Abstract
Innate immune surveillance mechanisms lie at the heart of the antiviral response. A growing number of germ-line encoded pattern recognition receptors have been identified which protect the host from infection by sensing the presence of viral molecules and inducing antiviral defenses. Most compartments that viruses gain access to are under active surveillance by one or more pattern recognition receptors. Members of the Toll-like receptor family guard the extracellular milieu and endosomal compartment where they are activated by viral glycoproteins or nucleic acids, respectively. More recently, the cytosolic compartment has emerged as the frontline in the arsenal of the host’s antiviral defenses. Families of receptors in the cytosol recognize viral RNA or DNA or perturbations of cellular homeostasis and orchestrate effector responses to eliminate the invader. Here, we review this expanding area of innate immunity by focusing on the molecular mechanisms of cytosolic host-defenses.
Introduction
The innate immune system is composed of receptors that collectively serve as a pathogen sensor to monitor the extracellular, vacuolar, and cytosolic compartments for signs of infection. Viruses interact with all of these compartments. The cytosol in particular represents a critical subcellular niche in the life cycle of the majority of RNA viruses and a limited number of DNA viruses such as poxviruses. Furthermore, herpes viruses traverse the cytosol en route to the nucleus, the site of their replication. During these processes, virions and/or their components accumulate in the cytosol. Intensive investigation over the last five years or so has unveiled new receptors that patrol the cytosolic compartment [1]. These cytosolic receptors include the retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), the nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs), the more recently identified absent in melanoma 2(AIM2)-like receptors (ALRs) and an expanding family of DEXDc helicases (DLRs). During viral infection the cytosol accumulates viral RNAs or DNAs that originate from the incoming viral genome, viral transcripts, or transcription and replication intermediates. Consequently, the cytosolic sensing of viruses relies largely on viral nucleic acids as major viral pathogen-associated molecular pattern (PAMPs) [2]. The recognition of viral PAMPs by cytosolic sensors leads to the elaboration of a robust program of gene expression that involves the production of antiviral inflammatory cytokines, chemokines, and interferons (IFNs). Most extensively studied in this context are the type I IFNs and the IL-1 family of cytokines [3].
Cytosolic sensors that induce type I IFN responses
The production of the type I IFNs (IFNα/β) represents one of the pivotal responses mediating the antiviral immune response. Type I IFNs exert antiviral effects by acting on immune cells (both innate and adaptive immune cells) as well as non-immune cells such as epithelial cells. The production of IFNα/β at the initial stages of viral infection not only establishes an early antiviral state in non-immune cells but also primes for the subsequent development of optimal antigen-specific T cell and antibody responses. Type I IFN production is triggered by several different classes of receptors particularly those in the cytosol that primarily sense viral RNA or DNA.
Core signaling pathway mediates type I IFN production
Although a multitude of receptors exist in the cytosol that induce IFNα/β gene transcription, the majority converge on the inhibitor of kappa B (IkB) kinase related kinases TRAF family member-associated NF-κB activator (TANK), (TNF receptor-associated factor (TRAF) family member-associated nuclear factor (NF)-κB activator)-binding kinase 1 (TBK1) and IkB kinase epsilon [4,5]. In contrast to the classical IkB kinases, IKKα and β, these kinases phosphorylate and activate the transcription factors IFN regulatory factor 3 (IRF3) and IRF7 [3,6,7]. Phosphorylation of IRF3 and/or IRF7 leads to their dimerization, nuclear translocation and association with CREB binding protein (CBP)/p300. Activated IRF3 and IRF7 assemble an enhanceosome with the transcription factors NF-κB, activating transcription factor 2 (ATF-2), and c-jun all of which function cooperatively to drive IFNα/β gene transcription [8] (Figure 1).
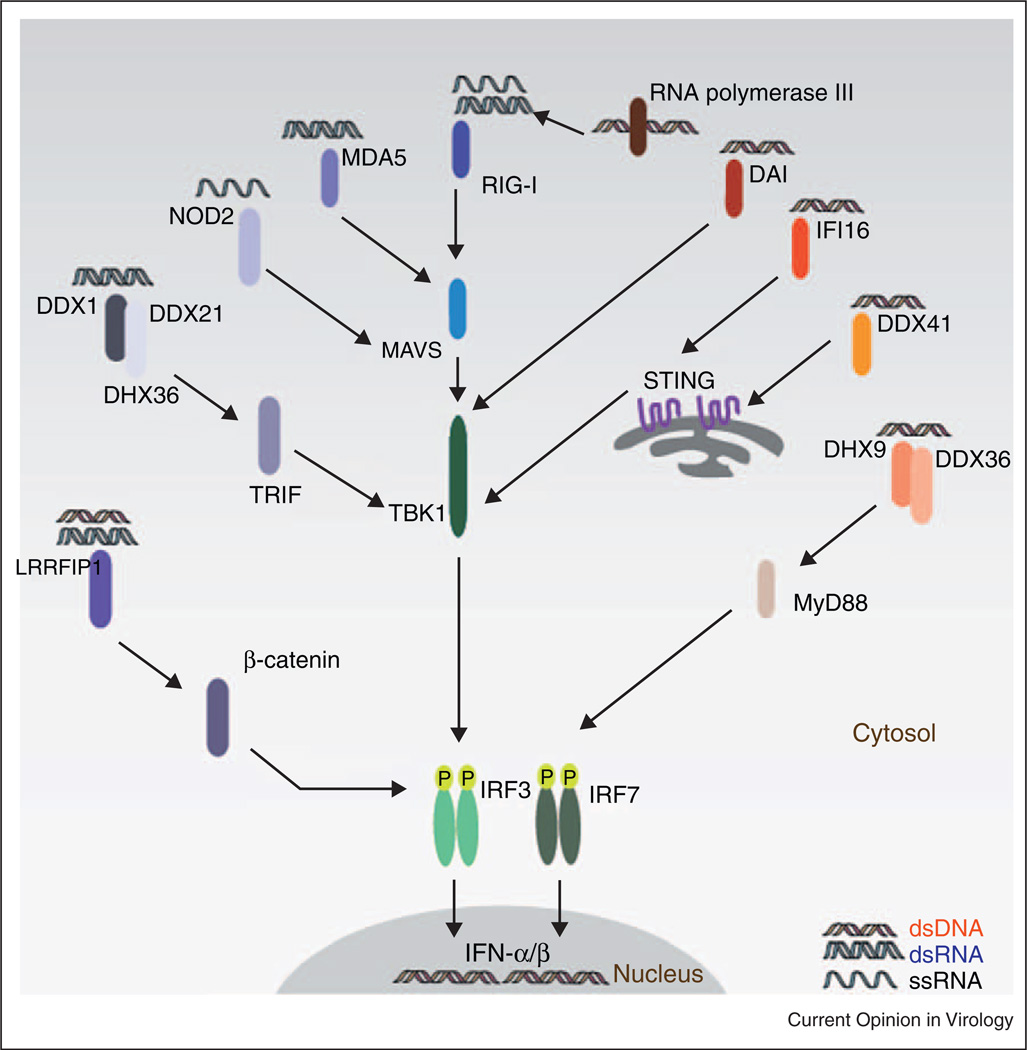
Figure 1.
