Abstract
High-risk individuals of familial pancreatic cancer (FPC) families are considered to be good candidates for screening programs to detect early PC or its high-grade precursor lesions, especially pancreatic intraepithelial neoplasia (PanIN) 2/3 lesions. There is a definite need for diagnostic markers as neither reliable imaging methods nor biomarkers are available to detect these lesions. On the basis of a literature search, the potential serum markers neutrophil gelatinase-associated lipocalin (LCN2), metallopeptidase inhibitor 1 (TIMP1), chemokine (C-X-C motif) ligand 16 (CXCL16), IGFBP4, and iC3a, which were first tested in transgenic KrasLSL.G12D/+;p53R172H/+;Pdx1-Cre mice, were identified. ELISA analyses of LCN2, TIMP1, and CXCL16 revealed significantly higher levels in mice with PanIN2/3 lesions or PC compared to mice with normal pancreata or PanIN1 lesions. Analysis of preoperative human serum samples from patients with sporadic PC (n = 61), hereditary PC (n = 24), chronic pancreatitis (n = 28), pancreatic neuroendocrine tumors (n = 11), and FPC patients with histologically proven multifocal PanIN2/3 lesions (n = 3), as well as healthy control subjects (n = 20), confirmed significantly higher serum levels of LCN2 and TIMP1 in patients with PC and multifocal PanIN2/3 lesions. The combination of LCN2 and TIMP1 as a diagnostic test for the detection of PC had a sensitivity, specificity, and positive predictive value of 100% each. Although this preliminary finding needs to be validated in a large series of individuals at high risk for FPC, serum measurement of LCN2 and TIMP1 might be a promising screening tool.
Introduction
Familial pancreatic cancer (FPC) is a rare but established inherited tumor predisposition syndrome that accounts for about 3% of all PC cases [1,2]. FPC defines families with at least two first-degree relatives with confirmed PC that do not fulfill the criteria of other inherited tumor syndromes with an increased risk of the development of PC, such as Peutz-Jeghers syndrome or hereditary pancreatitis [3,4]. The major gene defect is yet to be identified, although germ line mutations in the BRCA2, PALB2, and ATM are causative in about 15% of FPC families [5 6 7 8, reviewed in 4]. A 2003 consensus conference considered it appropriate to perform PC screening for those individuals who are considered at high risk of developing the disease [3]. Individuals with at least a 10-fold increased risk of PC, such as members of FPC families with two or more affected first-degree relatives, are deemed good candidates for screening. Although reliable imaging tools as well as biomarkers for the early detection of PC or, even better, its high-grade precursor lesions, pancreatic intraepithelial neoplasia (PanIN) 2/3, are still lacking, magnetic resonance imaging (MRI) and endoscopic ultrasonography are currently felt to be the best imaging modalities for screening [4]. Thus, there is a definite need for biomarkers that facilitate PC screening in this setting. We performed a literature search that led to the selection of five markers: IGFBP4 [9], chemokine (C-X-C motif) ligand 16 (CXCL16) [10], metallopeptidase inhibitor 1 (TIMP1) [9], iC3b [11], and neutrophil gelatinase-associated lipocalin (LCN2/NGAL) [12–14]. Sporadic and familial PC are characterized by progression from PanIN with different grades of dysplasia (PanIN 1 to 3) toward invasive cancer. Pancreatic specimens of FPC patients often reveal a so-called “PanIN disease” [15]. The stepwise progression comprises activating mutations of the Kras oncogene and inactivation of the ARF-p53 tumor suppressor pathway in the great majority of cases [16]. Nowadays, genetically engineered mouse models of PC that closely recapitulate the histopathogenesis of the human disease and include the KrasLSL.G12D/+;p53R172H/+;Pdx1-Cre (KPC) mice that develop a spectrum of premalignant PanIN lesions and ultimately invasive carcinoma in 5 to 10 months [17–19] are available. Because pancreatic specimens of FPC patients often reveal a so-called “PanIN disease” [15], this mouse model was valuable for the study presented.
