Abstract
Background
Nicotiana benthamiana has been widely used for transient gene expression assays and as a model plant in the study of plant-microbe interactions, lipid engineering and RNA silencing pathways. Assembling the sequence of its transcriptome provides information that, in conjunction with the genome sequence, will facilitate gaining insight into the plant’s capacity for high-level transient transgene expression, generation of mobile gene silencing signals, and hyper-susceptibility to viral infection.
Methodology/Results
RNA-seq libraries from 9 different tissues were deep sequenced and assembled, de novo, into a representation of the transcriptome. The assembly, of16GB of sequence, yielded 237,340 contigs, clustering into 119,014 transcripts (unigenes). Between 80 and 85% of reads from all tissues could be mapped back to the full transcriptome. Approximately 63% of the unigenes exhibited a match to the Solgenomics tomato predicted proteins database. Approximately 94% of the Solgenomics N. benthamiana unigene set (16,024 sequences) matched our unigene set (119,014 sequences). Using homology searches we identified 31 homologues that are involved in RNAi-associated pathways in Arabidopsis thaliana, and show that they possess the domains characteristic of these proteins. Of these genes, the RNA dependent RNA polymerase gene, Rdr1, is transcribed but has a 72 nt insertion in exon1 that would cause premature termination of translation. Dicer-like 3 (DCL3) appears to lack both the DEAD helicase motif and second dsRNA binding motif, and DCL2 and AGO4b have unexpectedly high levels of transcription.
Conclusions
The assembled and annotated representation of the transcriptome and list of RNAi-associated sequences are accessible at www.benthgenome.com alongside a draft genome assembly. These genomic resources will be very useful for further study of the developmental, metabolic and defense pathways of N. benthamiana and in understanding the mechanisms behind the features which have made it such a well-used model plant.
Introduction
Nicotiana benthamiana is a native tobacco that grows in isolated communities in remote regions of northern Australia, ranging from Western Australia to Queensland. It is an allo-tetraploid with 19 chromosome pairs and has 26 exclusively Australian native tobacco relatives [1]. An isolate of N. benthamiana from a single collection appears to have been passed from lab to lab over the last 80 years [2] and has been used widely in the study of plant-microbe interactions. N. benthamiana has been particularly useful to plant virologists as it is susceptible to over 500 different viruses [3] and more recently as a host for Virus Induced Gene Silencing (VIGS). VIGS technology is based on infecting plants with a virus that has been engineered to contain a fragment of the target plant gene. As the virus replicates and spreads the plant’s RNA interference mechanism generates short interfering RNAs (siRNAs) from the engineered viral genome, including the inserted fragment, and with these target silencing of the intended gene. This approach has helped to elucidate the functions of many different plant genes [4]. Over the last fifteen years, N. benthamiana has been increasingly adopted by the plant molecular biology community as the host for transient transgene expression by Agroinfiltration. In this method, Agrobacterium tumefaciens, containing a transgene construct, is infiltrated into the leaf using a needle-less syringe. The cells of the leaf are transiently transformed and the transgene, from within the T-DNA borders, is highly and rapidly expressed. This system has been particularly useful for examining the functions of proteins, their intracellular localization [3], [5], [6], as a design tool for metabolic engineering [7], [8] and for functional confirmation of forward and reverse genetics discoveries made in A. thaliana [9].
In addition to its role in VIGS, N. benthamiana has played a major part in characterisation of RNA silencing processes. It has become the species of choice in which to visualise silencing and silencing suppression, using GFP reporter transgenes. This includes showing the interactions between the plants’ silencing system and viral suppressors [10], [11], and the propagation and spread of mobile RNA silencing signals. However, almost all of genes in the plants’ RNA silencing pathways have been discovered and characterised in Arabidopsis. With this regard the visualisation and study of the processes in N. benthamiana has mainly relied on over-expression of Arabidopsis genes or constructs using N. benthamiana gene fragments that have been amplified from the genome using primers designed on partial ESTs or sequences from heterologous species.
To provide a better foundation upon which to use N. benthamiana as a model plant, we have assembled transcript sequences from 9 tissue types to provide a representation of the N. benthamiana transcriptome. This should enhance the annotation and use of the draft genome sequences of N. benthamiana [7], [12]. We have also focused on identifying genes involved in the RNA silencing and associated pathways, and provide an overview of their relative abundances in the different tissues.
Materials and Methods
Tissues and Total RNA Isolation
Nicotiana benthamiana plants were grown at 21°C under a 16-h photo-period and an 8-h dark period in an environmentally controlled glasshouse. Apex, capsule, flower, leaf, roots, seedling and stem samples from 6-week old plants were collected and immediately frozen in liquid nitrogen before storage at −80°C until further RNA extraction. For plants submitted to drought stress the water supply was stopped one week before the collection of the leaves (herein called Dstress). Tissues culture samples were grown from sterile N. benthamiana leaf pieces sub-cultured on MSN media. Green calli, approximately 4 weeks old, were collected and stored in the same conditions as the other samples (herein called tissue culture (TC)).
Total RNA was isolated from 500 mg of the selected plant tissues using the CTAB RNA extraction method [13]. Briefly, 5 ml of preheated (65°C) total RNA extraction buffer (2% (w/v) CTAB (Sigma), 2% (w/v) polyvinylpyrrolidone (PVP-40) (Sigma), 100 mM Tris HCl (pH 8.0), 25 mM EDTA (pH 8.0), 2 M NaCl and 2% β-mercaptoethanol) was added to each sample grounded in liquid nitrogen. Each sample was then extracted twice with an equal volume of Chloroform: Isoamylalcohol (24∶1), mixed and centrifuged at 4560 g for 20 minutes at room temperature. The resulting supernatant was carefully transferred into a new tube, mixed with LiCl at a final concentration of 2 M and incubated overnight at 4°C. After the incubation the samples were centrifuged at 4000 g for 20 min at 4°C and the resulting pellets were dissolved in 500 µl of preheated SSTE buffer (1 M NaCI, 0.5% SDS, 10 mM Tris HCl (pH 8.0), and 1 mM EDTA (pH 8.0)), extracted again with an equal volume of Chloroform: Isoamylalcohol and then washed with 75% ethanol and vacuum dried. Dried RNA pellets were diluted in RNAse free sterile water and stored at −80°C until used. The integrity of total RNA was determined by running samples on 1% denaturing agarose gel. The concentration and quality was initially assessed using a spectrophotometer (NanoDrop, Technologies Inc.) at an absorbance ratio of A260/230 and A260/280 nm. A Bioanalyzer (Agilent) was used to perform a final assessment on the quality prior to deep sequencing.
Deep Sequencing
RNA-seq libraries for each tissue were prepared from total RNA using the Illumina Truseq v1 RNA sample prep protocol (not strand specific) at the Australian Genome Research Facility (service provider). Briefly, poly-A mRNA were purified using oligo (dT) primed magnetic beads and chemically fragmented into smaller pieces. Cleaved fragments were converted to double-stranded cDNA using random hexamer primers. After purification and end-repair, the cDNA fragments were ligated to Illumina paired-end sequencing adapters which contain sample specific indexes to allow for sequencing of multiple libraries on a single lane of an Illumina flowcell. Following this, fragments were purified, size selected on a gel, and then amplified by PCR to obtain the final library. The libraries representing 9 tissues were then pooled in equal quantities into a single aliquot for sequencing on the Illumina HiSeq-2000 platform at the Australian Genome Research Facility, according to the manufacturers’ protocols. The libraries were sequenced as paired-end 100 nt reads. After sequencing, the reads were pre-processed as described below. All reads have been deposited in the Sequence Read Archive (SRA) at NCBI, under accession number SRA066161.
De novo Transcriptome Assembly, Annotation and Assessment of Completeness
Deep sequencing reads were quality assessed with the quality assessment software FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). For every tissue sample, reads were quality filtered (Q25) and trimmed (12 nt from 5′ end, and 5 nt from 3′ end) with the FastX-toolkit software suite (http://hannonlab.cshl.edu/fastx_toolkit/). The quality filtering of paired-end reads results in some reads losing its partner read. Further processing to extract properly-paired reads and orphaned (singleton) reads was performed with a script from the SHRIMP package [14].
All post-processed reads from the 9 tissue samples were pooled together and assembled de novo with Abyss v1.3 [15] with k-mer sizes ranging from 58 to 80 with a step size of 2. The k-mer assemblies were then merged with Trans-Abyss v1.1 [16]. Computational and storage resources were provided by Intersect Australia Ltd (http://www.intersect.org.au), and each k-mer assembly was allowed 16 CPUs and 60 GB of RAM. Each assembly typically completed within 24 hours.
De novo transcripts were annotated using Plant and Food Researchs’ in-house BioView Sequence Analysis and Annotation pipeline. BioView annotated the de novo transcripts by searching the SwissProt [17], Uniref90 (http://www.uniprot.org/downloads) [18], RefSeq (release 54) [19], and A. thaliana proteins (TAIR10) (http://www.arabidopsis.org/) databases using BLASTx (version 2.2.25) [20]. Searching against the NCBI non-redundant (NR) DNA database (ftp://ftp.ncbi.nlm.nih.gov/BLAST/db/) was performed using BLASTn (version 2.2.25) [20]. InterPro and Gene Ontology (GO) terms were derived following motif searching using InterProScan (version 4.8) [21] and InterPro Release 38 (http://www.ebi.ac.uk/interpro/). Each transcript was given an ID such as Nbv3K745626388, which depicts the assembly version (Nbv3), k-mer size from which the transcript was assembled (K74), and a 7 digit ID assigned by BioView.
The final “description” for each de novo transcript was based on a series of searches as well as the ratio lengths to the HSP (highest-scoring segment pairs) from BLAST. For each transcript, the best description and BLAST metrics for the best HSP to SwissProt was taken. The process was repeated with UniRef90 if no SwissProt matches were found, repeated with RefSeq if no UniRef matches were found, repeated with TAIR if no RefSeq matches were found, and repeated with the non-redundant (NR) databases in NCBI. Next, the HSP to sequence length ratios for query and subject was determined (qRatio = HSP length as percentage of query length; sRatio = HSP length as percentage of subject length). If the qRatio and sRatio are > = 90% and the description contains any of hypothetical, unknown, unclassified or uncharacterised terms, then the description is set to “highly conserved hypothetical protein”. If the qRatio and sRatio are > = 90% and identity is greater or equal to 50% but less than 90%, then it is called a “conserved hypothetical protein” if the description contains hypothetical, unknown, unclassified or uncharacterised terms. Otherwise “putative” is appended to the description. If the qRatio and sRatio are less than 90% but above 50% then “similar to” is appended. Otherwise “probable” is appended unless it is hypothetical, in which case the transcript is annotated a “hypothetical protein”.
For the creation of a unigene set of the raw assembly, cd-hit-est from the CD-HIT package [22] with a identity parameter of 95% was utilised. This will subsume all shorter sequences into a longer transcript if they have a sequence identity of 95% or greater.
To assess the completeness of the transcriptome assembly, the CEGMA (Core Eukaryotic Genes Mapping Approach) software was applied to identify the presence of a core protein set consisting of 248 highly conserved proteins that are found in a wide range of eukaryotes [23]. This software is usually used to assess the completeness of a genome assembly, but should also enable the assessment of a transcriptome under different interpretations. The transcriptome assembly was also compared to the tomato (Solanum lycopersicum) predicted proteins database (SL2.4) and the N. benthamiana unigene dataset v1 build from the Solgenomics database (http://solgenomics.net) using BLAST (threshold E-value of 1e−3).
Mapping statistics were obtained using standalone Bowtie v0.12.8 [24], mimicking parameters used in RSEM, except for the reporting of unique and multi-mapping reads (the options ‘-a -m 1’ were used).
The transcriptome assembly with annotations can be downloaded as a fasta file at http://www.benthgenome.com.
