Abstract
Selective signatures in whole genome can help us understand the mechanisms of selection and target causal variants for breeding program. In present study, we performed Extended Haplotype Homozygosity (EHH) tests to identify significant core regions harboring such signals in Chinese Holstein, and then verified the biological significance of these identified regions based on commonly-used bioinformatics analyses. Results showed a total of 125 significant regions in entire genome containing some of important functional genes such as LEP, ABCG2, CSN1S1, CSN3 and TNF based on the Gene Ontology database. Some of these annotated genes involved in the core regions overlapped with those identified in our previous GWAS as well as those involved in a recently constructed candidate gene database for cattle, further indicating these genes under positive selection maybe underlie milk production traits and other important traits in Chinese Holstein. Furthermore, in the enrichment analyses for the second level GO terms and pathways, we observed some significant terms over represented in these identified regions as compared to the entire bovine genome. This indicates that some functional genes associated with milk production traits, as reflected by GO terms, could be clustered in core regions, which provided promising evidence for the exploitability of the core regions identified by EHH tests. Findings in our study could help detect functional candidate genes under positive selection for further genetic and breeding research in Chinese Holstein.
Introduction
When a novel beneficial mutation have been under the force of selection over a long period of time, it will show some obvious features such as unusually long-range linkage disequilibrium (LD) and a high population frequency, which therefore represents a detectable " signature of selection" [1], [2]. Identifying signatures of recent positive selection could help us target causal variants for breeding and be potential to provide straightforward insights into the mechanisms of evolution. Furthermore, it can possibly highlight the genetic basis of phenotypic diversity for complex traits [3], [4].
Many methods have been developed to detect such selection signals hitherto, such as the integrated Haplotype Homozygosity Score (iHS) and the extended haplotype homozygosity (EHH) test [5], [6] based on haplotype lengths; FST test [7] for population differentiation detection. These methods have been successfully implemented in many studies to exploit strong signatures of mutations under positive selection in humans and in domestic animals, e.g., genes resistance to malaria [6], [8], in response to milk yield in dairy cattle [5], [9] and related to pigmentation [10].
In this study, we applied the EHH test to identify selective signatures in Chinese Holstein. The method of the EHH test, firstly introduced by Sabeti et al. (2002), is based on the relationship of an allele’s frequency and the extent of LD surrounding it. When under positive selection, it can make the allele frequency to increase at a very fast speed, such that the recombination does not totally break down the haplotype where the selected mutation occurs [6]. According to this, EHH is defined as the probability that two randomly chosen chromosomes carrying the core haplotype of interest are identical by descent for the entire interval from the core region to a certain point [6]. It has been reported that recombination rates are heterogeneous among chromosomal regions, and it may be a potential factor of false positives in the EHH detection [9]. REHH (relative EHH), proposed by Sabeti (2002), is supposed to overcome this limitation. The idea of REHH is to compare the EHH of the tested core haplotype with that of other core haplotypes for adjusting for local variation in recombination rates. With rectificatory recombination rates, we can readily use the REHH value to screen EHH at matched genetic distances, and haplotypes with excessive REHH values and high frequencies could indicate signal of positive selection [5], [6]. The superiorities of the EHH test are that it can incorporate information on both the allele frequency and the association among SNPs, and it is designed to work with SNP markers rather than sequencing data [9].
Since different populations are undergoing different climates, geographic environment and selective forces [11], the frequencies of alleles are largely different among populations as well as the selective signals [10]. Aiming at exploring specific selective signatures in Chinese Holstein, we performed a genome-wide detection for selective signatures employing EHH tests in present study. Our findings herein as well as incorporating those findings in our previous genome-wide association studies (GWAS) for milk production traits could possibly verify some specifically biologic functions reflected by these identified core regions, and provide foundations for further pinpointing causal variants for economically important traits in dairy cattle.