Materials and Methods
Literature Search
Protein markers overexpressed at the protein and RNA levels in human PC and mouse models of PC were compiled by searching the PubMed and MEDLINE databases for articles published from 1 January 1990 to 31 March 2010. The search terms “pancreatic cancer” or “familial pancreatic cancer” and “protein markers” or “biomarker”, or “early detection”, or “diagnostic test” were used. A second-level manual search included the reference list of the articles considered to be of interest. The literature search and study selection were performed by two authors (D.K.B. and E.P.S.). More recent publications were prioritized. This literature search led to the selection of five markers: IGFBP4 [9], CXCL16 [10], TIMP1 [9], iC3b [11], and LCN2/NGAL [12–14].
Transgenic Mouse Lines
Conditional LsL-Trp53R172H, LsL-KrasG12D, and Pdx1-Cre [14] strains were interbred to obtain LsL-KrasG12D;LsL-Trp53R172H; Pdx1-Cre triple mutant animals on a mixed 129/SvJae/C57Bl/6 background as described before [16]. The generation of RIP1-Tag2 mice as a model of pancreatic islet cell carcinogenesis has been previously reported [20]. All experiments were approved by the local committee for animal care and use. Animals were maintained in a climate-controlled room kept at 22°C, exposed to a 12:12-hour light-dark cycle, fed standard laboratory chow, and given water ad libidum.
For genotyping, genomic DNA was extracted from tail cuttings using the REDExtract-N-AmpTM Tissue Polymerase Chain Reaction (PCR) Kit (Sigma-Aldrich, St Louis, MO). Three PCRs were carried out for each animal to test for the presence of the oncogenic Kras (using LoxP) primers, p53, and Pdx1-Cre transgene constructs (using Crespecific primers), respectively, or one PCR to test for the presence of Tag2 in the endocrine mice.
Mice were killed, blood was collected from the thoracic cavity for serum, and the pancreas was removed and inspected for grossly visible tumors and preserved in 10% formalin solution (Sigma-Aldrich) for histology. Formalin-fixed, paraffin-embedded tissues were sectioned (4 µm) and stained with hematoxylin and eosin (H&E). Six sections (100 µm apart) of pancreatic tissues were histologically evaluated by an experienced pathologist (A.R.) blinded to the experimental groups. mPanIN lesions were classified according to histopathologic criteria as recommended elsewhere [21].
The pancreata of all KPC mice sacrificed were histologically analyzed and graded according to the highest lesions found before conducting the ELISAs [17,18]. The mice analyzed exhibited a range of PanINs (1 to 3; n = 27), as well as invasive carcinoma (n = 9). Wild-type mice of the same background and age served as controls (n = 20).
Human Samples
Serum samples from patients were obtained at the time of diagnosis following informed consent according to the ethics protocol from our university (No. 36/1997; 5/2003). Samples were stored at -80°C.
ELISAs of the three biomarkers LCN2, TIMP1, and CXCL16 were performed in the preoperative sera of patients who underwent potential curative pancreatic surgery for sporadic PC (n = 61), FPC (n = 24), chronic pancreatitis (CP; n = 28), and pancreatic endocrine tumors (n = 11), respectively. In addition, three FPC patients who underwent prophylactic surgery and revealed multifocal PanIN2/3 lesions without invasive PC upon histology were analyzed. Twenty healthy subjects aged between 38 and 63 years served as controls. The study was approved by the local ethics committee and all patients included in the analysis gave their informed consent (No. 36/1997; last amendment 5/2009).