Cytosolic receptors that sense viruses and induce type I interferons. Most cytosolic receptors that recognize RNAs such as RIG-I, MDA5, NOD2 activate TBK1–IRF3 via the mitochondria-associated MAVS. A recently identified receptor complex consisting of DDX1, DDX21, and DHX36 signals via TRIF to activate TBK1 after sensing RNA. On the other hand, DNA sensing in the cytosol triggers signaling via STING–TBK1–IRF3 axis to induce type I interferon production. RNA polymerase III recognizes cytosolic DNA and generates stimulatory RNA, which is subsequently detected by RIG-I. DHX9 and DDX36 sense cytosolic CpG and activate IRF7 via MyD88.
Although both TBK1 and IKKε can phosphorylate and activate IRF3, TBK1, which is ubiquitously expressed, appears to be the more important of the two kinases. Upstream of TBK1, adapter molecules integrate signals between different classes of sensing receptors and TBK1 kinase activity. To date, three distinct adapter molecules have been identified and include TRIF; a Toll/Interleukin-1 receptor (TIR) domain containing adapter molecule that is recruited either directly or indirectly to TIR domains of TLRs or some members of the DLRs, Mitochondrial antiviral-signaling protein (MAVS) (also called CARDif, Virus-Induced Signaling Adapto (VISA) and interferon promoter stimulating protein 1 (IPS1)); a caspase activation and recruitment domain (CARD) containing adapter molecule which is localized to the mitochondrion and relays signals from the RLRs or stimulator of IFNs (STING, also called transmembrane protein 173 (TMEM173),mediator of IRF-3 activation (MITA), endoplasmic reticulum IFN stimulator (ERIS) and membrane tetraspanner associated with MHC class II (MPYS)) an endoplasmic reticulum resident protein that relays signals from the ALRs or some of the DLRs. The RLRs, ALRs and DLRs are discussed further below.
IFN-inducing RNA sensors
The RLRs
RIG-I and melanoma differentiation-associated gene 5 (MDA5) were the first set of receptors identified to sense viral products in the cytosol [9]. RIG-I senses the nascent 5′ triphosphate moiety of viral genomes or virus derived transcripts of negative-sense ssRNA viruses [10]. In addition to the uncapped 5′ triphosphorylated RNA, base pairing at the 5′-end allows RIG-I to discriminate between viral and host mRNAs, which are capped with a 7-methyl-guanosine group [11–13]. In contrast, MDA5 is activated by longer dsRNA, a typical intermediate of the replication of plus-sense ssRNA viruses. Both RIG-I and MDA5 have a DExD/H-box RNA helicase domain and two N-terminal CARDs [10]. The binding of RNA by RIG-I and MDA5 results in the recruitment of the adapter protein MAVS via CARD–CARD interactions [11–14]. Recently, structural insights have provided important clues into the molecular mechanism of RIG-I activation [15••–18••]. The structure reveals that the CARDs of RIG-I are secured in a closed conformation by the helicase domains in the resting state. Once the C-terminal repressor and helicase domains bind RNA and ATP, a rapid and robust conformational change ensues releasing the CARDs, which are then capable of associating with the E3 ubiquitin ligase, tripartite motif protein 25 (TRIM25). K63-linked ubiquitination of the CARD by TRIM25 enables recruitment of MAVS. The MAVS protein plays a critical role in the assembly of an antiviral signaling complex on mitochondrial membranes. Intriguing recent evidence suggests that a conformational change in MAVS during virus infection, leads to the prion-like formation of functional self-aggregates that provide a sensitive trigger for antiviral signaling to the TBK1–IRF3 axis [19]. MAVS also resides on peroxisomes from where it exerts a rapid albeit blunted antiviral response that appears to be independent of autocrine IFN signaling to curb viral replication while production of type I IFNs, IFN signaling and induction of antiviral interferon-stimulated gene (ISGs) get underway [20]. Genetic evidence has revealed that RIG-I and MDA5 discriminate between different classes of RNA [21,22]. RIG-I is required for triggering antiviral responses against several Flaviviridae, Paramyxoviridae, Orthomyxoviridae and Rhabdoviridae, whereas MDA5 is required for the response against picornaviruses like encephalomyocarditis virus (EMCV) [23,24]. Both RIG-I and MDA5 are required for type I IFN responses to a certain group of viruses including the rotavirus and human rhinovirus [25,26]. LGP2 is the third member of RLR family and is structurally similar to RIG-I and MDA5 but lacks CARDs. LGP2 has been shown to bind RNA and compete with RIG-I and MDA5 thereby acting as a negative regulator of the RLRs [27,28]. Additional evidence, however, has indicated that LGP2 also acts in a positive manner and is required for RNA virus recognition by RIG-I and MDA5 [29].