Materials and Methods
Animal Resource and Genotyping
A total of 2019 daughters and 87 sires from 15 Holstein cattle farms in Beijing were collected as experimental population. Samples were collected as part of the regular quarantine inspection of the farms, and used in this study with the permission of the animals' owners. The population structure has been detailed in our previous study [12]. DNA was extracted from blood (for daughters) and semen (for sires), and all individuals were genotyped using the Illumina BovineSNP50 BeadChip containing 54001 SNPs, with a mean distance of 48.75 kb between markers.
Before analyses, we used Beagle [13] to impute the missing genotypes and to construct haplotypes. Besides, quality control for SNP markers was performed to remove those with minor allele frequency (MAF) less than 0.03.
Detection of Selection Signatures
For the EHH test, we firstly identified core regions in the genome using the software Sweep v.1.1 [6], and then calculated EHH values for haplotypes in each core region. The core region is characterized by SNPs with strong LD and consists of some core haplotypes [9]. To compare REHH values across many core features, genetic distance of 0.5 cM was chosen as the matched distance to determine the REHH value for each core region. The distance of 0.5 cM was an appropriate setting for the longer extent LD in cattle, as compared with that in humans [9], [14], [15].
Bioinformatics Analyses
After carrying out EHH tests, we further performed multi-level bioinformatics analyses to explore potential biology significance of genes harbored in the identified core regions. The following two aspects were involved in the analyses:
Genome annotation
In the analysis, each significant core region was extended 1 Mb towards both sides as the target region for annotation. In addition, genes involved in the core regions were compared with those in our previous GWAS [12] as well as those involved in a database of candidate genes for milk production performance and mastitis [16].
Enrichment analyses
Two different types of enrichment analyses relevant to the significant core regions were performed herein, i.e., gene ontology (GO) term enrichment analyses and pathway enrichment analyses. GeneMerge1.2 software tool [17] was employed to perform enriched GO term analyses and enriched pathway analyses.
Results
A Descriptive Statistics for Markers and Core Haplotype
After the quality control for SNP genotypes and excluding the SNPs on chromosome X, 40130 SNPs were finally retained in the analyses. Table 1 gives an overall description for the distribution of SNPs across whole genome. The numbers of SNPs for every chromosome ranged from 748 to 2610, with an average interval of 63.4 kb between adjacent markers.
Table 1. Summary of genome-wide marker and core region (CR) distribution in Chinese Holstein.