Results
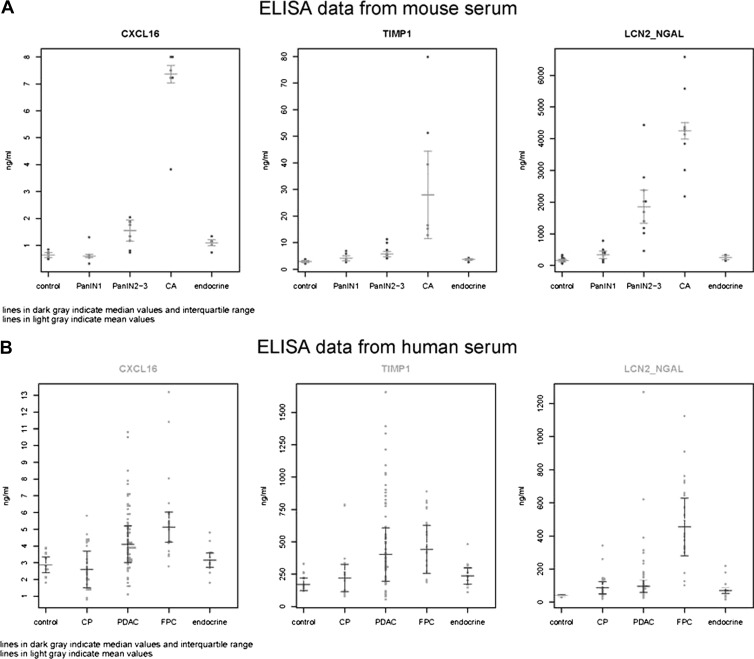
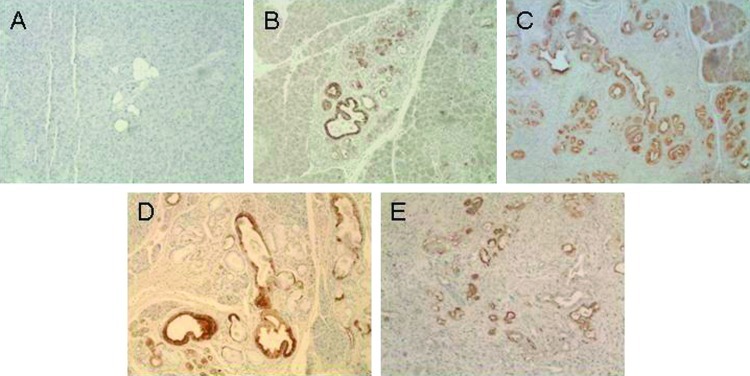
In a first step, the potential serum markers IGFBP4, CXCL16, TIMP1, iC3a, and LCN2 were tested in the serum of the KPC mice to determine their expression in mice with normal pancreas, different grades of PanINs (1 to 3), and invasive cancer. The serum drawn from mice at the time of sacrifice was tested in commercial ELISA tests specific for each of the five chosen markers. Following preliminary testing, the markers IGFBP4 and iC3a were excluded from further analysis as the values did not discriminate between affected and unaffected animals. In contrast, mouse CXCL16, mouse TIMP1, and mouse LCN2 ELISAs (R&D Systems, Wiesbaden, Germany) showed significantly higher serum levels in mice with PanIN2/3 lesions or invasive PC compared to controls. The results were also significant when comparing PC to PanIN1 levels. In addition, the serum levels of both CXCL16 and LCN2 were also higher in PanIN2/3 compared to PanIN1 mice. The results of the mouse ELISAs are shown in Table 1 and Figure 1A. On the basis of these data, LCN2 was the most promising serum biomarker for the early detection of PC in mice. These findings for LCN2 raised the question as to the cellular source of the antigen. To confirm that the serum LCN2 detected by the ELISA methodology was derived from pancreatic lesions, we performed immunohistochemical analysis with an anti-LCN2 antibody (R&D Systems) in pancreatic tissue from affected and unaffected mice. The level of expression of LCN2 was higher in PanIN lesions and PC than in control tissue (Figure 2). All mice with an increased level of serum LCN2 also showed strong positive immunostaining of high-grade PanIN lesions and cancer cells (data not shown). The promising results in KPC mice prompted us to perform ELISAs of the three biomarkers LCN2, TIMP1, and CXCL16 in human samples. These results are shown in Figure 1B.
Table 1.
ELISA Results from Mouse Serum.
| Marker | Control | PanIN1 | P Value | PanIN2/3 | P Value (vs Con) | P Value (vs PanIN1) | Invasive CA | P Value (vs Con) | P Value (vs PanIN1) |
| CXCL16 | 0.65 ng/ml (N = 9) | 0.69 ng/ml (N = 5) | .753* | 1.45 ng/ml (N = 8) | .0004 | .0152 | 6.96 ng/ml (N = 6) | .0001 | .0001 |
| TIMP1 | 2.9 ng/ml (N = 6) | 4.6 ng/ml (N = 6) | .035 | 6.6 ng/ml (N = 10) | .0021 | .0805* | 35.8 ng/ml (N = 6) | .0123 | .0162 |
| LCN2 | 165 ng/ml (N = 17) | 337 ng/ml (N = 16) | .0008 | 1937 ng/ml (N = 10) | .0001 | .0001 | 4249 ng/ml (N = 9) | <.0001 | .0001 |
ELISA tests were performed according to the recommendations by the manufacturer for CXCL16, TIMP1, and LCN2/NGAL (R&D Systems). The ELISA plate was measured on an Emax plate reader (Molecular Devices, Munich Germany) and then analyzed with SOFTmax Pro (Version 3-0) software.