NOD2
NOD2 is a well-documented cytosolic receptor for peptidoglycan from bacterial pathogens. Surprisingly, NOD2 has also been shown to recognize single stranded RNA (ssRNA) and mediate the type I IFN response to some viruses [30•]. Upon recognizing ssRNA, NOD2 uses a central nucleotide-binding domain, and a C-terminal LRR domain, instead of the CARD, to recruit MAVS. Immune cells and mice lacking NOD2 have been shown to be defective in eliciting type I IFN responses to vesicular stomatitis virus (VSV) and respiratory syncytial virus (RSV). Whether NOD2 detects ssRNA in a manner similar to RIG-I or a distinct feature of ssRNA is unknown.
DDX1, DDX21, and DHX36
Recently, additional cytosolic sensors for dsRNA have been identified in myeloid-derived DCs. Proteomic approaches aimed at identifying dsRNA-binding proteins revealed three members of the DExD/H-box helicase family, DDX1, DDX21, and DHX36 which act as a triad complex to sense polyI:C [31]. DDX1 binds dsRNA and DDX21 and DHX36 which then recruit the TIR-adapter molecule TRIF to relay downstream signaling. shRNA-mediated attenuation of expression of these helicases reduced IFNα/β induction in response to influenza A and reoviruses. The relative role ofDDX1, 21 and DHX36 versus the RLRs remains unclear. In contrast to RIG-I and MDA5 which are present at low levels in cells and require type I IFN signaling to upregulate their expression, DDX1, DDX21, and DHX36 proteins are constitutively present and may be ready and available to prime antiviral responses in the early stages of infection before RIG-I and MDA5 come into play [31].
IFN-inducing DNA sensors
As outlined above, in addition to RNA sensing mechanisms a growing number of DNA sensing pathways have been identified which signal in response to DNA that gains access to the cytosolic compartment. The first evidence for the existence of a TLR9-independent pathway for detecting DNA was provided by Stetson and Medzhitov [32] and Ishii et al. [33] who reported the ability of dsDNA to induce type I IFNs in cells lacking TLR signaling. Subsequent research efforts have unveiled a plethora of molecules that detect DNA in the cytosolic compartment. Unlike the world of RNA sensors, where each pathway is relatively non-redundant, cytosolic DNA sensing that trigger type I IFN response involves several receptors with largely overlapping and cell-type specific functions.
DAI
DNA-dependent activator of IRFs (DAI) was the first molecule that was identified as a DNA sensor [34]. DAI was shown to trigger TBK1–IRF3-dependent activation of IFNα/β genes in response to synthetic DNA and herpes simplex virus 1 (HSV1) infection. However, the generation of a mouse strain lacking this gene clearly demonstrated that DAI plays a redundant and cell-type specific role in cytosolic DNA sensing [35].
RNA polymerase III
RNA polymerase III senses cytosolic AT-rich DNA. Instead of directly activating downstream signaling, however, RNA polymerase III signals via generation of an immunostimulatory RNA intermediate with an uncapped 5′ triphosphate group that is subsequently sensed by RIG-I. This pathway contributes to the expression of IFNα/β genes in response to adenovirus and Epstein–Barr virus [36••,37••].
IFI16
Most recently, the IFN inducible protein, IFI16 was identified as a DNA-binding protein which could bind dsDNA through hematopoietic IFN-inducible nuclear proteins with 200-amino acids (HIN) domains and associate with an endoplasmic reticulum resident protein STING leading to activation of the TBK1–IRF3 pathway. During HSV1 infection or following treatment with DNA derived from either the HSV1 or Vaccinia virus genomes type I IFN induction was compromised when IFI16 or its mouse ortholog (Ifi204, p204) were subjected to shRNA knockdown in human or mouse macrophages respectively [38•].
LRRFIP1
LRRFIP1 was found to detect both dsRNA and dsDNA in the cytosol. The unique feature of this pathway is that it contributes to type I IFN production not through the core STING–TBK1–IRF3 axis but via a β-catenin-dependent coactivator pathway. LRRFIP1 activates β-catenin phosphorylation and nuclear translocation promoting the recruitment of the p300-histone acetyltransferase to IRF3 at the Ifnb1 promoter, events which ultimately enhance transcription of the IFN-β gene [39].
DHX36 and DHX9
Like the DExD/H-box helicases outlined above which bind dsRNA two members of this large family, DHX36 and 9 bind cytosolic CpG-A and CpG-B DNA, respectively, in the cytosol of human pDCs. Upon recognizing CpG DNA, DHX36 and DHX9 trigger MyD88-dependent activation of IRF7 and NF-κB to initiate the transcription of proinflammatory genes [40•].