Transcript Abundance Analyses
Transcript quantification of the de novo assembly was carried out with RSEM, which allows for an assessment of transcript abundances based on the mapping of RNA-seq reads to the assembled transcriptome [25]. Briefly, RSEM calculates maximum likelihood abundance estimates as well as posterior mean estimates and 95% credibility intervals for genes/isoforms. Abundance estimates are generated in the form of two measures, one which gives an estimate of the number of fragments that can be derived from an isoform or gene (the expected counts (EC)), and the other which is the estimated fraction of transcripts within the sample that is represented by the given isoform or gene.
To adjust for library sizes and skewed expression of transcripts, the EC values were normalized using the Trimmed Mean of M-values (TMM) normalization method [26] utilising an edgeR script from the Trinity package [27], [28]. This method calculates the effective library size of each sample which was then used to normalize the EC values (FPKM (Fragments Per Kilobase per Million) values were not calculated). The second metric from RSEM was multiplied by one million to obtain a measure given as transcripts per million (TPM). To assess the distribution of the EC and TPM values for each tissue, basic statistics and residuals (the difference between a data point and the median) were calculated and plotted with the statistics package, R [29]. The TPM value is preferred over other metrics such as FPKM [30] and RPKM (Reads Per Kilobase per Million) [31] as it is independent of the mean expressed transcript length and thus more comparable between different species and samples [25]. Due to the nature of how EC and TPM measures are derived and applied [25], [26], [28], [32], [33], for comparisons of abundance TPM values were used, whereas other comparisons made use of normalized EC values.
Identification of RNAi Genes
Thirty one well-characterized RNAi associated genes (RNAi genes from here on) from A. thaliana (Table S4) were used to search for homologues in N. benthamiana. Protein sequences were retrieved from TAIR, and first screened against the tomato predicted protein database (SL2.40) at Solgenomics using BLASTp [34]. Both A. thaliana and tomato RNAi protein sequences were then used to screen the transcriptome assembly of N. benthamiana using tBLASTn. The most significant matches to A. thaliana and tomato queries were manually assessed and in cases of multiple matches, the query coverage (%) and identity (%) of high-scoring segment pairs were analysed further. Where possible, a full length sequence was manually reconstituted from the shorter transcript sequences, and where this was not possible, sequence data from our draft genome assembly [7] and/or from the N. benthamiana sequences deposited in the Solgenomics database [12] was used. All putative transcripts were subsequently translated and reciprocally checked against the TAIR and tomato databases to compare and confirm their identities. Domain searches of the translated sequences of the RNAi genes was performed with InterProScan (http://www.ebi.ac.uk/Tools/pfa/iprscan/), using all the default databases. An E-value of 1e−3 was used as the cut-off threshold, and Pfam results were given precedence. Abundances of the transcripts constituting an RNAi gene are reported as TPM values as described above.
The bioinformatically identified N. benthamiana RNAi CDS and protein sequences can be downloaded at http://www.benthgenome.com.
Phylogeny Analysis
Sequences of homologous RNAi proteins from Oryza sativa (http://rice.plantbiology.msu.edu/), Populas trichocarpa (http://www.phytozome.net/poplar), tomato, A. thaliana, Nicotiana attenuata, Nicotiana tabacum, Nicotiana glutinosa, and previously reported N. benthamiana entries (all Nicotiana species were retrieved from NCBI Genbank), were retrieved for phylogeny analyses. Accession numbers are reported in the figures. Alignments and tree construction were performed in the Geneious Pro version program (http://www.geneious.com), using the Muscle algorithm for alignments, and Neighbour Joining method with bootstrapping of 1000 for consensus tree construction.
Confirmation of an Insertion Sequence in Rdr1 in N. benthamiana
To confirm the insertion sequence in Rdr1 in N. benthamiana, PCR was carried out on a 1,035 nt region containing the insertion, using the forward primer 5′-CACCATGCAAAGTTTATTTTTCTGGTCCAG and reverse prime 5′-GCCCGGAAAGTTTGCAGCATCATTGAAAGA. These primers were also used to amplify the region from another commonly used N. benthamiana variant, 16C. The region was also amplified from N. tabacum. All sequences were subcloned and determined using conventional BigDye 3.1 chemistry.
Results and Discussion
De novo Transcriptome Assembly
The sequences of the N. benthamiana transcriptome were captured by preparing RNA-seq libraries from seven different tissues grown under normal conditions (apex, capsule, flower, leaf, roots, seedling and stem), and from two samples of tissues under stress (drought stressed leaf and tissue culture callus). To counter the inherent sequencing biases incurred in RNA-seq library preparation such as a reduced 5′ sequence complexity in the reads which could affect the assembly ([35]–[39], http://seqanswers.com/forums/showthread.php?t=11843), and to maximise the quality, the pooled 368,674,918 raw 100 nt reads from the libraries were trimmed by 12 nt and 5 nt at their 5′ and 3′ends, respectively, and reads containing bases below a quality score of 25 were discarded. This resulted in a total of 197,872,501 (76,224,365 paired-end and 45,423,771 singleton reads) post-processed 83 nt RNA-seq reads that were used for de novo transcriptome assembly. This very stringent filtering gave high quality sequences, but at the cost of discarding approximately 50% of the raw reads. Abyss v1.3 [15] was used to make assemblies from the reads, using increasing k-mer sizes from 58 to 80 (step size of 2), and the k-mer assemblies were merged using Trans-Abyss [16].
A summary of the assembly statistics is given in Table 1, and shows that the complete assembly yielded 237,340 contigs with a median contig size of 510 nt and a maximum contig size of 7969 nt. The total size of the assembly is ∼188 Mb with an average of about 56-fold coverage. The raw assembly contigs were clustered into a unigene dataset, using a threshold nucleotide identity of 95%, to produce 119,014 contigs (a reduction of ∼50%) with a total size of 89.6 Mb (Table 1). This is substantially more than the recently reported 73,041 unigenes (total size of 37.8 Mb) from a fungus-infected N. benthamiana transcriptome obtained using a Velvet/Oases assembly [40]. However, the Abyss/Trans-Abyss assembly pipeline tends to generate a large total number of contigs with a high proportion of them under 500 nt, especially from polyploid plant transcriptomes [41]–[43], and 50% of the unigenes in our assembly are between 100 and 500 nt (Table 1). Nonetheless, Trans-Abyss is reported to generate a better overall representation of transcripts over a broad range of expression levels [16], [41].
Table 1. Statistics of the de novo transcriptome assembly.
| Total number of input reads (as singletons) | 197872501 | |
| Paired | 76224365 | |
| Orphaned | 45423771 | |
| Read length | 83 | |
| Full | Unigenes | |
| Total size of contigs | 1.88E+08 | 89550506 |
| Number of contigs | 237340 | 119014 |
| Mean contig size | 794 | 752 |
| Median contig size | 510 | 486 |
| Min contig size | 81 | 109 |
| Max contig size | 7969 | 7969 |
| Number of contigs >100 nt | 237335 | 119014 |
| Number of contigs >500 nt | 119944 | 58396 |
| Number of contigs >1000 nt | 67089 | 30076 |
The sizes of the libraries used for the assembly are given in Table 2. While the number of reads from each tissue varied, especially for seedling, approximately 80 to 85% of paired reads from each tissue were able to map back to the assembled transcriptome. As also discussed later, the contribution of reads to the assembly from all tissues appeared to be quite similar.
Table 2. Mapping statistics of reads from each tissue to the transcriptome assembly.
| Apex | Capsule | Dstress | Flower | Leaf | Root | Seedling | Stem | TC | |
| Total reads | 7982949 | 16946882 | 13511245 | 13904896 | 15492665 | 14801396 | 90081852 | 14188920 | 10961696 |
| Number of singleton reads | 1794295 | 3942278 | 2956639 | 3215352 | 3570565 | 3421770 | 20764812 | 3237574 | 2520486 |
| Number of properly paired reads | 3094327 | 6502302 | 5277303 | 5344772 | 5961050 | 5689813 | 34658520 | 5475673 | 4220605 |
| Properly paired read mappings to all contigs | |||||||||
| Unique mapping pairs | 636942 | 1333015 | 1068272 | 1187332 | 1181070 | 1183725 | 6843574 | 1086467 | 889774 |
| Multi-mapping pairs | 1846302 | 4047038 | 3160791 | 3198648 | 3718383 | 3681321 | 22002639 | 3377221 | 2627857 |
| Un-mappable pairs | 611083 | 1122249 | 1048240 | 958792 | 1061597 | 824767 | 5812307 | 1011985 | 702974 |
| Percentage of mappable reads in library | 80.25 | 82.74 | 80.14 | 82.06 | 82.19 | 85.50 | 83.23 | 81.52 | 83.34 |
| Properly paired read mappings to >500 nt contigs | |||||||||
| Unique mapping pairs | 658098 | 1380468 | 1079509 | 1204381 | 1192679 | 1237355 | 6845002 | 1102866 | 902369 |
| Multi-mapping pairs | 1668747 | 3786276 | 2975615 | 2888572 | 3392882 | 3441306 | 19113437 | 3150478 | 2442318 |
| Un-mappable pairs | 767482 | 1335558 | 1222179 | 1251819 | 1375489 | 1011152 | 8700081 | 1222329 | 875918 |
| Percentage of mappable reads in library | 75.20 | 79.46 | 76.84 | 76.58 | 76.93 | 82.23 | 74.90 | 77.68 | 79.25 |
| Properly paired read mappings to >1000 nt contigs | |||||||||
| Unique mapping pairs | 552755 | 1417269 | 1041322 | 1045083 | 1157024 | 1111401 | 6319347 | 1060379 | 841743 |
| Multi-mapping pairs | 1133912 | 2848170 | 2349818 | 2208774 | 2586029 | 2549677 | 12603350 | 2474767 | 1868513 |
| Un-mappable pairs | 1407660 | 2236863 | 1886163 | 2090915 | 2217997 | 2028735 | 15735823 | 1940527 | 1510349 |
| Percentage of mappable reads in library | 54.51 | 65.60 | 64.26 | 60.88 | 62.79 | 64.34 | 54.60 | 64.56 | 64.21 |
During our search for RNAi genes (described below), we observed that sequence variants of some genes showed a high nucleotide sequence identity (∼95%), and solely using the unigene transcript set would not have led to the identification and reconstitution of putative full length CDS sequences. We therefore considered both the raw transcriptome assembly and unigene dataset, where relevant, for subsequent analyses.
Assessment of Assembly
The quality and completeness of our N. benthamiana transcriptome assembly was assessed in three different ways: using CEGMA, by comparison with publicly available N. benthamiana sequences, and by comparison with tomato sequences from Solgenomics.
The CEGMA software [23] can be used to assess the completeness of a transcriptome assembly by evaluating the presence and completeness of a widely conserved set of 248 eukaryotic proteins, as has been applied elsewhere [40], [44]. These proteins are mostly from housekeeping genes and therefore can be expected to be expressed [45]. Analysis of our raw transcriptome assembly identified 236 out of the 248 core proteins (95%) as ‘complete’ (defined as >70%, alignment length with core protein). In addition, there was an average of ∼4 orthologues per core protein, with 219 of those detected having more than 1 orthologue. Repeating the analysis on the unigene dataset detected 237 core proteins, with an average of ∼3 orthologues per core protein and 194 having more than 1 orthologue. Compared to A. thaliana which has on average 2 orthologues per core protein [23], N. benthamiana appears to have about 3 to 4 orthologues per core protein (based on transcriptome data). It is tempting to speculate that this is due to its allo-tetraploidy, but ancestral whole genome duplication and allelic variation are also likely events. It will be interesting to see if these results are consistent with a genomic assessment, the assemblies of which are still in draft stages [7], [12].