| Chr | Chr length (Mbp) | SNP (n) | Mean distance (kb) | CR (n) | Total CR Length(kb) | Max CR Length(kb) | Mean CR Length(kb) | CR SNPs(n)a | Max CR SNPs(n) |
| 1 | 161.06 | 2610 | 61.7 | 292 | 40305.4 | 628.64 | 138.0±90.2 | 1099 | 11 |
| 2 | 140.63 | 2109 | 66.7 | 228 | 32967.5 | 696.32 | 144.6±102.5 | 839 | 9 |
| 3 | 127.91 | 2025 | 63.2 | 222 | 32430.1 | 1073.97 | 146.1±117.3 | 820 | 9 |
| 4 | 124.13 | 1935 | 64.1 | 196 | 26006.6 | 509.61 | 132.7±90.2 | 709 | 9 |
| 5 | 125.80 | 1681 | 74.8 | 152 | 23513.1 | 561.02 | 154.7±100.6 | 554 | 12 |
| 6 | 122.54 | 1982 | 61.8 | 206 | 28214.8 | 558.29 | 137.0±99.9 | 774 | 10 |
| 7 | 112.06 | 1759 | 63.7 | 176 | 24310.3 | 1131.54 | 138.1±117.7 | 631 | 8 |
| 8 | 116.94 | 1851 | 63.2 | 190 | 26709.9 | 600.82 | 140.6±92.1 | 716 | 9 |
| 9 | 108.07 | 1570 | 68.8 | 155 | 20279.0 | 588.08 | 130.8±85.6 | 563 | 9 |
| 10 | 106.20 | 1683 | 63.1 | 173 | 20536.7 | 347.15 | 118.7±63.2 | 624 | 8 |
| 11 | 110.17 | 1780 | 61.9 | 194 | 23677.6 | 780.69 | 122.0±91.4 | 684 | 10 |
| 12 | 85.28 | 1290 | 66.1 | 120 | 16387.3 | 618.68 | 136.6±104.5 | 421 | 7 |
| 13 | 84.34 | 1373 | 61.4 | 142 | 18376.8 | 517.75 | 129.4±87.1 | 517 | 11 |
| 14 | 81.32 | 1360 | 59.8 | 152 | 21804.0 | 548.68 | 143.4±98.6 | 549 | 10 |
| 15 | 84.60 | 1324 | 63.9 | 136 | 19048.2 | 780.38 | 140.1±100.9 | 484 | 7 |
| 16 | 77.82 | 1231 | 63.2 | 134 | 17809.6 | 730.69 | 132.9±85.0 | 502 | 11 |
| 17 | 76.45 | 1250 | 61.2 | 127 | 15106.7 | 368.96 | 119.1±66.1 | 442 | 8 |
| 18 | 66.12 | 1072 | 61.7 | 84 | 10677.9 | 690.26 | 127.1±89.3 | 289 | 6 |
| 19 | 65.21 | 1106 | 59.0 | 91 | 11736.3 | 622.59 | 129.0±98.6 | 347 | 6 |
| 20 | 75.71 | 1250 | 60.6 | 115 | 14453.6 | 313.29 | 125.7±65.6 | 413 | 8 |
| 21 | 69.17 | 1072 | 64.5 | 100 | 12443.5 | 490.57 | 124.4±75.0 | 347 | 6 |
| 22 | 61.83 | 980 | 63.1 | 108 | 12539.8 | 487.54 | 116.1±66.6 | 364 | 6 |
| 23 | 53.33 | 870 | 61.3 | 79 | 9709.7 | 502.67 | 122.9±87.3 | 267 | 8 |
| 24 | 64.95 | 993 | 65.4 | 89 | 10595.9 | 518.43 | 119.1±86.6 | 302 | 7 |
| 25 | 44.02 | 790 | 55.7 | 76 | 7983.1 | 359.94 | 105.0±61.4 | 254 | 5 |
| 26 | 51.73 | 834 | 62.0 | 83 | 9855.6 | 463.48 | 118.7±67.7 | 288 | 6 |
| 27 | 48.73 | 774 | 63.0 | 64 | 7273.0 | 385.59 | 113.6±66.7 | 210 | 6 |
| 28 | 46.00 | 748 | 61.5 | 50 | 6432.7 | 324.42 | 127.8±61.7 | 167 | 5 |
| 29 | 51.98 | 828 | 62.8 | 62 | 7923.9 | 321.19 | 127.8±61.7 | 215 | 6 |
| Total | 2544.1 | 40130 | 63.4 | 3996 | 529108.6 | 16521.24 | 129.7±85.6 | 14364 | 12 |
: the number of SNPs involved in core regions of each chromosome.
Genome-wide EHH Tests
A total of 3,996 core regions were identified and corresponding 31,182 EHH tests have been performed. These identified core regions contained 14,363 SNPs, with a range of 3–12 SNPs per core. The total length of these cores was 529,108.6 kb, with a mean length of 129.7±85.6 kb.