DDX41
The latest addition to the growing number of cytosolic DNA sensors isDDX41, a DExD/H helicase. A screening approach targeting 59 DExD/H helicases via small interfering RNA (siRNA) revealed that knockdown of DDX41 expression abrogated type I IFN production in response to cytosolic dsDNA in myeloid DCs and human monocytes [41•]. Importantly, HSV1 and adenovirus induced production of IFN-β was also found to be dependent on DDX41.DDX41, likeDDX1, 21 and DHX36 in the RNA sensing pathway, appears to be responsible for sensing DNA at the very early stages of viral infections before additional sensors such as IFI16 begin to play a role in this process.
A common feature of many of these cytosolic DNA sensing pathways is the involvement of STING. STING has been linked to IFI16 and DDX41 signaling, but extensive evidence points to a central role for STING in the antiviral DNA response. In the presence of cytosolic DNA, STING translocates from the ER to perinuclear microsomes containing Sec5 (EXOC2) and associates with TBK1 [42••,43••]. Genetic studies have revealed an absolute requirement for STING in the IFNα/β response to cytosolic DNA, several DNA viruses as well as a growing number of bacterial pathogens including the cytosolic pathogen Listeria monocytogenes.
Cytosolic sensors that induce IL-1 family of cytokines
Although the type I IFN response is the predominant antiviral signature associated with immunity to viruses, the cytokines belonging to the IL-1 family such as IL-1β and IL-18 also play an important role in the antiviral response [44]. These cytokines have potent proinflammatory functions and act in a number of ways to enhance antiviral immunity. IL-1β and IL-18 exert antiviral effects through distinct mechanisms. While IL-18 is mainly involved in coordinating IFN-γ production from NK cells and T cells at the early and late phases of infection, respectively, IL-1β governs the recruitment of inflammatory cells such as neutrophils to the site of infection and is important in the generation of optimal adaptive immunity.
Cytosolic sensing pathways contribute to the generation of IL-1β and IL-18 during virus infection. IL-18 and IL-1β are synthesized as inactive forms either constitutively or following NF-κB activation, respectively. The processing of these zymogens into biologically active cytokines is facilitated by the cysteinyl aspartate protease caspase-1, which is also present in the cell as an inactive zymogen. Caspases are responsible for crucial aspects of inflammation and cell death and can be broadly divided into two classes based on their substrate specificity: those that are pro-apoptotic and those that are proinflammatory. Caspase-1 is part of this latter group, which also includes caspase-4, caspase-5, caspase-11, and caspase-12 (reviewed in [45]). The requirement for two distinct stimuli to regulate IL-1β production ensures that IL-1β is not inappropriately released, which could have deleterious consequences for the host. Indeed, excess production of IL-1β is associated with a number of hereditary periodic fever syndromes as well as autoimmune and inflammatory diseases such as gout and rheumatoid arthritis [46,47].
A multiprotein complex known as the inflammasome catalyzes the conversion of procaspase-1 to active caspase-1, which in turn catalyzes the conversion of proIL-1β and proIL-18 into the mature cytokines [44]. Inflammasome complexes form in the cytosol in response to a variety of both pathogenic as well as environmental and endogenous danger signals (reviewed extensively in [48]). Two distinct families of proteins can form inflammasomes including members of the NLR and ALR (also called pyrin and HIN domain (PYHIN)) families (Figure 2).The NLRs are a large family of cytosolic sensors (23 members in humans, 34 members in mice) whose crucial role in the immune system is now well accepted. The NLRs have a tripartite structure, consisting of a C-terminal leucine-rich repeat domain, a central nucleotide-binding oligomerization (NOD or NACHT) domain, and a variable N-terminal protein–protein interaction domain, which can be either a CARD, a Pyrin domain (PYD) or a baculovirus inhibitor of apoptosis repeat domain (BIR) [49,50]. The PYD, CARD or BIR domains facilitate downstream signaling through protein–protein interactions. The PYD domain (also known as a DAPIN or PAAD domain) is a death domain (DD) protein fold, which forms homotypic interactions with other PYD-containing proteins to form higher complexes with known roles in inflammation, apoptosis, and the cell cycle [51]. The best understood role of PYD domains relates to their ability to form ‘inflammasomes’. Inflammasome complexes assemble upon activation by an appropriate stimulus leading to the multimerization of the adaptor molecule ASC. Subsequently, procaspase-1 is recruited to ASC by means of interactions between the CARDs of ASC and that of caspase-1. These events lead to the auto-cleavage of caspase-1. The two resulting subunits p10 and p20 assemble into the active caspase-1 that then cleaves IL-1β and IL-18.