To determine whether our assays show differences between healthy (wild-type) and affected (transgenic) animals, we evaluated serum protein levels from each of the ELISAs using an unpaired t test.
Values of P < .05 were considered to be statistically significant.
Not statistically significant.
Figure 1.
Scatter plots. (A) ELISA data from mouse serum. The results of mouse CXCL16, mouse TIMP1, and mouse LCN2/NGAL ELISAs were tested on mouse serum from control and transgenic mice with PanIN1, PanIN2/3, invasive pancreatic carcinoma (CA), or endocrine tumors. The dark gray lines indicate median values and the interquartile range. The light gray lines indicate mean values. (B) ELISA data from human serum. The results of human CXCL16, human TIMP1, and human LCN2/NGAL ELISAs were tested on human serum from control and patients with CP, sporadic PC (PDAC), FPC, or pancreatic endocrine tumors (endocrine). The dark gray lines indicate median values and the interquartile range. The light gray lines indicate mean values.
Figure 2.
Immunochemical staining of sections from wild-type (WT, A) and KPC mice with PanIN1B-2 (B) and PanIN2/3 (C) lesions and carcinoma (D, E) using anti-LCN2 antibody (AF 1857 [23]; R&D Systems, 1:1000 dilution). Strong, positive (brown) staining is apparent in the ductal lesions, whereas the normal/wild-type tissue is only weakly stained. For immunolabeling, formalin-fixed and paraffin-embedded archived tumor samples and corresponding normal tissues were stained as previously described [19]. Briefly, slides were heated to 60°C for 1 hour, deparaffinized using xylene, and hydrated by a graded series of ethanol washes. Antigen retrieval was accomplished by microwave heating in 10mM sodium citrate buffer of pH 6.0 for 10 minutes. For immunohistochemistry, endogenous peroxidase activity was quenched by 10-minute incubation in 3% H2O2. Nonspecific binding was blocked with 10%serum. Sections were then probed with lipocalin antibody (R&D Systems; AF1857) affinity-purified polyclonal goat IgG (1:1000) overnight at 4°C. Bound antibodies were detected using the avidinbiotin complex peroxidase method (ABC Elite Kit; Vector Labs, Burlingame, CA). Final staining was developed with the Sigma FAST DAB Peroxidase Substrate Kit (Sigma, Deisenhofen, Germany).
Receiver operating characteristic (ROC) curves were calculated to determine the ability of each individual marker, as well as of logistic models fitted with combinations of ELISA markers as predictors, to distinguish between pairs of different entities or so-called diagnostic groups. The results of the ROC curve analyses, including sensitivity, specificity, and cutoff value to distinguish between the two groups with the highest discrimination, are presented in Table 2. Due to the limited number of patients with multifocal PanIN2/3 lesions from FPC families (n = 3), results from these patients could not be included in the statistical analysis. They rather served as a first test whether the model correctly classified them as high-risk individuals. As in the KPC mice, LCN2 was the most sensitive and specific marker for the discrimination between normal individuals and FPC (sensitivity, 100%; specificity, 100%) or sporadic PC (sensitivity, 92%; specificity, 100%). LCN2 had its lowest sensitivity, 73% and 82%, in discrimination between normal individuals and individuals with endocrine tumors and CP, respectively. However, this is not a major drawback for the clinical setting for several reasons: 1) endocrine neoplasms rarely occur within the setting of FPC and also present an indication for operation itself, 2) patients with FPC rarely present advanced changes of CP on histopathologic examination [22], and 3) high-risk individuals of FPC families do not have as severe CP as the patients with end-stage CP that were used here for serum collection. Overall, the combination of markers LCN2 and TIMP1 could discriminate PC, FPC, and CP from controls, as well as FPC from CP with an area under the curve (AUC) of 92% to 100%, whereas the AUC (82%) was slightly lower for the discrimination between individuals with endocrine pancreatic tumors and healthy individuals. This might be partially explained by the low number (n = 11) of benign and malignant endocrine tumors that were included in the study (Table 2). A better discrimination could be achieved between FPC and endocrine tumors (AUC of 98.5%). A sufficient number of FPC patients with histologically confirmed multifocal PanIN2/3 lesions for a true validation of the proposed marker combination were not available, because these patients are extremely rare worldwide. However, preoperative and postoperative serum samples of three high-risk individuals from FPC families who underwent prophylactic total pancreatectomy for intraductal papillary mucinous neoplasm (IPMN)-like lesions on MRI and endoscopic ultrasonography were tested. All three patients had histologically confirmed multifocal PanIN2/3 lesions but no invasive cancers. The LCN2 and TIMP1 serum levels were above the calculated cutoff values (LCN2, 102 ng/ml; TIMP1, 273 ng/ml) in all three patients. The postoperative serum levels of both markers, taken within 4 weeks after surgery, dropped below the cutoff values in all three patients. This suggests that the proposed biomarkers may indeed be suitable for early detection of preneoplastic lesions of PC in high-risk individuals.