Comparing our N. benthamiana unigene set with the N. benthamiana unigenes from the Solgenomics database (total of 16,024 sequences as of November 2012, based on predictions from genomic sequences and ESTs) using BLASTn and an E-value filter of 1e−3, returned 15,039 (93.9%) matching to our unigene set, of which 14,826 (92.5%) have >90% sequence identity, and 14,166 (88.4%) have >95% sequence identity. Using sequence sizes between 100 and 500 nt, 501 and 1000 nt, and >1001 nt, gave 88.9%, 96.2% and 99.3% matching to our unigene dataset, respectively. The GC content of both datasets was approximately 41%.
The raw N. benthamiana transcriptome assembly was compared with the Solgenomics tomato genome predicted proteins database, using BLASTx (E-value <1e−3), and showed that 69,429 out of 237,340 transcripts (29.3%) have a match with a sequence identity greater than 90%, while 152,838 (64.4%) match with a sequence identity greater than 70%. In total, 174,421 (73.5%) exhibited a match to the tomato protein database. For the unigene dataset, a total of 74,492 out of 119,014 transcripts (62.6%) matched against the tomato protein database. This is in comparison to 43.8% and 85.4% of unigenes from other reported transcriptomes of N. benthamiana and N. tabacum, respectively, matching to the same tomato protein database [40], [46].
Taken altogether, these three analyses suggest that our N. benthamiana protein-coding transcriptome is a broad representation of the plant’s gene expression potential and that, while the N. benthamiana homologues of tomato proteins are identifiable, there is quite some amino acid sequence diversity between the counterparts in the two species.
Read Mapping to the N. benthamiana and Tomato Genomes
When the RNA-seq reads from each library were mapped back to the tomato and the two available N. benthamiana draft genomes [7], [12], [47], only up to 5% of reads could map back to the tomato genome compared to approximately 75% of all reads that could map back to the two versions of the N. benthamiana genomes (Table 3). While the metrics reported here are dependent on the mapping parameters (e.g. read length, seed length, number of mismatches allowed), the low read mapping percentage reflects a high nucleotide sequence divergence between tomato and N. benthamiana. It appears that there is a 1 in 12 base difference in gene-rich regions between the tomato and potato genomes [47], and given that tomato and potato are much more closely related, at least in the transcript space [48], the low mapping percentage of our RNA-seq reads to the tomato genome is not unexpected. Interestingly, there was a higher percentage of unique reads mapping to our draft N. benthamiana genome [7] compared to the one available in the Solgenomics database [12], although the overall proportions was very similar. This is likely due to differences in the ‘completeness’ of the assemblies, and perhaps some nucleotide differences accumulated by different lines passed down in different laboratories.
Table 3. Percentage of total reads from 9 tissues that mapped uniquely and to multiple locations on the tomato (Solgenomics, SL2.40) and two versions of the draft N. benthamiana genomes.
| N. benthamiana (ANZ consortium) | N. benthamiana (Solgenomics) | Tomato | |||||||
| % unique | % multi | % of total | % unique | % multi | % of total | % unique | % multi | % of total | |
| Apex | 51.52 | 23.33 | 74.85 | 46.88 | 26.70 | 73.58 | 1.30 | 0.43 | 1.73 |
| Capsule | 53.88 | 22.55 | 76.43 | 49.19 | 25.84 | 75.03 | 1.08 | 0.21 | 1.30 |
| Dstress | 56.63 | 20.04 | 76.67 | 53.03 | 22.19 | 75.21 | 1.04 | 0.22 | 1.26 |
| Flower | 50.98 | 25.95 | 76.93 | 46.64 | 28.91 | 75.56 | 2.70 | 2.20 | 4.90 |
| Leaf | 50.97 | 25.36 | 76.33 | 46.91 | 27.72 | 74.64 | 2.09 | 1.69 | 3.78 |
| Root | 52.14 | 23.44 | 75.58 | 47.93 | 26.46 | 74.39 | 1.31 | 0.24 | 1.55 |
| Seedling | 49.47 | 25.24 | 74.71 | 43.87 | 29.32 | 73.20 | 1.34 | 0.24 | 1.58 |
| Stem | 51.52 | 23.92 | 75.44 | 47.33 | 26.62 | 73.95 | 1.47 | 0.30 | 1.77 |
| TC | 51.72 | 23.64 | 75.36 | 47.45 | 26.41 | 73.87 | 1.33 | 0.28 | 1.61 |
Annotation
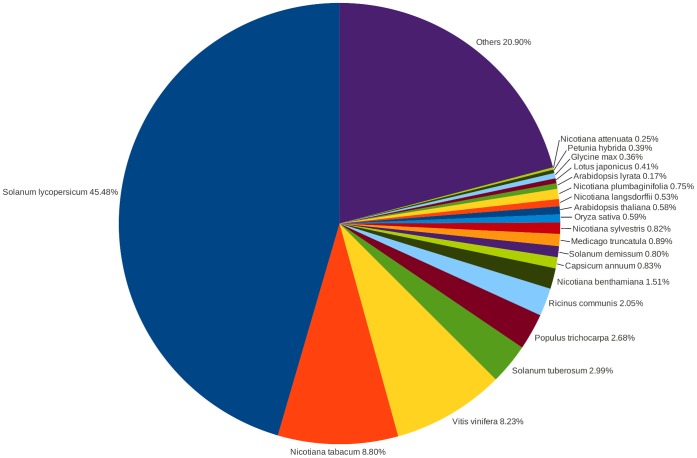
The assembled N. benthamiana transcriptome was annotated from comparisons with entries described in SwissProt, RefSeq, UniProt, TAIR, and Genbank databases. The proportion of the 119,014 unigenes showing matches with records in these databases ranged from 41.2% with SwissProt, to 68.8% with Genbank (Table 4). Not unexpectedly, the matches between the raw transcriptome and entries in Genbank’s NR protein database showed 45.5% of transcripts having significant similarity to tomato sequences, followed by 8.8% with N. tabacum (Figure 1). The species with the next most hits (8.2%) was Vitis vinifera (Grape seed), followed by smaller percentages of hits with other members of the Solanaceae. Clearly, the number of hits, per se, is not an absolute measure of relatedness between the species but rather a composite of the relatedness and the scope of available sequence data. This is well illustrated by only 1.5% of transcripts matching other available N. benthamiana entries, reflecting the prior lack of N. benthamiana sequences.
Table 4. Number of matches from complete transcriptome (237,340 transcripts) and unigene set (119,014 transcripts) to five databases.
| SwissProt | RefSeq plant | Uniprot | TAIR proteins | Genbank | |
| Total matches to database | 204575 | 214605 | 208399 | 224225 | 237271 |
| Unique matches (E-value <1e−3) | 122003 | 166440 | 166216 | 155596 | 185243 |
| Percentage of all transcripts | 51.40 | 70.13 | 70.03 | 65.56 | 78.05 |
| Unique unigene matches (E-value <1e−3) | 49013 | 66484 | 68473 | 64051 | 81920 |
| Percentage of all unigenes | 41.18 | 55.86 | 57.53 | 53.82 | 68.83 |
Figure 1. Species distribution of the transcriptome assembly.
A BLASTx using a cutoff value of E-value <1e−3 was used to search the transcriptome assembly against Genbank’s non-redundant database.
Gene ontology (GO) terms could be assigned to 41,016 (17.3%) of the 237,340 raw transcripts and 16,169 (13.6%) of the 119,014 unigene transcripts. This is comparable to the 15.3% of 95,916 unigenes annotated with GO terms in N. tabacum [49]. The N. benthamiana unigene transcripts were further refined to GO slim terms, annotating 25.5% as having a biological process (GO:008150), 24.3% to being a cellular component (GO:005575), and 24.3% to having a molecular function (GO:0003674). The distribution of the unigenes into GO slim categories is provided in Figure S1.
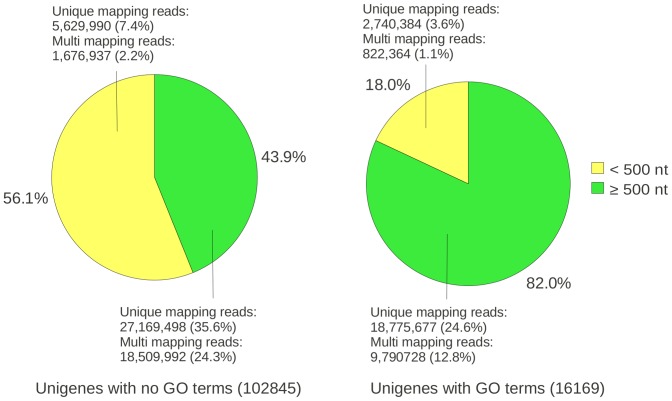
To better understand why only such a relatively small proportion of unigenes could be annotated with GO terms, the transcriptome mapping statistics were examined. This revealed that 82% of the GO-assignable unigene transcripts were >500 nt in length and 56% of the GO-unassignable transcripts were in the <500 nt size range (Figure 2). Moreover, read mapping statistics indicated that coverage was 30-fold lower for GO-unassignable transcripts that were <500 nt in size compared to those >500 nt in length (Figure 2). This showed that a high proportion of the assembly is comprised of short transcripts that make a relatively small contribution to the protein-coding transcriptome, and could be the representation of lowly expressed genes and/or from high-level transcription of non-coding RNAs. Such observations mirror transcriptome assembly studies of other polyploid plants, which also report high percentages of unassigned transcripts [40], [46], [50], [51].
Figure 2. Proportion of unigenes with and without GO annotations.
The figure shows the percentage of unigenes larger (green) and smaller (yellow) than 500 nt that could be annotated with (right) or without (left) GO terms. Mapping metrics show the number of uniquely and multi-mapping reads as a proportion of total number of reads used in the transcriptome assembly. For unigenes without GO terms, the total number of bases of transcripts under and over 500 nt was 14,954,024 nt and 54,714,622 nt, respectively. For unigenes assigned GO terms, the total number of bases under and over 500 nt was 890,086 nt and 18,991,774 nt, respectively.
Transcript Abundance in Individual Tissues and Mapping Statistics
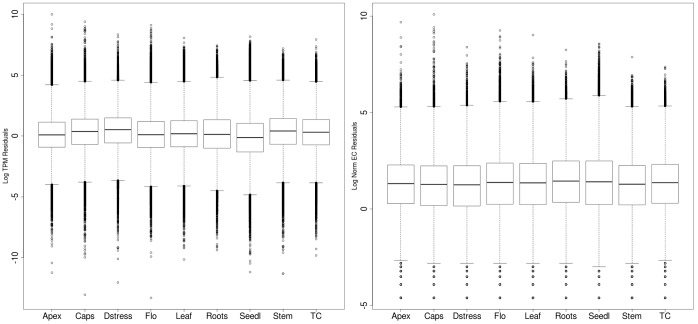
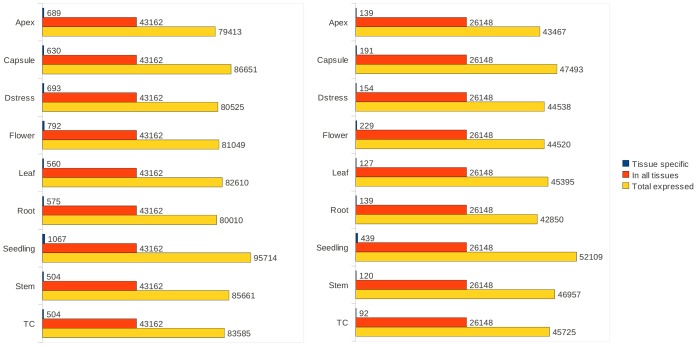
The representation of transcripts from each of the 9 tissues in the assembled transcriptome was evaluated in terms of expected counts (EC) and transcripts per million (TPM) generated by the RSEM software. For all tissues, the variation about the median was more uniform for TPM than for EC values, but overall the transcript expression profiles and the contribution of read data towards the assembly from each of the tissues were very similar (Figure 3). The median TPM values across all tissues ranged from 0.47 to 1.74, while the median for normalized ECs ranged between 1.13 and 1.98 (Table S1). However, a small proportion of transcripts appeared to be very highly expressed as shown by the difference in the 3rd quartile and maximum EC and TPM values for each tissue (Table S1). The different tissues had a common set of about 26,000 unigene transcripts that were greater than 500 nt but each tissue also uniquely expressed a number of transcripts (Figure 4). The undifferentiated callus cells grown in tissue culture produced the fewest transcripts exclusive to that tissue (92 transcripts) whereas the sample with the most unique transcripts came from seedlings (439 transcripts). This abundance of unique transcripts in seedlings may reflect the extra diversity of proteins required for rapid growth and differentiation, however, it may also be a measure of the depth of reads that were obtained for the sample, because its RNA-seq library had at least 5 times more reads than any other sample. Nonetheless, the proportion of all mappable reads (unique and multi-mapping) was similar for all tissues (Table 2), and taken together with the assessment of the distribution of transcript abundance values, the contribution of reads to the assembly does not appear to be skewed to any one tissue.