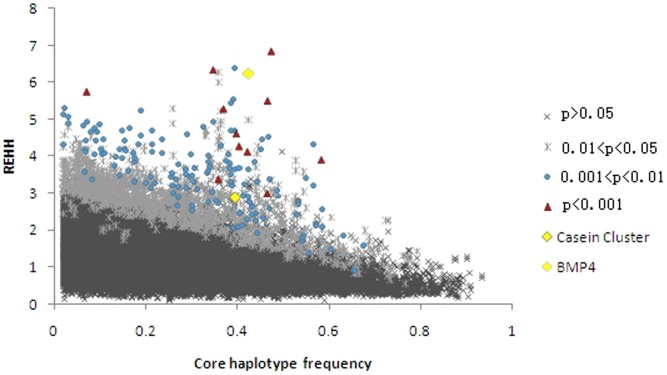
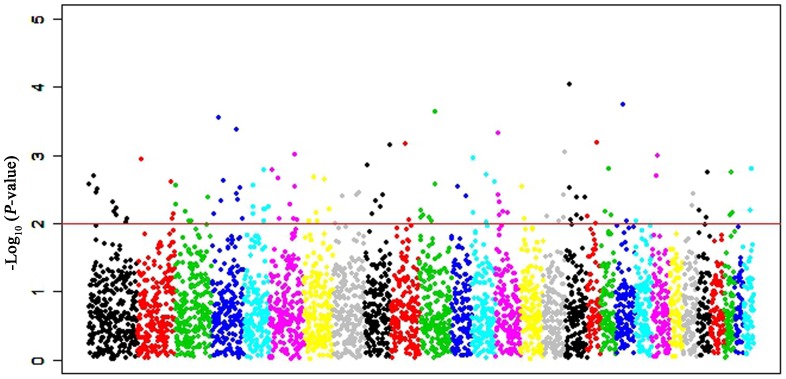
An intuitive scatter plot is presented in Fig. 1, showing the distribution of REHH values vs. haplotype frequencies. Different colors represented different ranges of p values. Moreover, a plot diagram is given in Fig. 2 to visualize the distribution of the REHH values across whole genome. Based on the concept of selective signature aforementioned, one character of such signals is the high frequency of a beneficial selected gene involved in a core haplotype. Hence we further discarded those core haplotypes with frequency <20%. Accordingly, 16,035 EHH tests remained for all core haplotypes after filtering (See Table 2), with a maximum of 1,177 tests on chromosome 1. Among these EHH tests, 541 and 125 out of them achieved significant level corresponding to the significance levels of 0.05 and 0.01, respectively.
Figure 1. Distribution of the REHH vs. the core haplotype frequency.
Different P values are marked by different color symbols presented in the right of this figure.
Figure 2. The distribution of the P values of haplotypes with frequency≥0.20 on the whole genome.
Table 2. Summary of whole genome extended haplotype homozygosity tests.
| Chr | Tests on CH (n)a | P-value<0.05 (n) | P-value<0.01(n) |
| 1 | 1177 | 29 | 10 |
| 2 | 895 | 30 | 5 |
| 3 | 898 | 36 | 7 |
| 4 | 807 | 27 | 9 |
| 5 | 594 | 19 | 8 |
| 6 | 822 | 31 | 9 |
| 7 | 681 | 17 | 7 |
| 8 | 784 | 32 | 5 |
| 9 | 610 | 19 | 6 |
| 10 | 640 | 21 | 2 |
| 11 | 766 | 27 | 8 |
| 12 | 456 | 22 | 2 |
| 13 | 579 | 20 | 5 |
| 14 | 614 | 15 | 6 |
| 15 | 569 | 25 | 2 |
| 16 | 540 | 12 | 5 |
| 17 | 511 | 14 | 7 |
| 18 | 345 | 15 | 3 |
| 19 | 369 | 14 | 3 |
| 20 | 466 | 14 | 2 |
| 21 | 401 | 16 | 1 |
| 22 | 436 | 16 | 2 |
| 23 | 326 | 15 | 0 |
| 24 | 360 | 15 | 2 |
| 25 | 333 | 11 | 3 |
| 26 | 325 | 8 | 0 |
| 27 | 264 | 6 | 3 |
| 28 | 214 | 7 | 0 |
| 29 | 253 | 8 | 3 |
| Total | 16035 | 541 | 125 |
: Core haplotypes involved in each core region determined by Sweep across genome.