Table 2.
Results of ROC Curves of ELISA Analyses.
| LCN2/NGAL | TIMP1 | CXCL16 | TL* | CTL† | ||
| con-FPC | Sensitivity | 1 | 0.80 | 0.81 | 1 | 1 |
| Specificity | 1 | 0.85 | 1 | 1 | 1 | |
| AUC | 1 | 0.885 | 0.934 | 1 | 1 | |
| Cutoff | 42.3–102 | 273 | 4.17 | |||
| con-PC | Sensitivity | 0.92 | 0.75 | 0.62 | 0.93 | 0.93 |
| Specificity | 1 | 0.85 | 0.85 | 1 | 1 | |
| AUC | 0.956 | 0.812 | 0.778 | 0.967 | 0.971 | |
| Cutoff | 43.4 | 282 | 3.7 | |||
| con-Endo | Sensitivity | 0.73 | 0.60 | 0.70 | 0.82 | 0.73 |
| Specificity | 1 | 0.69 | 0.54 | 0.82 | 1 | |
| AUC | 0.811 | 0.600 | 0.612 | 0.818 | 0.802 | |
| Cutoff | 69 | 231 | 2.9 | |||
| con-CP | Sensitivity | .825 | 0.47 | 0.35 | 0.89 | .89 |
| Specificity | 1 | 0.69 | 1 | 1 | 1 | |
| AUC | 0.859 | 0.456 | 0.473 | 0.912 | 0.918 | |
| Cutoff | 48.4 | 243 | 4 | |||
| CP-FPC | Sensitivity | 0.90 | 0.80 | 0.95 | 0.92 | 0.92 |
| Specificity | 0.92 | 0.71 | 0.68 | 1 | 1 | |
| AUC | 0.958 | 0.775 | 0.864 | 0.983 | 0.981 | |
| Cutoff | 176 | 273 | 3.4 | |||
| FPC-Endo | Sensitivity | 0.85 | 0.70 | 0.86 | 0.92 | 0.92 |
| Specificity | 1 | 0.80 | 0.80 | 1 | 1 | |
| AUC | 0.968 | 0.810 | 0.871 | 0.985 | 0.985 | |
| Cutoff | 301 | 318 | 3.69 |
For the human samples, ROC curves were used to determine the ability of each individual marker to distinguish between the diagnostic groups. The discrimination was determined for each of the following pairs with the group to the right defined as affected: control < endocrine < CP < duAdCa or FPC. This allowed for the determination of sensitivity (true-positive rate) and specificity (true-negative rate) as well as the determination of the AUC as a performance index. For the combinations of two or three markers, a logistic regression for binary outcome, affected or not, was performed as a first step to obtain a model with regression coefficients, which reflect the strength of influence for each marker on the probability that one of the two outcomes is present in an individual. The resulting logistic model was applied to determine the probability for each person to have one of the two outcomes. These individual probabilities based on the marker combination were then used to create an ROC curve, similar to the serum protein levels when looking only at a single marker.
TIMP1 plus LCN2.
CXCL16 plus TIMP1 plus LCN2.
