role in regulating growth and maintenance of CSC/CSLC-enriched colonies derived from various human colon cancer cells such as colonospheres [20]. The colonospheres are found to express LGR5, CD44, CD166, Musashi-1, and epithelial-specific antigen, which are markers of CSCs/CSLCs, and show elevated levels of total β-catenin causing exaggerated transcriptional activation of TCF/LEF gene responsible for the progression of cancer stem cells [20]. Colon cancer HCT-116 cells are heterozygous for β-catenin, harboring one wild-type allele and one mutant allele with inactivation of SER45, one of the residues phosphorylated by GSK3β [24,26]. Since one of alleles is wild type, we believe that this might be responsible for siRNA-induced down-regulation of β-catenin.
More interestingly, our studies also show that miR-21, whose levels are greatly increased in colon tumors, is also greatly elevated in CR colon cancer cells [9]. Overexpression of miR-21 led to increased TCF/LEF activity, increased sphere forming ability in vitro, and tumor formation in SCID mice and that down-regulation of miR-21 in CR colon cancer cells markedly decreased their sphere-forming ability, a phenomenon similar to that noted for β-catenin downregulated cells, supporting the contention that miR-21 regulates some of the functional properties of colon CSCs [9].
These observations prompted us to examine whether down-regulation of miR-21 in CR colon cancer cells would induce differentiation of CSCs/CSLCs and render them susceptible to the conventional or nonconventional chemotherapeutic regimens. The basis for this investigation is that while CSCs/CSLCs are resistant to conventional chemotherapy, differentiated or differentiating cells that form the bulk of the tumor are sensitive to chemotherapy. In the current investigation, we have observed that down-regulation of miR-21 in CR HCT-116 cells, which are highly enriched in CSCs/CSLCs, leads to differentiation as evidenced by increased expression of gastrointestinal differentiation marker CK-20 [23] and an increase in alkaline phosphatase activity [27,28] and may also be causally related to decreased levels of CD44, pri-miR-21, and EGFR.
We have also hypothesized that down-regulation of miR-21 in CR cells that leads to differentiation would render them susceptible to conventional or nonconventional chemotherapeutics. CDF, a newly developed analog of curcumin with a greater bioavailability than the parent compound, have been shown to exert a greater growth inhibitory property than curcumin [7]. Recent studies also showed that CDF was much more superior in the killing of gemcitabine-resistant pancreatic cancer cells with epithelial-to-mesenchymal phenotype that is reminiscent of CSCs [29]. We have reported that CDF in combination with FUOX causes growth inhibition in CR colon cancer cells in vitro, decreases the proportion of CSCs/CSLCs, and inhibits expression of miR-21 [7]. Our current observation that following down-regulation of miR-21 in CR colon cancer HCT-116 cells they become highly susceptible to the growth inhibitory properties of FUOX, CDF, and CDF + FUOX clearly indicate increased vulnerability of CSC/CSLC-enriched CR colon cancer cells not only to conventional therapeutic FUOX but also to nonconventional therapeutics such as CDF or the combination of FUOX and CDF. Furthermore, the fact that downregulation of CK-20 by the corresponding siRNA produced no significant change in cellular growth of CR colon cancer cells in the absence or presence of FUOX suggest that CK-20 per se is not involved in regulating the growth of CR colon cancer cells but acts as a marker of differentiation.
In conclusion, the results of our current investigation suggest that down-regulation of miR-21 is an effective therapeutic strategy for CR colon cancer by regulating cancer stem cell differentiation and subsequently rendering them susceptible to therapeutic agents.
Footnotes
This work was supported by grants (APNM) from the National Institutes of Health/National Institute on Aging (AG014343) and the Department of Veterans Affairs. The authors declare that they have no competing interests.