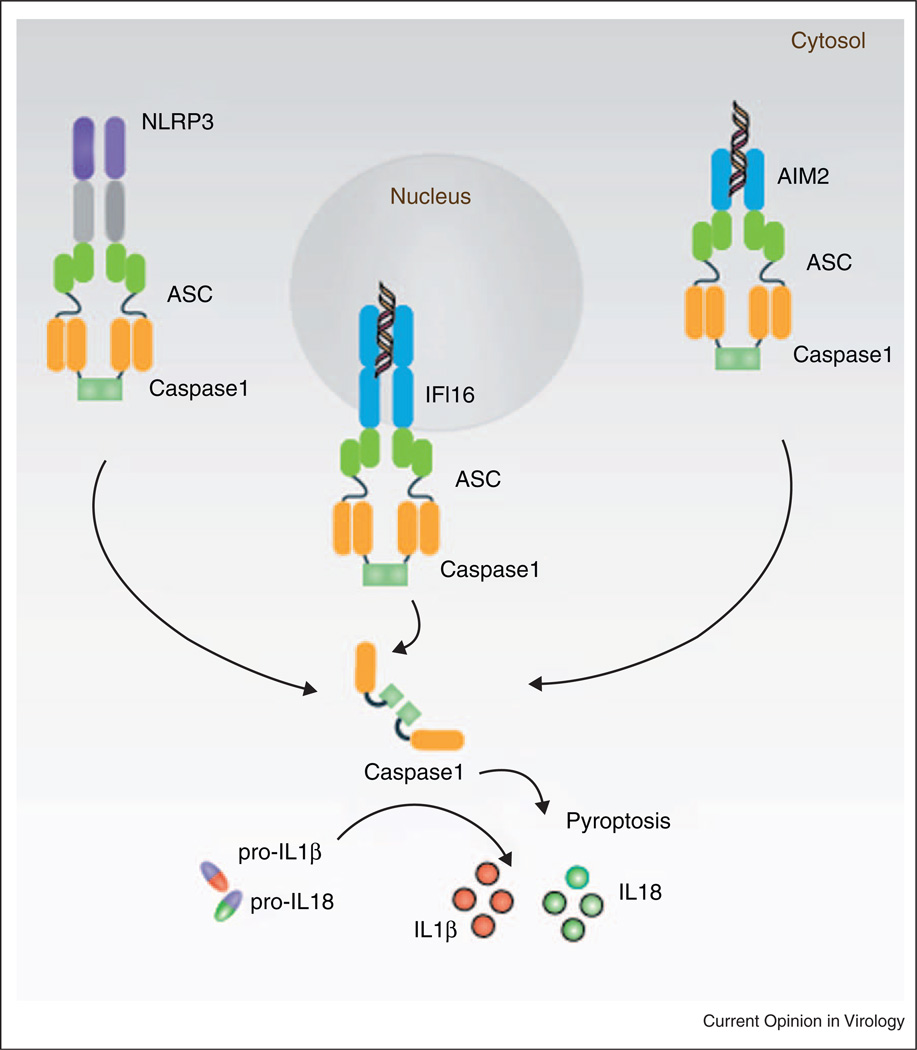
Figure 2.
Cytosolic receptors that sense viruses and induce inflammasome activation. NLRP3, AIM2, IFI16 sense viral infections and recruit ASC to form caspase-1-containing inflammasome complexes. The autoproteolytic processing of procaspase-1 results in the generation of active caspase-1, which then processes proIL-1β and proIL-18 to IL-1β and IL-18, respectively. The active caspase-1 also mediates an inflammatory form of cell death called pyroptosis.
Like the NLRs, the PYHIN proteins AIM2 and IFI16 have a pyrin domain and associate with ASC via pyrin domain interactions to activate caspase-1 and IL-1β/IL-18 processing (Figure 2). Six verified PYHIN proteins have been uncovered in mice (p202a, p202b, p203, p204, MNDAL, and AIM2) and four in humans (IFI16, AIM2, MNDA, and IFIX). Sequence analysis of the mouse genome predicts that seven additional members of this family likely exist [52]. The N-terminus of the PYHIN proteins, with the notable exception of p202a and p202b, contains a pyrin (PYD) domain. In addition to a PYD domain, the PYHIN proteins contain a DNA-binding HIN-200 domain.
The NLRP3 inflammasome
Although NLRP3 is the best-studied inflammasome pathway, the precise mechanism of NLRP3 activation remains elusive. NLRP3 is activated by diverse signals including microbial products (bacterial pore forming toxins and mRNA), endogenous products (uric acid and ATP) as well as crystalline particles (silica, asbestos and alum). All of these agonists trigger the assembly of the NLRP3 inflammasome (reviewed in detail in [53,54]). NLRP3 has been implicated in the recognition of both DNA and RNA viruses. Several groups have shown that NLRP3 contributes to antiviral responses in influenza A virus infection to varying extents [55–59]. Additionally, NLRP3 is involved in recognizing DNA viruses such as adenovirus and modified vaccinia virus Ankara strain [60,61]. Two potential mechanisms for the NLRP3 activation by viruses have been proposed. One of these proposes a model whereby viral RNAs lead to NLRP3-dependent IL-1β processing [55]. Additionally, an ion channel protein M2 of influenza A virus has been shown to trigger the NLRP3 inflammasome by causing perturbations in intracellular ion homeostasis [57]. How NLRP3 is activated during virus infection therefore remains unclear.
The AIM2 inflammasome
AIM2 forms a caspase-1 activating inflammasome upon sensing dsDNA from the cytosolic bacterial pathogens Fransicella tularensis and L. monocytogenes or the DNA viruses vaccinia and mouse cytomegalovirus (mCMV) [62–65,66•]. AIM2 does not appear to sense particular sequences of DNA, rather the length of the DNA is the determining feature. Double stranded DNA of less than ~50 base pairs is a poor agonist for AIM2. AIM2 is essential for caspase-1-dependent maturation of IL-1β and IL-18 in response to murine cytomegalovirus and vaccinia virus. In the case of mCMV infection, the ability of AIM2 to regulate IL-18 processing is particularly important. AIM2-deficient mice fail to make optimal amounts of IL-18 and as a consequence have compromised IFN-γ production by NK cells. Accordingly, mice lacking AIM2 are compromised in their ability to control mCMV early during infection. Surprisingly AIM2 is not involved in sensing other DNA viruses such as HSV1. While the mechanistic basis for this observation is unclear it suggests that certain viruses may have evolved mechanisms to escape AIM2-mediated surveillance.