Figure 3. Distribution of TPM and EC residuals.
Boxplots of the residuals (see methods) of TPM (Transcripts Per Million) (left) and normalized EC (Expected Counts) (right) values in different tissues showing the distribution of these values about the median. For the right panel, only EC values greater than 0 were used.
Figure 4. Number of transcripts detected in the 9 tissues.
The figure shows the number of tissue-specific transcripts, and transcripts that were detected in all 9 tissues, that are over 500 nt in the transcriptome assembly. Left panel: number of transcripts from the full assembly. Right panel: number of transcripts from the unigene transcript set. Only transcripts with EC (Expected Counts) values of more than zero were used.
Each tissue had a similar GO category distribution of expressed transcripts (Table S2), but the most highly expressed transcripts varied according to tissue type (Table 5 and Table S3). The apex and flower tissues had a prevalence of transcripts encoding defence related genes while the transcripts in seedling, leaf, and stem tissues were predominantly from photosynthesis and cell structure genes. The most highly expressed transcripts in capsules, which include developing seeds, were for seed storage proteins (globulins) and, unsurprisingly, the most highly expressed transcripts in the drought stressed leaves were from genes associated with stress.
Table 5. Top 10 transcripts expressed transcripts (based on TPM values) in the 9 tissues used forthe transcriptome assembly.
| TPM | Description | TPM | Description | ||
| Apex | Root | ||||
| Nbv3K765635181 | 21909.14 | Protein TAP1 precursor | Nbv3K765637499 | 1669.75 | Phosphatidylserine decarboxylase beta chain |
| Nbv3K765636475 | 9619.12 | Defensin-like protein, Precursor | Nbv3K805659615 | 1632.48 | Tubulin alpha chain |
| Nbv3K805655645 | 6757.63 | Ethylene-responsive proteinase inhibitor 1, | Nbv3K605746981 | 1510.15 | Putative lipid-transfer protein DIR1, Precursor |
| Precursor | |||||
| Nbv3K725839522 | 3424.63 | Wound-induced proteinase inhibitor 2, Precursor | Nbv3K805655878 | 1229.00 | MLP-like protein 43 |
| Nbv3K785653169 | 3318.90 | Non-specific lipid-transfer protein (LTP), | Nbv3K785643634 | 1218.77 | Tubulin alpha-2 chain |
| Precursor | |||||
| Nbv3K785643755 | 1899.95 | hypothetical protein | Nbv3K805664031 | 1190.27 | Fruit-specific protein |
| Nbv3K785644141 | 1894.17 | Organ-specific protein S2 | Nbv3K605748986 | 1135.91 | Glycine-rich protein, Precursor |
| Nbv3K725616268 | 1756.37 | defensin-like protein | Nbv3K605747902 | 1105.61 | Glucan endo-1,3-beta-glucosidase, basic vacuolar isoform GGIB50 ((1->3)-beta-glucanase), Precursor |
| Nbv3K765632833 | 1519.04 | serine protease inhibitor 2 | Nbv3K725841908 | 1097.43 | Foot protein 1 variant 1, Precursor |
| Nbv3K805664026 | 1492.69 | hypothetical protein | Nbv3K765632492 | 1093.49 | Metallothionein-like protein type 2 |
| Capsule | Seedling | ||||
| Nbv3K805662064 | 11957.53 | 11S globulin seed storage protein 2 basic chain, | Nbv3K805662530 | 3493.49 | Ribulose bisphosphate carboxylase small chain |
| Precursor | 8B, chloroplastic (RuBisCO small subunit 8B), | ||||
| Precursor | |||||
| Nbv3K785648979 | 8103.38 | Legumin B basic chain, Precursor | Nbv3K805661563 | 2485.08 | Oxygen-evolving enhancer protein 3-2, |
| chloroplastic (OEE3), Precursor | |||||
| Nbv3K805658849 | 6989.87 | 11S globulin seed storage protein G3 basic chain, | Nbv3K805660593 | 2351.66 | Vesicular integral-membrane protein VIP36, |
| Precursor | Precursor | ||||
| Nbv3K705821994 | 6752.20 | 2S albumin large chain, Precursor | Nbv3K725839270 | 2297.67 | Ribulose bisphosphate carboxylase small chain |
| 8B, chloroplastic, Precursor | |||||
| Nbv3K785649564 | 6163.69 | Legumin B basic chain, Precursor | Nbv3K805660696 | 2274.73 | Ferredoxin-1, chloroplastic, Precursor |
| Nbv3K785654438 | 5910.78 | 11S globulin seed storage protein G3 basic chain, | Nbv3K785653197 | 2172.34 | Ribulose bisphosphate carboxylase small chain |
| Precursor | 8B, chloroplastic, Precursor | ||||
| Nbv3K805657565 | 5520.25 | Glutelin type-B 4 basic chain, Precursor | Nbv3K805658276 | 2041.68 | chlorophyll a-b binding protein |
| Nbv3K705825953 | 5461.75 | 11S globulin delta chain, Precursor | Nbv3K805656982 | 2027.77 | chlorophyll a-b binding protein |
| Nbv3K685810384 | 5417.70 | 2S albumin large chain, Precursor | Nbv3K785643703 | 1958.04 | Chlorophyll a-b binding protein CP29.1, |
| chloroplastic, Precursor | |||||
| Nbv3K625771317 | 4444.27 | Legumin B basic chain, Precursor | Nbv3K805658200 | 1952.76 | chlorophyll a-b binding protein |
| Dstress | Stem | ||||
| Nbv3K805657258 | 4158.01 | Vignain, Precursor | Nbv3K805659615 | 1313.56 | Tubulin alpha chain |
| Nbv3K805662978 | 3399.45 | E3 ubiquitin ligase interacting with arginine | Nbv3K785653169 | 1105.28 | Non-specific lipid-transfer protein (LTP), Precursor |
| methyltransferase (ISS) | |||||
| Nbv3K785647202 | 2390.34 | Kirola | Nbv3K785643634 | 1101.55 | Tubulin alpha-2 chain |
| Nbv3K785646208 | 2241.37 | Retrotransposon-derived protein PEG10 (MyEF-3) | Nbv3K805662029 | 916.63 | Putative uncharacterized protein ART2 |
| Nbv3K785647827 | 1959.69 | Vignain, Precursor | Nbv3K805660095 | 868.53 | Tubulin alpha chain |
| Nbv3K805661569 | 1726.16 | Abscisic acid and environmental stress | Nbv3K785651507 | 827.10 | hypothetical protein |
| inducible protein TAS14 | |||||
| Nbv3K725841908 | 1557.96 | Foot protein 1 variant 1, Precursor | Nbv3K805664610 | 798.36 | Thiazole biosynthetic enzyme, chloroplastic, |
| Precursor | |||||
| Nbv3K745620329 | 1495.50 | hypothetical protein | Nbv3K805658269 | 781.03 | Metallothionein-like protein type 2 |
| Nbv3K785651507 | 1340.51 | hypothetical protein | Nbv3K805663773 | 770.90 | Metallothionein-like protein type 2 B |
| Nbv3K745627524 | 1319.71 | Male-specific sperm protein Mst84Dc | Nbv3K805656914 | 764.39 | Ribulose bisphosphate carboxylase/oxygenase |
| activase 1, chloroplastic (RA 1), Precursor | |||||
| Flower | TC | ||||
| Nbv3K805662029 | 8996.18 | Putative uncharacterized protein ART2 | Nbv3K805655878 | 2769.66 | MLP-like protein 43 |
| Nbv3K805658722 | 7047.10 | Heme A synthase (HAS) | Nbv3K805663311 | 1571.49 | Kirola |
| Nbv3K805664066 | 6114.56 | Neurophysin IT 1, Precursor | Nbv3K785644141 | 1329.37 | Organ-specific protein S2 |
| Nbv3K805656915 | 5895.86 | TRAF2 and NCK-interacting protein kinase | Nbv3K805664026 | 1002.15 | hypothetical protein |
| Nbv3K805663996 | 5179.72 | RRNA intron-encoded homing endonuclease | Nbv3K785645902 | 867.50 | Probable aquaporin PIP-type pTOM75 (RAMP) |
| Nbv3K805663381 | 4649.96 | Laminin subunit beta-1, Precursor | Nbv3K805661363 | 842.94 | Non-specific lipid-transfer protein 1 (LTP 1), |
| Precursor | |||||
| Nbv3K705822805 | 4617.65 | Regulator of rDNA transcription protein 15 | Nbv3K605747902 | 828.55 | Glucan endo-1,3-beta-glucosidase, basic |
| vacuolar isoform GGIB50 ((1->3)-beta- | |||||
| glucanase), Precursor | |||||
| Nbv3K805662548 | 4026.96 | Protein YLR162W | Nbv3K785647691 | 816.97 | hypothetical protein |
| Nbv3K805661757 | 3965.65 | Putative uncharacterized protein YLR154W-F | Nbv3K785643634 | 763.45 | Tubulin alpha-2 chain |
| Nbv3K705831290 | 3536.86 | Aconitate hydratase | Nbv3K805662530 | 748.55 | Ribulose bisphosphate carboxylase small chain |
| 8B, chloroplastic, Precursor | |||||
| Leaf | |||||
| Nbv3K805662029 | 3158.40 | Putative uncharacterized protein ART2 | |||
| Nbv3K805656915 | 2137.05 | TRAF2 and NCK-interacting protein kinase | |||
| Nbv3K805663381 | 1551.93 | Laminin subunit beta-1, Precursor | |||
| Nbv3K705822805 | 1497.99 | Regulator of rDNA transcription protein 15 | |||
| Nbv3K805663773 | 1402.41 | Metallothionein-like protein type 2 B | |||
| Nbv3K805662548 | 1367.96 | Protein YLR162W | |||
| Nbv3K705831290 | 1350.36 | Aconitate hydratase | |||
| Nbv3K805658722 | 1329.29 | Heme A synthase (HAS) | |||
| Nbv3K785653197 | 1203.91 | Ribulose bisphosphate carboxylase small chain | |||
| 8B, chloroplastic, Precursor | |||||
| Nbv3K805663996 | 1194.86 | RRNA intron-encoded homing endonuclease | |||
The A. thaliana genome has recently been shown to have unsuspected transcriptome complexity from alternative splicing. A total of 57,447 transcripts representing 23,901 genes were detected by RNA-seq, and more than 60% of the genes containing introns displayed some form of alternative splicing [52]. Our unigene set of 119,014 N. benthamiana transcripts (Table 1) is possibly an overestimate. However, there are 76,379 predicted transcripts in the draft N. benthamiana genome in the Solgenomics database (http://solgenomics.net/tools/BLAST/dbinfo.pl), and the number of unigenes of a fungus-infected N. benthamiana was reported to be 73,041 [40]. It will be interesting to see how important and widespread differential splicing is in this allopolyploid transcriptome.