References
- 1.American Cancer Society, author. Colorectal Cancer Facts & Figures 2011–2013. Atlanta, GA: American Cancer Society Inc; 2011. [Google Scholar]
- 2.Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5:275–284. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- 3.Sanders MA, Majumdar AP. Colon cancer stem cells: implications in carcinogenesis. Front Biosci. 2011;16:1651–1662. doi: 10.2741/3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dalerba P, Dylla SJ, Park IK, Liu R, Wang X, Cho RW, Hoey T, Gurney A, Huang EH, Simeone DM, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci USA. 2007;104:10158–10163. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patel BB, Yu Y, Du J, Levi E, Phillip PA, Majumdar AP. Age-related increase in colorectal cancer stem cells in macroscopically normal mucosa of patients with adenomas: a risk factor for colon cancer. Biochem Biophys Res Commun. 2009;378:344–347. doi: 10.1016/j.bbrc.2008.10.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jordan CT, Guzman ML, Noble M. Cancer stem cells. N Engl J Med. 2006;355:1253–1261. doi: 10.1056/NEJMra061808. [DOI] [PubMed] [Google Scholar]
- 7.Kanwar SS, Yu Y, Nautiyal J, Patel BB, Padhye S, Sarkar FH, Majumdar AP. Difluorinated-curcumin (CDF): a novel curcumin analog is a potent inhibitor of colon cancer stem-like cells. Pharm Res. 2011;28:827–838. doi: 10.1007/s11095-010-0336-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu Y, Kanwar SS, Patel BB, Nautiyal J, Sarkar FH, Majumdar AP. Elimination of colon cancer stem-like cells by the combination of curcumin and FOLFOX. Transl Oncol. 2009;2:321–328. doi: 10.1593/tlo.09193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu Y, Kanwar SS, Patel BB, Oh PS, Nautiyal J, Sarkar FH, Majumdar AP. MicroRNA-21 induces stemness by downregulating transforming growth factor beta receptor 2 (TGFβR2) in colon cancer cells. Carcinogenesis. 2012;33:68–76. doi: 10.1093/carcin/bgr246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 11.Sassen S, Miska EA, Caldas C. MicroRNA: implications for cancer. Virchows Arch. 2008;452:1–10. doi: 10.1007/s00428-007-0532-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lussier YA, Stadler WM, Chen JL. Advantages of genomic complexity: bioinformatics opportunities in microRNA cancer signatures. J Am Med Inform Assoc. 2012;19:156–160. doi: 10.1136/amiajnl-2011-000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu Z, Liu M, Stribinskis V, Klinge CM, Ramos KS, Colburn NH, Li Y. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene. 2008;27:4373–4379. doi: 10.1038/onc.2008.72. [DOI] [PubMed] [Google Scholar]
- 14.Jazbutyte V, Thum T. MicroRNA-21: from cancer to cardiovascular disease. Curr Drug Targets. 2010;11:926–935. doi: 10.2174/138945010791591403. [DOI] [PubMed] [Google Scholar]
- 15.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 16.Nautiyal J, Kanwar SS, Yu Y, Majumdar AP. Combination of dasatinib and curcumin eliminates chemo-resistant colon cancer cells. J Mol Signal. 2011;6:7. doi: 10.1186/1750-2187-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patel BB, Sengupta R, Qazi S, Vachhani H, Yu Y, Rishi AK, Majumdar AP. Curcumin enhances the effects of 5-fluorouracil and oxaliplatin in mediating growth inhibition of colon cancer cells by modulating EGFR and IGF-1R. Int J Cancer. 2008;122:267–273. doi: 10.1002/ijc.23097. [DOI] [PubMed] [Google Scholar]
- 18.Patel BB, Gupta D, Elliott AA, Sengupta V, Yu Y, Majumdar AP. Curcumin targets FOLFOX-surviving colon cancer cells via inhibition of EGFRs and IGF-1R. Anticancer Res. 2010;30:319–325. [PMC free article] [PubMed] [Google Scholar]
- 19.Xu H, Yu Y, Marciniak D, Rishi AK, Sarkar FH, Kucuk O, Majumdar AP. Epidermal growth factor receptor (EGFR)-related protein inhibits multiple members of the EGFR family in colon and breast cancer cells. Mol Cancer Ther. 2005;4:435–442. doi: 10.1158/1535-7163.MCT-04-0280. [DOI] [PubMed] [Google Scholar]
- 20.Kanwar SS, Yu Y, Nautiyal J, Patel BB, Majumdar AP. The Wnt/β-catenin pathway regulates growth and maintenance of colonospheres. Mol Cancer. 2010;9:212. doi: 10.1186/1476-4598-9-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 22.Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 23.Chan CW, Wong NA, Liu Y, Bicknell D, Turley H, Hollins L, Miller CJ, Wilding JL, Bodmer WF. Gastrointestinal differentiation marker Cytokeratin 20 is regulated by homeobox gene CDX1. Proc Natl Acad Sci USA. 2009;106:1936–1941. doi: 10.1073/pnas.0812904106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler KW. Activation of β-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997).;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 25.Kolligs FT, Bommer G, Goke B. Wnt/beta-catenin/tcf signaling: a critical pathway in gastrointestinal tumorigenesis. Digestion. 2002;66:131–144. doi: 10.1159/000066755. [DOI] [PubMed] [Google Scholar]
- 26.Sparks AB, Morin PJ, Vogelstein B, Kinzler KW. Mutational analysis of the APC/β-catenin/Tcf pathway in colorectal cancer. Cancer Res. 1998;58:1130–1134. [PubMed] [Google Scholar]
- 27.Speckmann B, Bidmon HJ, Pinto A, Anlauf M, Sies H, Steinbrenner H. Induction of glutathione peroxidase 4 expression during enterocytic cell differentiation. J Biol Chem. 2011;286:10764–10772. doi: 10.1074/jbc.M110.216028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oh PS, Patel VB, Sanders MA, Kanwar SS, Yu Y, Nautiyal J, Patel BB, Majumdar AP. Schlafen-3 decreases cancer stem cell marker expression and autocrine/juxtacrine signaling in FOLFOX-resistant colon cancer cells. Am J Physiol Gastrointest Liver Physiol. 2011;301:G347–G355. doi: 10.1152/ajpgi.00403.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ali S, Ahmad A, Banerjee S, Padhye S, Dominiak K, Schaffert JM, Wang ZW, Philip PA, Sarkar FH. Gemcitabine sensitivity can be induced in pancreatic cancer cells through modulation of miR-200 and miR-21 expression by curcumin or its analogue CDF. Cancer Res. 2010;70:3606–3617. doi: 10.1158/0008-5472.CAN-09-4598. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]