Genome Annotation
We annotated genes involved in all significant identified core regions based on the Gene Ontology database (http://www.geneontology.org/), and further found out those potential candidate genes relevant to milk production traits. Accordingly, a total of 9,829 genes had been identified, 6573 of them (66.87%) fall into the regions of QTL affecting milk production included in the cattle QTL database (http://www.animalgenome.org/cgi-bin/QTLdb/index).
In our previous GWAS [12], 75 genes were identified associated with one or multiple milk production traits in dairy cattle such as milk fat yield (FY), milk yield (MY), milk protein yield (PY), milk fat percentage (FP) and milk protein percentage (PP). Among these 75 genes, 45 out of them were contained in the core regions detected in present study, further indicating these genes under positive selection maybe underlie milk production traits in Chinese Holstein. Table 3 gives symbols of these genes as well as the closest core positions and corresponding p values of core haplotypes. Totally, 45 genes were involved in 19 core regions which contained 25 significant core haplotypes.
Table 3. Genes within the core regions overlapping with those by genome wide association studies by Jiang et al (2010).
| Gene Symbol | Chr. | Closest core position (bp) | Hap Freq(%) | P-value |
| LOC614166 | 1 | 148796434–148911817 | 0.286 | 0.0547 |
| 149189841–149242164 | 0.209 | 0.0365 | ||
| DIP2A | 1 | 148796434–148911817 | 0.286 | 0.0547 |
| 149189841–149242164 | 0.209 | 0.0365 | ||
| KBTBD10 | 2 | 27607855–27651437 | 0.382 | 0.0369/0.014 |
| SLC30A7 | 3 | 44239453–44433532 | 0.373 | 0.0211 |
| 45775675–45934869 | 0.418 | 0.00681 | ||
| LOC511240 | 5 | 76788487–76882812 | 0.325 | 0.0154 |
| 77546764–77805841 | 0.351 | 0.0587 | ||
| HERC3 | 6 | 37135013–37231101 | 0.572/0.0479 | 0.00220/0.0378 |
| PKD2 | 6 | 37135013–37231101 | 0.572/0.0479 | 0.00220/0.0378 |
| 38479643–38558526 | 0.216 | 0.0562 | ||
| NFIB | 8 | 31713520–31832256 | 0.235 | 0.00359/0.00573 |
| LOC788012 | 9 | 5992605–6049113 | 0.348/0.267 | 0.0334/0.0596 |
| C14H8orf33 | 14 | 260341–443937 | 0.248 | 0.0568 |
| FOXH1 | 14 | 260341–443937 | 0.248 | 0.0568 |
| CYHR1 | 14 | 260341–443937 | 0.248 | 0.0568 |
| VPS28 | 14 | 260341–443937 | 0.248 | 0.0568 |
| DGAT1 | 14 | 260341–443937 | 0.248 | 0.0568 |
| MAF1 | 14 | 260341–443937 | 0.248 | 0.0568 |
| LOC786966 | 14 | 260341–443937 | 0.248 | 0.0568 |
| GRINA | 14 | 260341–443937 | 0.248 | 0.0568 |
| 1889210–1967406 | 0.33 | 0.0454 | ||
| GML | 14 | 260341–443937 | 0.248 | 0.0568 |
| 1889210–1967406 | 0.330 | 0.0454 | ||