Identification of RNA Silencing Genes and Family Members
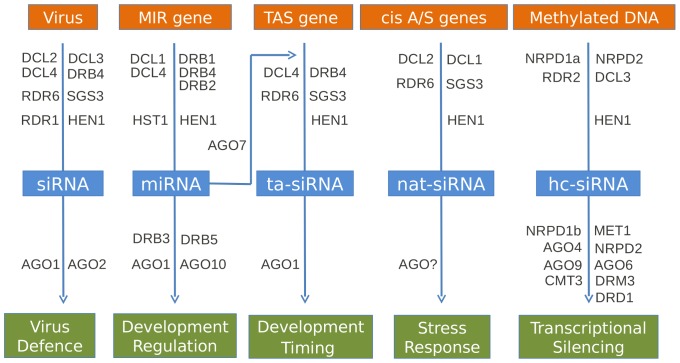
The gene silencing (including RNAi) machinery in plants generates small 21–24 nt RNAs and uses them to guide at least five interwoven pathways providing different forms of gene regulation and plant defence (Figure 5). The core components of the pathways are encoded by gene families of the dicer-like (Dcl) nucleases, argonaute (Ago) proteins, double stranded RNA binding (Drb) proteins, RNA polymerases and methyl-transferases. In association with other proteins, they give targeted RNA degradation, translational repression and heterochromatin modification [53]. Many of the effects of these processes have been studied in N. benthamiana but with little detail about the intrinsic pathway components and assuming that they are similar to those characterised in A. thaliana. We investigated the presence and characteristics of RNAi gene transcripts in our N. benthamiana transcriptome assembly to gain a better insight into whether the plant may be defective or unusual in some of these pathways, as has been suggested [54], [55], and also as another way to evaluate the transcriptome assembly’s ‘completeness’.
Figure 5. RNAi-associated pathways and their core components in plants.
The schematic depicts the major proteins involved in producing different small RNAs (blue boxes) from RNA transcripts of various templates (orange boxes) and using these sRNAs to regulate a spectrum of important biological processes (green boxes).
Using the protein sequences of 31 well characterised A. thaliana RNAi genes (Figure 5 and Table S4) we first scanned for homologues in the Solgenomics tomato (S. lycopersicum) predicted proteins database. Screening the N. benthamiana transcriptome with the A. thaliana query set, aided by the S. lycopersicum counterparts, identified homologues for each RNAi gene query (Table 6) except for AGO3 (returned as AGO2) and AGO9 (returned as a AGO4 variant). Of the identified homologues, 24 of them appeared to be fully assembled. The AGO1, AGO2, AGO6, AGO7, DCL1, NRPD1a, NRPD1b, NRPD2a and RDR2 queries resulted in multiple hits due to the possible presence of additional gene family members or to shorter than full-length contigs (Table 6). The inability of the assembler to integrate these smaller contigs into full-length CDS transcripts is probably due to the presence of nucleotide polymorphisms from different mRNA splice-forms or from close family members, sequencing errors, and/or low sequencing depth. However with manual intervention, full-length CDS homologues of all but one were generated (Table 6). This was greatly assisted by the N. benthamiana sequences having both high sequence identities and high query coverage (>90%) with their S. lycopersicum counterparts. The one remaining partial assembly was of Nrpd1a. However, a 1414 aa sequence was compiled which, although shorter than its homologue in A. thaliana (by an estimated ∼30 aa), is much longer than the 1206 aa ORF in S. lycopersicum. The identified sequences were also compared to other N. benthamiana RNAi gene entries from Genbank and showed almost perfect identity (Figure S2).
Table 6. List of RNAi associated genes identified and compared to tomato and A. thaliana counterparts.
| RNAi gene | Status | A.A. Length | Transcriptome Ids | Tomato ID; % identity to tomato; Query coverage | Athan ID; % identity to Athan; Query coverage |
| Ago1a | Full | 1058 | Nbv3K705826800 | Solyc06g072300.2.1; 92.8%; 100% | AT1G48410.1 (AGO1); 82.7%; 89.3% |
| Ago1b* | Full | 1110 | Nbv3K785652119; | Solyc03g098280.2.1; 92.3%; 84.3% | AT1G48410.1 (AGO1); 81.3%; 85.9% |
| Nbv3K785652117; | |||||
| Nbv3K705830082 | |||||
| Ago2* | Full | 1093 | Nbv3K585706870; | Solyc02g069260.2.1; 72.9%; 94% | AT1G31280.1 (AGO2); 50.1%; 84.4% |
| Nbv3K585676062 | |||||
| Ago4a | Full | 912 | Nbv3K585737054 | Solyc06g073540.2.1; 94.4%; 100% | AT2G27040.1 (AGO4); 74.3%; 96.4% |
| Ago4b | Full | 905 | Nbv3K745626388 | Solyc01g008960.2.1; 92.5%; 100% | AT2G27040.1 (AGO4); 74.6%; 96.7% |
| AT5G21150.1 (AGO9); 69.9%; 97.6% | |||||
| Ago5 | Full | 1013 | Nbv3K585731374 | Solyc06g074730.2.1; 67%; 95.1% | AT2G27880.1 (AGO5); 59.5%; 92.6% |
| Ago6* | Full | 903 | Nbv3K705827462; | Solyc07g049500.2.1; 86.4%; 100% | AT2G32940.1 (AGO6); 61.8%; 97.2% |
| Nbv3K645784811; | |||||
| Nbv3K645784061; | |||||
| Nbv3K585708242 | |||||
| Ago7*? | Full | 991 | Nbv3K585720936; | Solyc01g010970.2.1; 84.7%; 100% | AT1G69440.1 (AGO7); 68.2%; 87.4% |
| Nbv3K585735851; | |||||
| Nbv3K585719994; | |||||
| Nbv3K665804949 | |||||
| Ago10 | Full | 988 | Nbv3K585734208 | Solyc09g082830.2.1; 93.2%; 100% | AT5G43810.1 (AGO10); 80.6%; 100% |
| Cmt3a | Full | 855 | Nbv3K625768297 | Solyc01g006100.2.1; 70.9%; 100% | AT1G69770.1 (CMT3); 54.8%; 89.8% |
| Cmt3b | Full | 907 | Nbv3K585704505 | Solyc12g100330.1.1; 80.3%; 99.7% | AT1G69770.1 (CMT3); 54.9%; 85.1% |
| Dcl1* | Full | 1909 | Nbv3K605750463; | Solyc10g005130.2.1; 87.1%; 100% | AT1G01040.1 (DCL1); 71.6%; 100% |
| Nbv3K745628138; | |||||
| Nbv3K585722110 | |||||
| Dcl2 | Full | 1402 | Nbv3K725833766 | Solyc06g048960.2.1; 83%; 98.3% | AT3G03300.1 (DCL2); 55.8%; 99.6% |
| Dcl3 | Full | 1456 | Nbv3K585704110 | Solyc08g067210.2.1; 83.5%; 94.9% | AT3G43920.1 (DCL3); 47%; 97.7% |
| Dcl4 | Full | 1622 | Nbv3K625768999 | Solyc07g005030.2.1; 85.9%; 94.6% | AT5G20320.1 (DCL4); 52.9%; 97.3% |
| Drb1 | Full | 316 | Nbv3K605753726 | Solyc04g076420.2.1; 61.8%; 94.3% | AT1G09700.1 (DRB1); 59.8%; 66.5% |
| Drb2a | Full | 399 | Nbv3K585718488 | Solyc11g069460.1.1; 85.3%; 100% | AT2G28380.1 (DRB2); 64.7%; 87.5% |
| Drb2b | Full | 400 | Nbv3K605753084 | Solyc11g069460.1.1; 85.5%; 100% | AT2G28380.1 (DRB2); 64.7%; 87.5% |
| Drb3 | Full | 469 | Nbv3K725608215 | Solyc05g056100.2.1; 80.8%; 97% | AT3G26932.1 (DRB3); 70.3%; 39.5% |
| AT2G28380.1 (DRB2); 51.8%; 88% | |||||
| AT5G41070.1 (DRB5); 43.8%; 72.5% | |||||
| Drb4 | Full | 352 | Nbv3K725839976 | Solyc01g056620.2.1; 64.7%; 99.7% | AT3G62800.1 (DRB4); 55.4%; 45.1% |
| Drb5 | Full | 477 | Nbv3K585684100 | Solyc05g056100.2.1; 71.6%; 96.9% | AT5G41070.1 (DRB5); 71.9%; 38.8% |
| AT3G26932.1 (DRB3); 70.8%; 38.8% | |||||
| AT2G28380.1 (DRB2); 55%; 85.1% | |||||
| Drd1 | Full | 926 | Nbv3K585726931 | Solyc01g109970.2.1; 84.8%; 98.6% | AT2G16390.1 (DRD1); 54.7%; 94.2% |
| Drm3 | Full | 705 | Nbv3K745624408 | Solyc05g053260.2.1; 82.3%; 76.5% | AT3G17310.2 (DRM3); 38.7%; 96.6% |
| Hen1 | Full | 948 | Nbv3K645785075 | Solyc05g026050.2.1; 82.5%; 99.6% | AT4G20910.1 (HEN1); 49.7%; 98.4% |
| Hst1 | Full | 1199 | Nbv3K625768898 | Solyc01g098170.2.1; 91.3%; 100% | AT3G05040.1 (HST); 65.5%; 98.1% |
| Met1 | Full | 1558 | Nbv3K585692501 | Solyc11g030600.2.1; 87.2%; 99.6% | AT5G49160.1 (MET1); 60.4%; 95.8% |
| Nrpd1a* | Partial | 1414 | Nbv3K585708997; | Solyc08g080210.2.1; 81.4%; 84.7% | AT1G63020.1 (NRPD1a); 48.6%; 98.2% |
| Nbv3K585738556; | |||||
| Nbv3K585698167 | |||||
| Nrpd1b* | Full | 2047 | Nbv3K685818512; | Solyc01g096390.2.1; 86.1%; 72.1% | AT2G40030.1 (NRPD1b); 51.9%; 80% |
| Nbv3K685816071; | |||||
| Nbv3K585729430; | |||||
| Nbv3K585720083 | |||||
| Nrpd2a* | Full | 1212 | Nbv3K585720667; | Solyc03g110880.2.1; 92.8%; 100% | AT3G23780.1 (NRPD2a); 64.7%; 98.4% |
| Nbv3K685813559 | |||||
| Rdr1 | Full | 1140 | Nbv3K745620210 | Solyc05g007510.2.1; 83.1%; 99.9% | AT1G14790.1 (RDR1); 61.4%; 98.3% |
| Rdr2? | Full | 1120 | Nbv3K625766705 | Solyc03g114140.2.1; 87%; 99.7% | AT4G11130.1 (RDR2); 59.1%; 99% |
| Rdr6 | Full | 1197 | Nbv3K585707928 | Solyc04g014870.2.1; 85%; 100% | AT3G49500.1 (RDR6); 65.3%; 100% |
| Sgs3 | Full | 635 | Nbv3K785651293 | Solyc04g025300.2.1; 81.2%; 99.8% | AT5G23570.1 (SGS3); 49.2%; 99.1% |
Screening the N. benthamiana transcriptome assembly with AtAGO9 identified a potential counterpart transcript, Nbv3K745626388, but a reciprocal BLAST revealed it to be more similar to AtAGO4. However, scanning the N. benthamiana transcriptome with AtAGO4 had identified another transcript, Nbv3K585737054, as the most probable AGO4 homologue, and pair-wise comparison of these two N. benthamiana transcripts showed them to be 86.7% identical. From this we inferred that these two transcripts represent different Ago4 family members, which we designate Ago4a (Nbv3K585737054) and Ago4b (Nbv3K745626388). From a similar analysis, Ago1 also appears to have 2 gene family members (Table 6). These results confirm the previous identification of two variants of Ago1 and Ago4 in N. benthamiana [56]. Many of the other RNAi genes in N. benthamiana may also contain multiple family members. Multiple copies of several RNAi genes do exist in S. lycopersicum [57], and preliminary analyses of single nucleotide polymorphisms in our RNAi genes do suggest the presence of other members in some (data not shown).