The IFI16 inflammasome
Recently, IFI16 has been reported to form an ASC-containing inflammasome complex following infection with Kaposi sarcoma-associated herpesvirus (KSHV) in endothelial cells [67]. In contrast to other inflammasomes, IFI16 seems to trigger the inflammasome assembly in response to KSHV, most likely via recognition of its DNA, in the nucleus. However, the basis of differential recognition of viral and host DNA in the nucleus by IFI16 remains unknown.
Conclusions
The discovery of new classes of innate immune sensors, which patrol the cytosol for nucleic acids, or other components of viruses has provided enormous insights into the host–pathogen interface. However, despite these tremendous advances, key questions remain to be resolved before a comprehensive understanding of immunity to viruses can be obtained. With the exception of RIG-I and AIM2, the ligand specificity and molecular basis of ligand recognition are still unclear for most cytosolic sensors. While the contribution of cytosolic sensors to innate immune responses is relatively well-characterized at the cellular level, it is of the utmost importance from a therapeutic perspective that we understand the impact of these sensing and signaling pathways on disease pathogenesis and host resistance against viruses of clinical significance. Given the importance of innate immunity in vaccine adjuvancy and the potential for dysregulation of these pathways, understanding innate antiviral defenses is central in order to impact our ability to intervene in both infectious and non-infectious inflammatory and autoimmune diseases.
Acknowledgements
V.A.R is supported by a postdoctoral fellowship from the New England Regional Center of Excellence for Biodefense and Emerging Infectious Diseases (NERCE; NIH/NIAID AI057159). This work is also supported by NIH grant AI083713 to K.A.F.
Abbreviations
- AIM
absent in melanoma
- ATF
activating transcription factor
- CBP
CREB binding protein
- ERIS
endoplasmic reticulum IFN stimulator
- HIN
hematopoietic IFN-inducible nuclear proteins with 200-amino acids
- HSV
herpes simplex virus
- IkB
inhibitor of kappa B
- IPS
interferon promoter stimulating protein
- ISG
interferon-stimulated gene
- MAVS
Mitochondrial antiviral-signaling protein
- MITA
mediator of IRF-3 activation
- MPYS
membrane tetraspanner associated with MHC class II
- NF
nuclear factor
- PAMP
pathogen-associated molecular pattern
- PYHIN
pyrin and HIN domain
- RIG-I
retinoic acid-inducible gene I
- TANK
TRAF family member-associated NF-kappa-B activator
- TIR
Toll/Interleukin-1 receptor
- TMEM
transmembrane protein
- TRAF
TNF receptor-associated factor
- TRIM
tripartite motif protein
- VISA
Virus-Induced Signaling Adapto
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Barbalat R, Ewald SE, Mouchess ML, Barton GM. Nucleic acid recognition by the innate immune system. Annu Rev Immunol. 2011;29:185–214. doi: 10.1146/annurev-immunol-031210-101340. [DOI] [PubMed] [Google Scholar]
- 2.Rathinam VA, Fitzgerald KA. Innate immune sensing of DNA viruses. Virology. 2011;411:153–162. doi: 10.1016/j.virol.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hornung V, Latz E. Intracellular DNA recognition. Nat Rev Immunol. 2010;10:123–130. doi: 10.1038/nri2690. [DOI] [PubMed] [Google Scholar]
- 4.Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, Maniatis T. IKKε and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 5.Sharma S, tenOever BR, Grandvaux N, Zhou GP, Lin R, Hiscott J. Triggering the interferon antiviral response through an IKK-related pathway. Science. 2003;300:1148. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- 6.Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao S-M, Maniatis T. IKK[epsi] and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 7.Ishii K, Kawagoe T, Koyama S, Matsui K, Kumar H, Kawai T, Uematsu S, Takeuchi O, Takeshita F, Coban C. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008;451:725–729. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]
- 8.Panne D, Maniatis T, Harrison SC. An atomic model of the interferon-[beta] enhanceosome. Cell. 2007;129:1111–1123. doi: 10.1016/j.cell.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 10.Rehwinkel J, Reis e Sousa C. RIGorous detection: exposing virus through RNA sensing. Science. 2010;327:284. doi: 10.1126/science.1185068. [DOI] [PubMed] [Google Scholar]
- 11.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-[kappa] B and IRF3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 12.Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 13.Xu L-G, Wang Y-Y, Han K-J, Li L-Y, Zhai Z, Shu H-B. VISA is an adapter protein required for virus-triggered IFN-[beta] signaling. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 14.Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 15. Jiang F, Ramanathan A, Miller MT, Tang GQ, Gale M, Patel SS, Marcotrigiano J. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature. 2011;479:423–427. doi: 10.1038/nature10537.. In these studies the authors provide key insights into the structural basis of RIG-I activation.
- 16. Kowalinski E, Lunardi T, McCarthy AA, Louber J, Brunel J, Grigorov B, Gerlier D, Cusack S. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–435. doi: 10.1016/j.cell.2011.09.039. Same as ref. [15].
- 17. Luo D, Ding SC, Vela A, Kohlway A, Lindenbach BD, Pyle AM. Structural insights into RNA recognition by RIG-I. Cell. 2011;147:409–422. doi: 10.1016/j.cell.2011.09.023. Same as ref. [15].
- 18. Zeng W, Sun L, Jiang X, Chen X, Hou F, Adhikari A, Xu M, Chen ZJ. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–330. doi: 10.1016/j.cell.2010.03.029. Same as ref. [15].