| 2163275–2239116 | 0.268 | 0.0554 | ||
| GPIHBP1 | 14 | 1889210–1967406 | 0.330 | 0.0454 |
| 2163275–2239116 | 0.268 | 0.0554 | ||
| COL22A1 | 14 | 2805785–2849483 | 0.579 | 0.0556 |
| 3018726–3099635 | 0.260 | 0.0401/0.0246 | ||
| NKAIN3 | 14 | 28014144–28185224 | 0.322 | 0.0282 |
| OPLAH | 14 | 260341–443937 | 0.248 | 0.0568 |
| MAPK15 | 14 | 260341–443937 | 0.248 | 0.0568 |
| ZNF623 | 14 | 260341–443937 | 0.248 | 0.0568 |
| EEF1D | 14 | 260341–443937 | 0.248 | 0.0568 |
| 1889210–1967406 | 0.330 | 0.0454 | ||
| ZC3H3 | 14 | 260341–443937 | 0.248 | 0.0568 |
| 1889210–1967406 | 0.330 | 0.0454 | ||
| LYPD2 | 14 | 260341–443937 | 0.248 | 0.0568 |
| 1889210–1967406 | 0.330 | 0.0454 | ||
| RHPN1 | 14 | 1889210–1967406 | 0.330 | 0.0454 |
| 2163275–2239116 | 0.268 | 0.0554 | ||
| GPR20 | 14 | 1889210–1967406 | 0.330 | 0.0454 |
| 2163275–2239116 | 0.268 | 0.0554 | ||
| 2805785–2849483 | 0.579 | 0.0556 | ||
| PTK2 | 14 | 1889210–1967406 | 0.330 | 0.0454 |
| 2163275–2239116 | 0.268 | 0.0554 | ||
| 2805785–2849483 | 0.579 | 0.0556 | ||
| 3018726–3099635 | 0.260 | 0.0401/0.0246 | ||
| EIF2C2 | 14 | 1889210–1967406 | 0.330 | 0.0454 |
| 2163275–2239116 | 0.268 | 0.0554 | ||
| 2805785–2849483 | 0.579 | 0.0556 | ||
| 3018726–3099635 | 0.260 | 0.0401/0.0246 | ||
| KCNK9 | 14 | 2163275–2239116 | 0.268 | 0.0554 |
| 2805785–2849483 | 0.579 | 0.0556 | ||
| 3018726–3099635 | 0.260 | 0.0401/0.0246 | ||
| LOC618755 | 14 | 3018726–3099635 | 0.260 | 0.0401/0.0246 |
| ZNF7 | 14 | 260341–443937 | 0.248 | 0.0568 |
| EPPK1 | 14 | 260341–443937 | 0.248 | 0.0568 |
| CYP11B2 | 14 | 260341–443937 | 0.248 | 0.0568 |
| 1889210–1967406 | 0.330 | 0.0454 | ||
| 2163275–2239116 | 0.268 | 0.0554 | ||
| GLI4 | 14 | 1889210–1967406 | 0.330 | 0.0454 |
| 2163275–2239116 | 0.268 | 0.0554 | ||
| TRAPPC9 | 14 | 1889210–1967406 | 0.330 | 0.0454 |
| 2163275–2239116 | 0.268 | 0.0554 | ||
| 2805785–2849483 | 0.579 | 0.0556 | ||
| 3018726–3099635 | 0.260 | 0.0401/0.0246 | ||
| LOC782462 | 20 | 36691324–36837401 | 0.537/0.215 | 0.0187/0.0467 |
| LOC782833 | 20 | 36691324–36837401 | 0.537/0.215 | 0.0187/0.0467 |
| C9 | 20 | 36691324–36837401 | 0.537/0.215 | 0.0187/0.0467 |
| FYB | 20 | 36691324–36837401 | 0.537/0.215 | 0.0187/0.0467 |
| RICTOR | 20 | 36691324–36837401 | 0.537/0.215 | 0.0187/0.0467 |
| LOC100138964 | 20 | 36691324–36837401 | 0.537/0.215 | 0.0187/0.0467 |
| RAI14 | 20 | 40690535–40854433 | 0.341 | 0.0443 |
This table describes genes that associated with milk production traits when compared with genome wide association studies, Hap Freq describes the frequency of core haplotype involved in each core region determined by Sweep across genome.