In addition to our assessment of the assembly completeness, the identification of apparently full length RNAi gene homologues from 30 of 31 queries, the near to possibly full-length assembly of the remaining Nrpd1a homologue, and the identification of additional family members in Drb2, Ago1 and Ago4, suggests that our transcriptome assembly does provide a broad representation of expressed genes in N. benthamiana. With the exception of Rdr1, which is discussed below, if this plant is defective in any of the RNAi pathways it is not because of the absence of transcription of any of these 31 core genes.
Assessment of Nb-AGO, Nb-DCL, Nb-DRB and Nb-RDR Genes
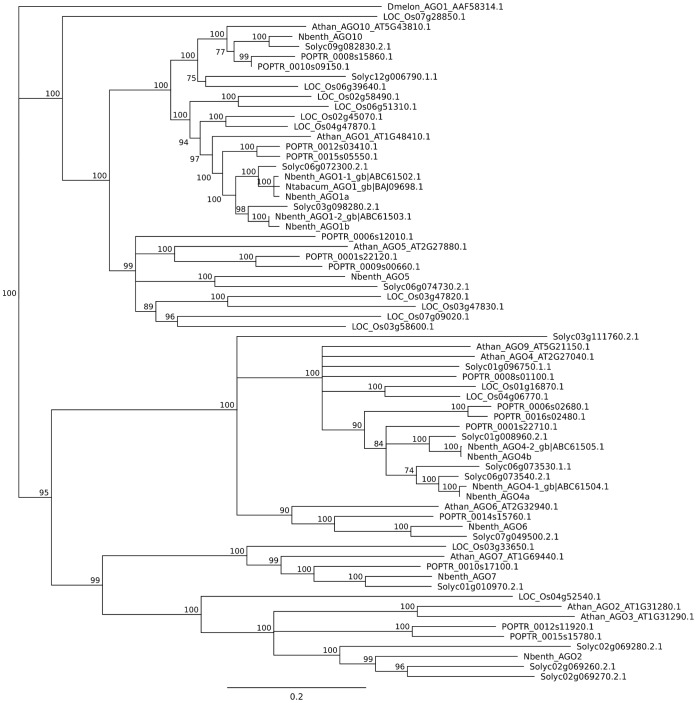
The AGO, DCL, DRB and RDR sequences from the N. benthamiana transcriptome, along with homologues from O. sativa, P. trichocarpa, S. lycopersicum, A. thaliana and some Nicotiana species, including previously reported N. benthamiana sequences from NCBI, were retrieved from their respective databases and analysed for phylogenetic relationships. Figure 6 shows the neighbour joining tree of AGO sequences (see Figure S3, S4, and S5 for trees of DCLs, DRBs and RDRs). As expected, sequences from the Nicotiana species formed their own clades, and are closely related to the S. lycopersicum sequences.
Figure 6. Neighbour joining tree of AGO proteins.
Sequences from N. benthamiana (Nbenth), A. thaliana (Athan), S. lycopersicum (Solyc), O. sativa (LOC_Os) and P. trichocarpa (POPTR) were aligned with the MUSCLE algorithm. Bootstrap values are shown at the nodes. The Athan sequences can be used to classify the AGO family. In all clades, the Nbenth sequences clearly group with Solyc sequences.
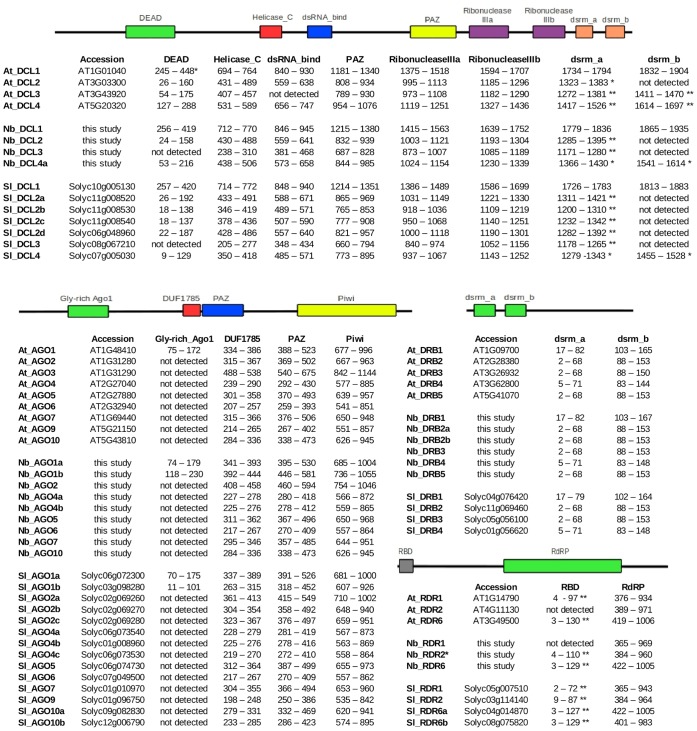
To further assess whether the AGO, DCL, DRB and RDR sequences identified in this study conformed to the characteristics representative of each category, they and their A. thaliana and S. lycopersicum counterparts were searched for protein domains using InterProScan. Most of the characteristic domains for the genes in each category were identified (Figure 7) but there were some notable exceptions. Nb-RDR1 contains two premature stop codons, from a 72bp insertion, within its RdRP domain (Figure 8A), in the first exon of the coding sequence. This insertion has been previously reported [54], [55] and we further verified its existence, in both our lab line and in a well-known GFP-expressing line (16C) [56], [58], using PCR with primers flanking the insertion site (Fig. 8B). Interestingly, PCR indicated the absence of this insertion in N. tabacum, and sequence comparison with the RDR of tomato and A. thaliana implied that this insertion is so far particular to N. benthamiana.
Figure 7. Domains of RNAi proteins.
The figure shows the domains of the RNAi proteins AGO, DCL, DRB and RDR from N. benthamiana (Nb), A. thaliana (At, from TAIR) and S. lycopersicum (Sl, from Solgenomics). Domains were detected with InterProScan against all its default databases, and defined according to the Pfam predictions unless otherwise annotated (*according to SMART database; **according to SUPERFAMILY database).
Figure 8. Insertion in the RDR1 sequence of N. benthamiana.

(A) Alignment of the RDR1 sequence from two lines of N. benthamiana (Nb) (16C and Lab), N. tabacum (Nt), S. lycopersicum (Sl) and A. thaliana (At). Only the Nb lines possess an insertion containing two stop codons. (B): PCR of region flanking the 72 base insert in Nb 16C and Lab lines, and Nt, indicating that the insertion is only present in N. benthamiana.
Two other notable exceptions are in Nb-DCL3, which appears to have only one double stranded RNA motif (dsrm) in its C-terminal region and to lack the DEAD motif in its N-terminal helicase region (Figure 7). A tandem pair of dsrms (a and b) in DCL1, DCL3 and DCL4, and a single dsrm in DCL2, seemed to be the canonical arrangement in plants [59]. However, both Nb-DCL3 and Sl-DCL3 lack dsrm-b. While DCL3 has been shown to generate 24 nt siRNAs from transposons to direct heterochromatin modification [60], [61], it also produces siRNAs from viruses and their satellites and, in concert with DCL4 and DCL2, plays a role in repression of replication [62]. Nicotiana benthamiana is hyper-susceptible to viruses and it is possible that, besides the inactivation of RDR1 by the 72bp insertion, the loss of dsrm-b and DEAD motif in Nb-DCL3 may also play a role in this. The same motifs are not present in Sl-DCL3 (Figure 7). There is no premature stop codon in Sl-RDR1 (Figure 8), and tomato is susceptible to over 130 different viruses. This raises the possibility that it may not (only) be the RDR1 insertion in N. benthamiana but rather the unusual DCL3, or a combination of several such traits that is engendering weaker viral defence.
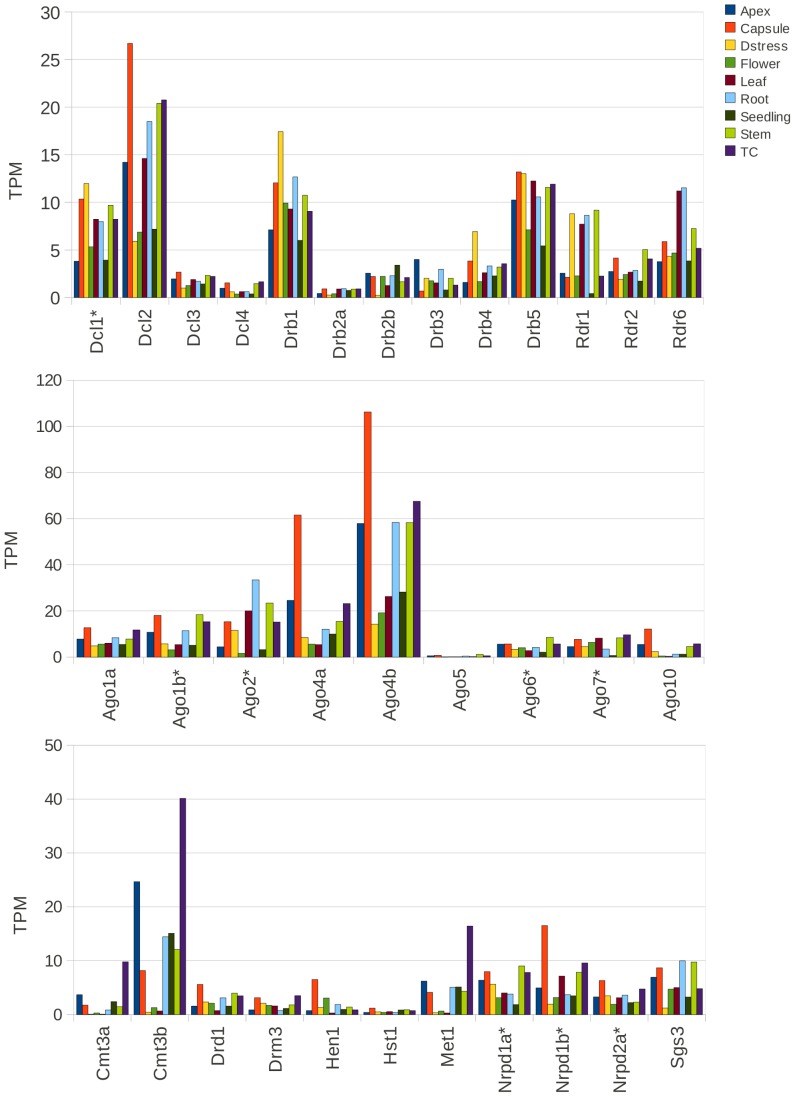
In these four gene families, there was a spectrum of expression levels within and across different tissues, as assessed by their TPM values (Figure 9 and Table S5). Overall, Ago4, Dcl1 and 2, Drb1 and 5, and Rdr1 and 6 had higher expression levels than other family members in all tissues. The higher abundance of Dcl1 compared to Dcl3, and Ago4a compared to Ago1a, reported here, does appear to correlate with the findings of other reports [56], [63]. In fact, an RSEM analysis using RNA-seq data from another N. benthamiana transcriptome [40] showed a very similar expression profile to our RNAi gene set (Figure S6), supporting the broad-level interpretation of transcript abundances based on our transcriptome.
Figure 9. Relative abundances of RNAi-associated genes.