Discussion
As there is a definite need to improve upon the screening of individuals at high risk of developing precursor lesions and ultimately PC, we tested the ability of ELISAs to detect PC first in a transgenic mouse model and then in human samples. The results of our study demonstrate that the KPC mouse models of PC can be used to identify and test diagnostic markers. Comparative analysis of candidate biomarkers clearly documented a similarity of serum levels in human and mice with distinct pancreatic lesions relative to normal specimens. The serum levels of LCN2 and TIMP1 correlated with the severity of the neoplastic changes in the pancreatic tissue in both mice and humans. The diagnostic model fitted for these two markers could reliably discriminate between normal and precancerous/cancerous pancreatic lesions in our training data set and thus might be a promising tool for PC screening in high-risk individuals of FPC families. The two markers might be most valuable for individuals from FPC families who present IPMN-like lesions or unclear lesions on imaging. However, these data have to be considered preliminary, especially due to the small number of FPC patients with histologically verified PanIN2/3 lesions. The validation of the diagnostic value of the serum marker combination LCN2 and TIMP1 for the early detection of PC in a large cohort of high-risk individuals of FPC families is now mandatory.
Acknowledgments
We thank Aninja Baier and Helena Honig for their excellent technical assistance. We express our appreciation to Günther Kloppel for the histopathologic evaluation of the resected pancreata of the three highrisk individuals and to all patients who participated in the study.
Footnotes
This work was supported by the Deutsche Krebshilfe (109126 to E.P.S., V.F., P.L., and D.K.B.). The authors declare no conflict of interest.
References
- 1.Bartsch DK, Kress R, Sina-Frey M, Grützmann R, Gerdes B, Pilarsky C, Heise JW, Schulte KM, Colombo-Benkmann M, Schleicher C, et al. Prevalence of familial pancreatic cancer in Germany. Int J Cancer. 2004;110:902–906. doi: 10.1002/ijc.20210. [DOI] [PubMed] [Google Scholar]
- 2.Hemminki K, Li X. Familial and second primary pancreatic cancers: a nationwide epidemiologic study from Sweden. Int J Cancer. 2003;103:525–530. doi: 10.1002/ijc.10863. [DOI] [PubMed] [Google Scholar]
- 3.Brand RE, Lerch MM, Rubinstein WS, Neoptolemos JP, Whitcomb DC, Hruban RH, Brentnall TA, Lynch HT, Canto MI. Advances in counselling and surveillance of patients at risk for pancreatic cancer. Gut. 2007;56:1460–1469. doi: 10.1136/gut.2006.108456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartsch DK, Gress TM, Langer P. Familial pancreatic cancer—current knowledge. Nat Rev Gastroenterol Hepatol. 2012;9:445–453. doi: 10.1038/nrgastro.2012.111. [DOI] [PubMed] [Google Scholar]
- 5.Hahn SA, Greenhalf W, Ellis I, Sina-Frey M, Rieder H, Korte B, Gerdes B, Kress R, Ziegler A, Raeburn JA, et al. BRCA2 germline mutations in familial pancreatic carcinoma. J Natl Cancer Inst. 2003;95:214–221. doi: 10.1093/jnci/95.3.214. [DOI] [PubMed] [Google Scholar]
- 6.Slater EP, Langer P, Fendrich V, Habbe N, Chaloupka B, Matthäi E, Sina M, Hahn SA, Bartsch DK. Prevalence of BRCA2 and CDKN2a mutations in German familial pancreatic cancer families. Fam Cancer. 2010;9:335–343. doi: 10.1007/s10689-010-9329-6. [DOI] [PubMed] [Google Scholar]
- 7.Slater EP, Langer P, Niemczyk E, Strauch K, Butler J, Habbe N, Neoptolemos JP, Greenhalf W, Bartsch DK. PALB2 mutations in European familial pancreatic cancer families. Clin Genet. 2010;78:490–494. doi: 10.1111/j.1399-0004.2010.01425.x. [DOI] [PubMed] [Google Scholar]