- 19.Hou F, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146:448–461. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dixit E, Boulant S, Zhang Y, Lee ASY, Odendall C, Shum B, Hacohen N, Chen ZJ, Whelan SP, Fransen M, et al. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 2010;141:668–681. doi: 10.1016/j.cell.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii K. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 22.Loo Y-M, Fornek J, Crochet N, Bajwa G, Perwitasari O, Martinez-Sobrido L, Akira S, Gill MA, Garcia-Sastre A, Katze MG, et al. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol. 2008;82:335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, Flavell RA, Diamond MS, Colonna M. Essential role of MDA-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci U S A. 2006;103:8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loo YM, Fornek J, Crochet N, Bajwa G, Perwitasari O, Martinez-Sobrido L, Akira S, Gill MA, Garcia-Sastre A, Katze MG. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol. 2008;82:335. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Broquet AH, Hirata Y, McAllister CS, Kagnoff MF. RIG-I/MDA5/MAVS are required to signal a protective IFN response in rotavirus-infected intestinal epithelium. J Immunol. 2011;186:1618. doi: 10.4049/jimmunol.1002862. [DOI] [PubMed] [Google Scholar]
- 26.Slater L, Bartlett NW, Haas JJ, Zhu J, Walton RP, Sykes A, Dahdaleh S, Clarke DL, Belvisi MG, Kon OM. Co-ordinated role of TLR3, RIG-I and MDA5 in the innate response to rhinovirus in bronchial epithelium. PLoS Pathog. 2010;6:e1001178. doi: 10.1371/journal.ppat.1001178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, Foy E, Loo YM, Gale M, Akira S. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol. 2005;175:2851. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 28.Rothenfusser S, Goutagny N, DiPerna G, Gong M, Monks BG, Schoenemeyer A, Yamamoto M, Akira S, Fitzgerald KA. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J Immunol. 2005;175:5260–5268. doi: 10.4049/jimmunol.175.8.5260. [DOI] [PubMed] [Google Scholar]
- 29.Satoh T, Kato H, Kumagai Y, Yoneyama M, Sato S, Matsushita K, Tsujimura T, Fujita T, Akira S, Takeuchi O. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc Natl Acad Sci U S A. 2010;107:1512. doi: 10.1073/pnas.0912986107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sabbah A, Chang TH, Harnack R, Frohlich V, Tominaga K, Dube PH, Xiang Y, Bose S. Activation of innate immune antiviral responses by NOD2. Nat Immunol. 2009;10:1073–1080. doi: 10.1038/ni.1782.. This study showed that NOD2 can sense ssRNA species originating from viral infections and initiate type I interferon responses.
- 31.Zhang Z, Kim T, Bao M, Facchinetti V, Jung SY, Ghaffari AA, Qin J, Cheng G, Liu YJ. DDX1, DDX21, and DHX36 helicases form a complex with the adaptor molecule TRIF to sense dsRNA in dendritic cells. Immunity. 2011;34:866–878. doi: 10.1016/j.immuni.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 33.Ishii KJ, Coban C, Kato H, Takahashi K, Torii Y, Takeshita F, Ludwig H, Sutter G, Suzuki K, Hemmi H, et al. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat Immunol. 2006;7:40–48. doi: 10.1038/ni1282. [DOI] [PubMed] [Google Scholar]
- 34.Takaoka A, Wang Z, Choi M, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 35.Wang Z, Choi M, Ban T, Yanai H, Negishi H, Lu Y, Tamura T, Takaoka A, Nishikura K, Taniguchi T. Regulation of innate immune responses by DAI (DLM-1/ZBP1) and other DNA-sensing molecules. Proc Natl Acad Sci U S A. 2008;105:5477. doi: 10.1073/pnas.0801295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald K, Hornung V. RIG-I-dependent sensing of poly (dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779.. These studies revealed a novel pathway of DNA sensing mediated by RNA polymerase III.
- 37. Chiu Y, MacMillan J, Chen Z. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. Same as Ref. [36].
- 38. Unterholzner L, Keating S, Baran M, Horan K, Jensen S, Sharma S, Sirois C, Jin T, Latz E, Xiao T. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932.. Identified IFI16 as an IFN-inducing sensor for HSV-1 and vaccinia virus DNA in the cytosol.
- 39.Yang P, An H, Liu X, Wen M, Zheng Y, Rui Y, Cao X. The cytosolic nucleic acid sensor LRRFIP1 mediates the production of type I interferon via a [beta]-catenin-dependent pathway. Nat Immunol. 2010;11:487–494. doi: 10.1038/ni.1876. [DOI] [PubMed] [Google Scholar]
- 40. Kim T, Pazhoor S, Bao M, Zhang Z, Hanabuchi S, Facchinetti V, Bover L, Plumas J, Chaperot L, Qin J. Aspartate-glutamate-alanine-histidine box motif (DEAH)/RNA helicase A helicases sense microbial DNA in human plasmacytoid dendritic cells. Proc Natl Acad Sci U S A. 2010;107:15181. doi: 10.1073/pnas.1006539107.. DExD/H-box helicases have been identified as critical molecules sensing nucleic acids in the cytosol and mediating type I interferon production.
- 41. Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu Y-J. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol. 2011;12:959–965. doi: 10.1038/ni.2091. Same as Ref. [40].
- 42. Ishikawa H, Barber GN. STING an endoplasmic reticulum adaptor that facilitates innate immune signaling. Nature. 2008;455:674. doi: 10.1038/nature07317.. Identified STING as a central molecule in the cytosolic DNA-driven IFN responses.
- 43. Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. Same as Ref. [42].