The figure shows the relative abundances of RNAi genes identified in this study in terms of TPM (Transcripts Per Million) values calculated from the RSEM software. Genes with an asterisk (*) were comprised of more than oneassembled transcript, and the TPM value in this case represents the TPM sum of all its component transcripts.
Ago4a and especially Ago4b transcripts were highly abundant in the apex and capsle tissues. The capsule tissue contains developing seeds and Ago9 has recently been shown to play an important role in transferring the maternal epigenetic overlay to the next generation in A. thaliana [64]. Given that AGO4B was most similar to AtAGO9 in our homologue analysis, it is tempting to speculate that this AGO4B might aid similar epigenetic transfer processes in N. benthamiana.
Curiously, the level of Nb-DCL4 was relatively low in all tissues whereas the Nb-DCL2 level was very high. This is in contrast to the situation in A. thaliana, in which all four DCLs have fairly similar, and relatively low, expression levels. At-DCL4 is the major producer of tasiRNAs and of secondary siRNAs for antiviral activity, whereas At-DCL2 has signalling and back-up anti-viral functions [62], [65]. Perhaps the low expression of NbDCL4 also plays a role in the plant’s susceptibility to viruses. It will be interesting to see how the expression profiles of these genes change upon virus infection.
Conclusion
We present here the transcriptome of a lab line of N. benthamiana, assembled from deep sequencing data from 9 different tissue samples. From comparisons with other databases, it appears to broadly represent the coding capacity of the genome, and should be a useful resource for researchers in the plant community. The annotated transcriptome is available for download and BLAST searches at http://www.benthgenome.com, where a draft genome is also available. The transcriptome has been mapped onto the genome to generate putative gene models, supported by RNA-seq read data of each tissue. These models and mapped reads can be visualised through a Gbrowse portal and may be interrogated by the provided tracks. We also provide on this site the largest list, to date, of N. benthamiana RNAi-associated gene sequences, which have been identified bioinformatically from this study. These may be of particular interest to researchers using this plant in RNA silencing studies.
Supporting Information
Distribution of GO slim terms. The figure shows the distribution unigene transcripts from the de novo N. benthamiana transcriptome assembly that could be annotated GO slim terms. The distributions are shown as categories of biological process (top panel), cellular component (middle panel), and molecular function (bottom panel). Only terms with more than 10 unigenes are shown.
(TIFF)
Alignment of previously reported DCL, AGO and RDR sequences against those identified in this study. The alignment view shows the position of publically available sequences with respect to the sequence identified in this study. The slight difference in sequence identity could be due to differences in N. benthamiana lines maintained in different laboratories.
(TIFF)
Neighbour joining tree of RDR proteins. Sequences from N. benthamiana (Nbenth), A. thaliana (Athan), S. lycopersicum (Solyc), O. sativa (LOC_Os) and P. trichocarpa (POPTR) were aligned with the MUSCLE algorithm. Bootstrap values are shown at the nodes.
(TIFF)
Neighbour joining tree of DRB proteins. Sequences from N. benthamiana (Nbenth), A. thaliana (Athan), S. lycopersicum (Solyc), O. sativa (LOC_Os) and P. trichocarpa (POPTR) were aligned with the MUSCLE algorithm. Bootstrap values are shown at the nodes.
(TIFF)
Neighbour joining tree of DCL proteins. Sequences from N. benthamiana (Nbenth), A. thaliana (Athan), S. lycopersicum (Solyc), O. sativa (LOC_Os) and P. trichocarpa (POPTR) were aligned with the MUSCLE algorithm. Bootstrap values are shown at the nodes.
(TIFF)
Relative abundances of RNAi-associated genes identified in this study using RNA-seq data from [40] (8DPI dataset). TPM values were calculated using the RSEM software. With the exception of Drb2b and Rdr1, the overall abundance profile is highly similar to that reported in Figure 9.
(TIFF)
Distribution statistics calculated by the R software (5-number summary) of TPM (top) and normalized EC (bottom) values of transcripts in the 9 tissues used for the transcriptome assembly.
(DOC)
Number of transcripts with normalized EC counts greater than 0 that could be annotated with GO slim terms in each tissue*. *Within the biological process category, biosynthetic process (GO:0009058), cellular nitrogen compound metabolic process (GO:0034641), small molecule metabolic process (GO:0044281), translation (GO:0006412) and transport (GO:0006810) were highly represented in all tissues. Similarly, within the cellular component category, cell (GO:0005623), cytoplasm (GO:0005737), intracellular (GO:0005622) and organelle (GO:0043226) were highly represented. Within the molecular function category, there was no one sub-category that stood out, but oxidoreductase activity (GO:0016491), DNA binding (GO:0003677), ion binding (GO:0043167) and RNA binding (GO:0003723) were noticeably represented categories. The highly similar GO profiles for all tissues may be due to the limited subset of transcripts that could actually be annotated with a GO classification.
(DOC)
Top 10 expressed transcripts (based on EC values) in the 9 tissues used for the transcriptome assembly.
(DOC)
List of RNAi-associated genes from Arabidopsis thaliana used to screen for orthologues in Nicotiana benthamiana . 1. Eamens A, Wang M-B, Smith NA, Waterhouse PM (2008) RNA Silencing in Plants: Yesterday, Today, and Tomorrow. Plant Physiol 147: 456–468. 2. Jaubert M, Bhattacharjee S, Mello AFS, Perry KL, Moffett P (2011) ARGONAUTE2 Mediates RNA-Silencing Antiviral Defenses against Potato virus X in Arabidopsis. Plant Physiol 156: 1556–1564. 3. Vaucheret H (2008) Plant ARGONAUTES. Trends Plant Sci 13: 350–358. 4. Havecker ER, Wallbridge LM, Hardcastle TJ, Bush MS, Kelly KA, et al. (2010) The Arabidopsis RNA-Directed DNA Methylation Argonautes Functionally Diverge Based on Their Expression and Interaction with Target Loci. Plant Cell 22: 321–334. 5. Zhu H, Hu F, Wang R, Zhou X, Sze S-H, et al. (2011) Arabidopsis Argonaute10 Specifically Sequesters miR166/165 to Regulate Shoot Apical Meristem Development. Cell 145: 242–256. 6. Eamens AL, Kim KW, Curtin SJ, Waterhouse PM (2012) DRB2 Is Required for MicroRNA Biogenesis in Arabidopsis thaliana. PLoS One 7: e35933. 7. Eamens AL, Kim KW, Waterhouse PM (2012) DRB2, DRB3 and DRB5 function in a non-canonical microRNA pathway in Arabidopsis thaliana. Plant Signal Behav 7: 1224–1229.
(DOC)
Relative abundances of RNAi-associated genes identified in this study, shown as TPM values calculation by the RSEM software (this is a numerical version of Figure 5 ). *Genes comprised of more than 1 transcript from the assembly; each TPM value represents the sum of all the constituent TPMs.
(DOC)
Acknowledgments
We thank Chloe Paul for performing the PCR experiments. We thank Dr. Jeremy Burdon for all of his encouragement.
Funding Statement
This work was funded collaboratively by the University of Sydney, the Commonwealth Scientific and Industrial Research Organisation, and The New Zealand Institute for Plant and Food Research. PMW is funded by the Australian Research Council Federation Fellowship FF0776510. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Marks CE, Newbigin E, Ladiges PY (2011) Comparative morphology and phylogeny of Nicotiana section Suaveolentes (Solanaceae) in Australia and the South Pacific. Aust Syst Bot 24: 61–86. [Google Scholar]
- 2. Goodin MM, Zaitlin D, Naidu RA, Lommel SA (2008) Nicotiana benthamiana: Its History and Future as a Model for Plant–Pathogen Interactions. Mol Plant Microbe Interact 21: 1015–1026. [DOI] [PubMed] [Google Scholar]
- 3.Clemente T (2006) Nicotiana (Nicotiana tobaccum, Nicotiana benthamiana). In: Wang K, editor. Agrobacterium Protocols: Humana Press. 143–154.
- 4. Purkayastha A, Dasgupta I (2009) Virus-induced gene silencing: A versatile tool for discovery of gene functions in plants. Plant Physiol Biochem 47: 967–976. [DOI] [PubMed] [Google Scholar]
- 5. Wydro M, Kozubek E, Lehmann P (2006) Optimization of transient Agrobacterium-mediated gene expression system in leaves of Nicotiana benthamiana . Acta Biochim Pol 53: 289–298. [PubMed] [Google Scholar]
- 6. Sparkes IA, Runions J, Kearns A, Hawes C (2006) Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protoc 1: 2019–2025. [DOI] [PubMed] [Google Scholar]
- 7.Naim F, Nakasugi K, Crowhurst RN, Hilario E, Zwart AB, et al. (2012) Advanced metabolic engineering in Nicotiana benthamiana using a draft genome and the V2 viral silencing-suppressor protein PLoS One: In press. [DOI] [PMC free article] [PubMed]
- 8. Wood CC, Petrie JR, Shrestha P, Mansour MP, Nichols PD, et al. (2009) A leaf-based assay using interchangeable design principles to rapidly assemble multistep recombinant pathways. Plant Biotechnol J 7: 914–924. [DOI] [PubMed] [Google Scholar]
- 9. Zhou Y, Ni M (2010) SHORT HYPOCOTYL UNDER BLUE1 Truncations and Mutations Alter Its Association with a Signaling Protein Complex in Arabidopsis . Plant Cell 22: 703–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Burgyán J, Havelda Z (2011) Viral suppressors of RNA silencing. Trends Plant Sci 16: 265–272. [DOI] [PubMed] [Google Scholar]
- 11.Csorba T, Pantaleo V, Burgyán J (2009) Chapter 2 - RNA Silencing: An Antiviral Mechanism. In: Gad L, John PC, editors. Adv Virus Res: Academic Press. 35–230. [DOI] [PubMed]
- 12. Bombarely A, Rosli HG, Vrebalov J, Moffett P, Mueller LA, et al. (2012) A Draft Genome Sequence of Nicotiana benthamiana to Enhance Molecular Plant-Microbe Biology Research. Mol Plant Microbe Interact 25: 1523–1530. [DOI] [PubMed] [Google Scholar]
- 13. Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11: 113–116. [Google Scholar]
- 14. Rumble SM, Lacroute P, Dalca AV, Fiume M, Sidow A, et al. (2009) SHRiMP: Accurate Mapping of Short Color-space Reads. PLoS Comput Biol 5: e1000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Birol I, Jackman SD, Nielsen CB, Qian JQ, Varhol R, et al. (2009) De novo transcriptome assembly with ABySS. Bioinformatics 25: 2872–2877. [DOI] [PubMed] [Google Scholar]
- 16. Robertson G, Schein J, Chiu R, Corbett R, Field M, et al. (2010) De novo assembly and analysis of RNA-seq data. Nat Methods 7: 909–912. [DOI] [PubMed] [Google Scholar]
- 17. Consortium TU (2012) Reorganizing the protein space at the Universal Protein Resource (UniProt). Nucleic Acids Res 40: D71–D75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Suzek BE, Huang H, McGarvey P, Mazumder R, Wu CH (2007) UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics 23: 1282–1288. [DOI] [PubMed] [Google Scholar]
- 19. Pruitt KD, Tatusova T, Klimke W, Maglott DR (2009) NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res 37: D32–D36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mulder N, Apweiler R (2007) InterPro and InterProScan. In: Bergman N, editor. Comparative Genomics: Humana Press. 59–70.
- 22. Li W, Godzik A (2006) Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22: 1658–1659. [DOI] [PubMed] [Google Scholar]
- 23. Parra G, Bradnam K, Ning Z, Keane T, Korf I (2009) Assessing the gene space in draft genomes. Nucleic Acids Res 37: 289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Langmead B, Trapnell C, Pop M, Salzberg S (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li B, Dewey C (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Robinson M, Oshlack A (2010) A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, et al. (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29: 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Team RC (2012) R: A Language and Environment for Statistical Computing. R J.