- 8.Bartsch DK, Sina-Frey M, Lang S, Wild A, Gerdes B, Barth P, Kress R, Grützmann R, Colombo-Benkmann M, Ziegler A, et al. CDKN2A germline mutations in familial pancreatic cancer. Ann Surg. 2002;236:730–737. doi: 10.1097/00000658-200212000-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faca VM, Song KS, Wang H, Zhang Q, Krasnoselsky AL, Newcomb LF, Plentz RR, Gurumurthy S, Redston MS, Pitteri SJ, et al. A mouse to human search for plasma proteome changes associated with pancreatic tumor development. PLoSMed. 2008;5:e123. doi: 10.1371/journal.pmed.0050123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wente MN, Gaida MM, Mayer C, Michalski CW, Haag N, Giese T, Felix K, Bergmann F, Giese NA, Friess H. Expression and potential function of the CXC chemokine CXCL16 in pancreatic ductal adenocarcinoma. Int J Oncol. 2008;33:297–308. [PubMed] [Google Scholar]
- 11.Märten A, Buchler MW, Werft W, Wente MN, Kirschfink M, Schmidt J. Soluble iC3b as an early marker for pancreatic adenocarcinoma is superior to CA19.9 and radiology. J Immunother. 2010;33:219–224. doi: 10.1097/CJI.0b013e3181bed29f. [DOI] [PubMed] [Google Scholar]
- 12.Moniaux N, Chakraborty S, Yalniz M, Gonzalez J, Shostrom VK, Standop J, Lele SM, Ouellette M, Pour PM, Sasson AR, et al. Early diagnosis of pancreatic cancer: neutrophil gelatinase-associated lipocalin as a marker of pancreatic intraepithelial neoplasia. Br J Cancer. 2008;98:1540–1547. doi: 10.1038/sj.bjc.6604329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tonack S, Aspinall-O'Dea M, Neoptolemos JP, Costello E. Pancreatic cancer: proteomic approaches to a challenging disease. Pancreatology. 2009;9:567–576. doi: 10.1159/000212083. [DOI] [PubMed] [Google Scholar]
- 14.Gronborg M, Bunkenborg J, Kristiansen TZ, Jensen ON, Yeo CJ, Hruban RH, Maitra A, Goggins MG, Pandey A. Comprehensive proteomic analysis of human pancreatic juice. J Proteome Res. 2004;3:1042–1055. doi: 10.1021/pr0499085. [DOI] [PubMed] [Google Scholar]
- 15.Sipos B, Frank S, Gress T, Hahn S, Klöppel G. Pancreatic intraepithelial neoplasia revisited and updated. Pancreatology. 2009;9:45–54. doi: 10.1159/000178874. [DOI] [PubMed] [Google Scholar]
- 16.Hezel AF, Kimmelman AC, Stanger BZ, Bardeesy N, DePinho RA. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2006;20:1218–1249. doi: 10.1101/gad.1415606. [DOI] [PubMed] [Google Scholar]
- 17.Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, Ross S, Conrads TP, Veenstra TD, Hitt BA, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 18.Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, Rustgi AK, Chang S, Tuveson DA. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7:469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 19.Fendrich V, Chen NM, Neef M, Waldmann J, Buchholz M, Feldmann G, Slater EP, Maitra A, Bartsch DK. The angiotensin-I-converting enzyme inhibitor enalapril and aspirin delay progression of pancreatic intraepithelial neoplasia and cancer formation in a genetically engineered mouse model of pancreatic cancer. Gut. 2010;59:630–637. doi: 10.1136/gut.2009.188961. [DOI] [PubMed] [Google Scholar]
- 20.Hanahan D. Heritable formation of pancreatic beta-cell tumours in transgenic mice expressing recombinant insulin/simian virus 40 oncogenes. Nature. 1985;315:115–122. doi: 10.1038/315115a0. [DOI] [PubMed] [Google Scholar]
- 21.Hruban RH, Adsay NV, Albores-Saavedra J, Anver MR, Biankin AV, Boivin GP, Furth EE, Furukawa T, Klein A, Klimstra DS, et al. Pathology of genetically engineered mouse models of pancreatic exocrine cancer: consensus report and recommendations. Cancer Res. 2006;66:95–106. doi: 10.1158/0008-5472.CAN-05-2168. [DOI] [PubMed] [Google Scholar]
- 22.Langer P, Kann PH, Fendrich V, Habbe N, Schneider M, Sina M, Slater EP, Heverhagen JT, Gress TM, Rothmund M, et al. Five Years of prospective screening of high-risk individuals from families with familial pancreatic cancer. Gut. 2009;58,:1410–1418. doi: 10.1136/gut.2008.171611. [DOI] [PubMed] [Google Scholar]
- 23.Viau A, El Karoui K, Laouari D, Burtin M, Nguyen C, Mori K, Pillebout E, Berger T, Mak TW, Knebelmann B, et al. Lipocalin 2 is essential for chronic kidney disease progression in mice and humans. J Clin Invest. 2010;120:4065–4076. doi: 10.1172/JCI42004. [DOI] [PMC free article] [PubMed] [Google Scholar]