- 44.Rathinam V, Fitzgerald K. Inflammasomes and anti-viral immunity. J Clin Immunol. 2010;5:632–637. doi: 10.1007/s10875-010-9431-4. [DOI] [PubMed] [Google Scholar]
- 45.Siegel RM. Caspases at the crossroads of immune-cell life and death. Nat Rev Immunol. 2006;6:308–317. doi: 10.1038/nri1809. [DOI] [PubMed] [Google Scholar]
- 46.McDermott MF, Tschopp J. From inflammasomes to fevers, crystals and hypertension: how basic research explains inflammatory diseases. Trends Mol Med. 2007;13:381–388. doi: 10.1016/j.molmed.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 47.Masters SL, Simon A, Aksentijevich I, Kastner DL. Horror autoinflammaticus: the molecular pathophysiology of autoinflammatory disease. Annu Rev Immunol. 2009;27:621. doi: 10.1146/annurev.immunol.25.022106.141627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martinon F, Mayor A, Tschopp J. The inflammasomes: guardians of the body. Annu Rev Immunol. 2009;27:229–265. doi: 10.1146/annurev.immunol.021908.132715. [DOI] [PubMed] [Google Scholar]
- 49.Martinon F, Tschopp J. NLRs join TLRs as innate sensors of pathogens. Trends Immunol. 2005;26:447–454. doi: 10.1016/j.it.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 50.Franchi L, Eigenbrod T, Munoz-Planillo R, Nunez G. The inflammasome: a caspase-1-activation platform that regulates immune responses and disease pathogenesis. Nat Immunol. 2009;10:241–247. doi: 10.1038/ni.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park HH, Lo Y-C, Lin S-C, Wang L, Yang JK, Wu H. The death domain superfamily in intracellular signaling of apoptosis and inflammation. Annu Rev Immunol. 2007;25:561–586. doi: 10.1146/annurev.immunol.25.022106.141656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ludlow LEA, Johnstone RW, Clarke CJP. The HIN-200 family: more than interferon-inducible genes? Exp Cell Res. 2005;308:1–17. doi: 10.1016/j.yexcr.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 53.Hornung V, Latz E. Critical functions of priming and lysosomal damage for NLRP3 activation. Eur J Immunol. 2010;40:620–623. doi: 10.1002/eji.200940185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Latz E. The inflammasomes: mechanisms of activation and function. Curr Opin Immunol. 2010;22:28–33. doi: 10.1016/j.coi.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Allen IC, Scull MA, Moore CB, Holl EK, McElvania-TeKippe E, Taxman DJ, Guthrie EH, Pickles RJ, Ting JP. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity. 2009;30:556–565. doi: 10.1016/j.immuni.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ichinohe T, Lee HK, Ogura Y, Flavell R, Iwasaki A. Inflammasome recognition of influenza virus is essential for adaptive immune responses. J Exp Med. 2009;206:79–87. doi: 10.1084/jem.20081667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ichinohe T, Pang IK, Iwasaki A. Influenza virus activates inflammasomes via its intracellular M2 ion channel. Nat Immunol. 2010;11:404–410. doi: 10.1038/ni.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Owen DM, Gale M., Jr Fighting the flu with inflammasome signaling. Immunity. 2009;30:476–478. doi: 10.1016/j.immuni.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 59.Thomas PG, Dash P, Aldridge JR, Jr, Ellebedy AH, Reynolds C, Funk AJ, Martin WJ, Lamkanfi M, Webby RJ, Boyd KL, et al. The intracellular sensor NLRP3 mediates key innate and healing responses to influenza A virus via the regulation of caspase-1. Immunity. 2009;30:566–575. doi: 10.1016/j.immuni.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Delaloye J, Roger T, Steiner-Tardivel QG, Le Roy D, Knaup Reymond M, Akira S, Petrilli V, Gomez CE, Perdiguero B, Tschopp J, et al. Innate immune sensing of modified vaccinia virus Ankara (MVA) is mediated by TLR2-TLR6, MDA-5 and the NALP3 inflammasome. PLoS Pathog. 2009;5:e1000480. doi: 10.1371/journal.ppat.1000480. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 61.Muruve D, PÈtrilli V, Zaiss A, White L, Clark S, Ross P, Parks R, Tschopp J. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature. 2008;452:103–107. doi: 10.1038/nature06664. [DOI] [PubMed] [Google Scholar]
- 62.Burckstummer T, Baumann C, Bluml S, Dixit E, Durnberger G, Jahn H, Planyavsky M, Bilban M, Colinge J, Bennett KL, et al. An orthogonal proteomic–genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 63.Fernandes-Alnemri T, Yu J, Datta P, Wu J, Alnemri E. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fernandes-Alnemri T, Yu JW, Juliana C, Solorzano L, Kang S, Wu J, Datta P, McCormick M, Huang L, McDermott E, et al. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat Immunol. 2010;11:385–393. doi: 10.1038/ni.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rathinam VA, Jiang Z, Waggoner SN, Sharma S, Cole LE, Waggoner L, Vanaja SK, Monks BG, Ganesan S, Latz E, et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. 2010;11:395–402. doi: 10.1038/ni.1864.. This study by generating AIM2-deficient mice demonstrated that AIM2 is an integral component of innate detection of certain DNA viruses.
- 67.Kerur N, Veettil MV, Sharma-Walia N, Bottero V, Sadagopan S, Otageri P, Chandran B. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi sarcoma-associated herpesvirus infection. Cell Host Microbe. 2011;9:363–375. doi: 10.1016/j.chom.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]