- 30. Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, et al. (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628. [DOI] [PubMed] [Google Scholar]
- 32. Bullard J, Purdom E, Hansen K, Dudoit S (2010) Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics 11: 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Oshlack A, Robinson M, Young M (2010) From RNA-seq reads to differential expression results. Genome Biol 11: 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, et al. (2009) BLAST+: architecture and applications. BMC Bioinformatics 10: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hansen KD, Brenner SE, Dudoit S (2010) Biases in Illumina transcriptome sequencing caused by random hexamer priming. Nucleic Acids Res. [DOI] [PMC free article] [PubMed]
- 36.Li W, Jiang T (2012) Transcriptome Assembly and Isoform Expression Level Estimation from Biased RNA-Seq Reads. Bioinformatics. [DOI] [PMC free article] [PubMed]
- 37.Schliesky S, Gowik U, Weber APM, Braeutigam A (2012) RNA-seq assembly – Are we there yet? Front Plant Sci 3. [DOI] [PMC free article] [PubMed]
- 38.Kozik A, Matvienko M, Michelmore RW (2010) Effects Of Filtering, Trimming, Sampling And K-mer Value On De Novo Assembly Of Illumina GA Reads; January 9–13; Plant & Animal Genomes XVIII Conference. San Diego, CA.
- 39.Li Z, Chen Y, Mu D, Yuan J, Shi Y, et al.. (2011) Comparison of the two major classes of assembly algorithms: overlap–layout–consensus and de-bruijn-graph. Brief Funct Genomics. [DOI] [PubMed]
- 40. Faino L, de Jonge R, Thomma BP (2012) The transcriptome of Verticillium dahliae-infected Nicotiana benthamiana determined by deep RNA sequencing. Intravital 7: 1065–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Duan J, Xia C, Zhao G, Jia J, Kong X (2012) Optimizing de novo common wheat transcriptome assembly using short-read RNA-Seq data. BMC Genomics 13: 392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gruenheit N, Deusch O, Esser C, Becker M, Voelckel C, et al. (2012) Cutoffs and k-mers: implications from a transcriptome study in allopolyploid plants. BMC Genomics 13: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, et al. (2011) Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics 12: S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hu H, Bandyopadhyay P, Olivera B, Yandell M (2011) Characterization of the Conus bullatus genome and its venom-duct transcriptome. BMC Genomics 12: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Parra G, Bradnam K, Korf I (2007) CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23: 1061–1067. [DOI] [PubMed] [Google Scholar]
- 46. Bombarely A, Edwards K, Sanchez-Tamburrino J, Mueller L (2012) Deciphering the complex leaf transcriptome of the allotetraploid species Nicotiana tabacum: A phylogenomic perspective. BMC Genomics 13: 406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tomato_Genome_Consortium (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rensink W, Lee Y, Liu J, Iobst S, Ouyang S, et al. (2005) Comparative analyses of six solanaceous transcriptomes reveal a high degree of sequence conservation and species-specific transcripts. BMC Genomics 6: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lu J, Du Z-X, Kong J, Chen L-N, Qiu Y-H, et al. (2012) Transcriptome Analysis of Nicotiana tabacum Infected by Cucumber mosaic virus during Systemic Symptom Development. PLoS One 7: e43447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Barakat A, DiLoreto D, Zhang Y, Smith C, Baier K, et al. (2009) Comparison of the transcriptomes of American chestnut (Castanea dentata) and Chinese chestnut (Castanea mollissima) in response to the chestnut blight infection. BMC Plant Biol 9: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Massa AN, Childs KL, Lin H, Bryan GJ, Giuliano G, et al. (2011) The Transcriptome of the Reference Potato Genome Solanum tuberosum Group Phureja Clone DM1–3 516R44. PLoS One 6: e26801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Marquez Y, Brown JWS, Simpson C, Barta A, Kalyna M (2012) Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis . Genome Res 22: 1184–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Eamens A, Wang M-B, Smith NA, Waterhouse PM (2008) RNA Silencing in Plants: Yesterday, Today, and Tomorrow. Plant Physiol 147: 456–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yang S-J, Carter SA, Cole AB, Cheng N-H, Nelson RS (2004) A natural variant of a host RNA-dependent RNA polymerase is associated with increased susceptibility to viruses by Nicotiana benthamiana . Proc Natl Acad Sci U S A 101: 6297–6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ying X-B, Dong L, Zhu H, Duan C-G, Du Q-S, et al. (2010) RNA-Dependent RNA Polymerase 1 from Nicotiana tabacum Suppresses RNA Silencing and Enhances Viral Infection in Nicotiana benthamiana. Plant Cell: 1358–1372. [DOI] [PMC free article] [PubMed]
- 56. Jones L, Keining T, Eamens A, Vaistij FE (2006) Virus-Induced Gene Silencing of Argonaute Genes in Nicotiana benthamiana Demonstrates That Extensive Systemic Silencing Requires Argonaute1-Like and Argonaute4-Like Genes. Plant Physiol 141: 598–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bai M, Yang G-S, Chen W-T, Mao Z-C, Kang H-X, et al. (2012) Genome-wide identification of Dicer-like, Argonaute and RNA-dependent RNA polymerase gene families and their expression analyses in response to viral infection and abiotic stresses in Solanum lycopersicum . Gene 501: 52–62. [DOI] [PubMed] [Google Scholar]
- 58. Brigneti G, Voinnet O, Li W-X, Ji L-H, Ding S-W, et al. (1998) Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana . EMBO J 17: 6739–6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 59. Margis R, Fusaro AF, Smith NA, Curtin SJ, Watson JM, et al. (2006) The evolution and diversification of Dicers in plants. FEBS Lett 580: 2442–2450. [DOI] [PubMed] [Google Scholar]
- 60. Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJM (2009) RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol 21: 367–376. [DOI] [PubMed] [Google Scholar]
- 61. Law JA, Jacobsen SE (2010) Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11: 204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fusaro AF, Matthew L, Smith NA, Curtin SJ, Dedic-Hagan J, et al. (2006) RNA interference-inducing hairpin RNAs in plants act through the viral defence pathway. EMBO Rep 7: 1168–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dadami E, Boutla A, Vrettos N, Tzortzakaki S, Karakasilioti I, et al. (2012) DICER-LIKE 4 but not DICER- LIKE 2 may have a positive effect on Potato spindle tuber viroid accumulation in Nicotiana benthamiana. Mol Plant. [DOI] [PubMed]
- 64. Olmedo-Monfil V, Duran-Figueroa N, Arteaga-Vazquez M, Demesa-Arevalo E, Autran D, et al. (2010) Control of female gamete formation by a small RNA pathway in Arabidopsis . Nature 464: 628–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Deleris A, Gallego-Bartolome J, Bao J, Kasschau KD, Carrington JC, et al. (2006) Hierarchical Action and Inhibition of Plant Dicer-Like Proteins in Antiviral Defense. Science 313: 68–71. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Distribution of GO slim terms. The figure shows the distribution unigene transcripts from the de novo N. benthamiana transcriptome assembly that could be annotated GO slim terms. The distributions are shown as categories of biological process (top panel), cellular component (middle panel), and molecular function (bottom panel). Only terms with more than 10 unigenes are shown.
(TIFF)
Alignment of previously reported DCL, AGO and RDR sequences against those identified in this study. The alignment view shows the position of publically available sequences with respect to the sequence identified in this study. The slight difference in sequence identity could be due to differences in N. benthamiana lines maintained in different laboratories.
(TIFF)
Neighbour joining tree of RDR proteins. Sequences from N. benthamiana (Nbenth), A. thaliana (Athan), S. lycopersicum (Solyc), O. sativa (LOC_Os) and P. trichocarpa (POPTR) were aligned with the MUSCLE algorithm. Bootstrap values are shown at the nodes.
(TIFF)
Neighbour joining tree of DRB proteins. Sequences from N. benthamiana (Nbenth), A. thaliana (Athan), S. lycopersicum (Solyc), O. sativa (LOC_Os) and P. trichocarpa (POPTR) were aligned with the MUSCLE algorithm. Bootstrap values are shown at the nodes.
(TIFF)
Neighbour joining tree of DCL proteins. Sequences from N. benthamiana (Nbenth), A. thaliana (Athan), S. lycopersicum (Solyc), O. sativa (LOC_Os) and P. trichocarpa (POPTR) were aligned with the MUSCLE algorithm. Bootstrap values are shown at the nodes.
(TIFF)
Relative abundances of RNAi-associated genes identified in this study using RNA-seq data from [40] (8DPI dataset). TPM values were calculated using the RSEM software. With the exception of Drb2b and Rdr1, the overall abundance profile is highly similar to that reported in Figure 9.
(TIFF)
Distribution statistics calculated by the R software (5-number summary) of TPM (top) and normalized EC (bottom) values of transcripts in the 9 tissues used for the transcriptome assembly.
(DOC)
Number of transcripts with normalized EC counts greater than 0 that could be annotated with GO slim terms in each tissue*. *Within the biological process category, biosynthetic process (GO:0009058), cellular nitrogen compound metabolic process (GO:0034641), small molecule metabolic process (GO:0044281), translation (GO:0006412) and transport (GO:0006810) were highly represented in all tissues. Similarly, within the cellular component category, cell (GO:0005623), cytoplasm (GO:0005737), intracellular (GO:0005622) and organelle (GO:0043226) were highly represented. Within the molecular function category, there was no one sub-category that stood out, but oxidoreductase activity (GO:0016491), DNA binding (GO:0003677), ion binding (GO:0043167) and RNA binding (GO:0003723) were noticeably represented categories. The highly similar GO profiles for all tissues may be due to the limited subset of transcripts that could actually be annotated with a GO classification.
(DOC)
Top 10 expressed transcripts (based on EC values) in the 9 tissues used for the transcriptome assembly.
(DOC)
List of RNAi-associated genes from Arabidopsis thaliana used to screen for orthologues in Nicotiana benthamiana . 1. Eamens A, Wang M-B, Smith NA, Waterhouse PM (2008) RNA Silencing in Plants: Yesterday, Today, and Tomorrow. Plant Physiol 147: 456–468. 2. Jaubert M, Bhattacharjee S, Mello AFS, Perry KL, Moffett P (2011) ARGONAUTE2 Mediates RNA-Silencing Antiviral Defenses against Potato virus X in Arabidopsis. Plant Physiol 156: 1556–1564. 3. Vaucheret H (2008) Plant ARGONAUTES. Trends Plant Sci 13: 350–358. 4. Havecker ER, Wallbridge LM, Hardcastle TJ, Bush MS, Kelly KA, et al. (2010) The Arabidopsis RNA-Directed DNA Methylation Argonautes Functionally Diverge Based on Their Expression and Interaction with Target Loci. Plant Cell 22: 321–334. 5. Zhu H, Hu F, Wang R, Zhou X, Sze S-H, et al. (2011) Arabidopsis Argonaute10 Specifically Sequesters miR166/165 to Regulate Shoot Apical Meristem Development. Cell 145: 242–256. 6. Eamens AL, Kim KW, Curtin SJ, Waterhouse PM (2012) DRB2 Is Required for MicroRNA Biogenesis in Arabidopsis thaliana. PLoS One 7: e35933. 7. Eamens AL, Kim KW, Waterhouse PM (2012) DRB2, DRB3 and DRB5 function in a non-canonical microRNA pathway in Arabidopsis thaliana. Plant Signal Behav 7: 1224–1229.
(DOC)
Relative abundances of RNAi-associated genes identified in this study, shown as TPM values calculation by the RSEM software (this is a numerical version of Figure 5 ). *Genes comprised of more than 1 transcript from the assembly; each TPM value represents the sum of all the constituent TPMs.
(DOC)