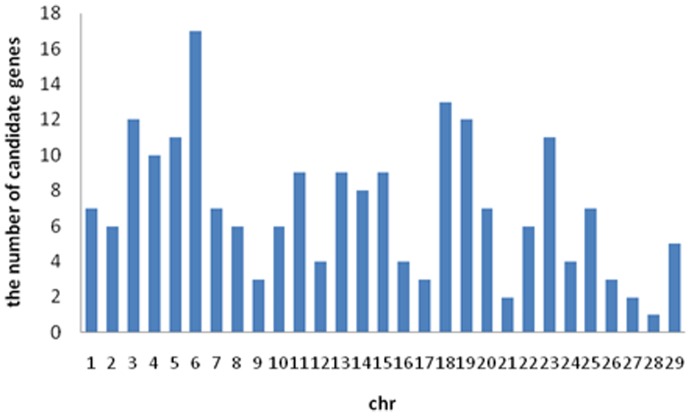
In addition, 203 genes within the core regions overlapped with those included in a database of candidate genes in cattle underlying milk production performance and mastitis [16]. A histogram showing the overall distribution of the candidate genes across the genome is given in Fig. 3. It can be seen that the largest number of overlapped candidate genes were located on chromosome 6. Among these 203 functional genes, 13 genes (LEP, ABCG2, CSN1S1, CSN3, IL8, TLR4, IL1B, LBP, DGAT1, TLR2, C5AR1, LTF, TNF), which were suggested as being under selection, have been frequently reported in different studies [16].
Figure 3. Distributions of candidate genes in whole genome.
GO Term Enrichment Analyses
By searching for over-represented terms in core regions compared to those across whole genome, we can detect those functional genes clustered in the identified core regions contributing to milk production traits. We analyzed the significant enriched terms for genes involved in the core region. As a result, 36 second level GO terms (64.3%) were found to be significantly over-represented (Table 4). For instance, the term ‘metabolic process‘(GO: 0008152) showed significantly enriched, which indicated genes associated with milk production traits involved in ‘metabolic process‘ were clustered in the core regions. Other terms such as ‘immune system process‘(GO: 0002376) and ‘reproductive process‘(GO: 0022414) were also showed clear biological association with mastitis and milk production traits.
Table 4. Enriched GO terms when comparing candidate genes to the whole genome using GeneMerge1.2.
| GO term | Description | Pop-frec | CR-frec | ratio | P value |
| GO:0000003 | reproduction | 210/27430 | 101/9829 | 1.34 | 9.08E-3 |
| GO:0044421 | extracellular region part | 361/27430 | 171/9829 | 1.32 | 2.12E-4 |
| GO:0016265 | death | 403/27430 | 182/9829 | 1.26 | 3.49E-3 |
| GO:0022414 | reproductive process | 208/27430 | 101/9829 | 1.36 | 5.76E-3 |
| GO:0032991 | macromolecular complex | 1327/27430 | 596/9829 | 1.25 | 1.03E-10 |
| GO:0005623 | cell | 5024/27430 | 2264/9829 | 1.26 | 8.171E-49 |
| GO:0048519 | negative regulation of biological process | 669/27430 | 306/9829 | 1.28 | 3.34E-06 |
| GO:0050896 | response to stimulus | 1500/27430 | 693/9829 | 1.30 | 7.77E-16 |
| GO:0044422 | organelle part | 1953/27430 | 885/9829 | 1.26 | 1.32E-17 |
| GO:0051234 | establishment of localization | 1094/27430 | 494/9829 | 1.26 | 3.30E-09 |
| GO:0031974 | membrane-enclosed lumen | 728/27430 | 340/9829 | 1.30 | 3.68E-08 |
| GO:0022610 | biological adhesion | 198/27430 | 94/9829 | 1.32 | 0.0261 |
| GO:0008152 | metabolic process | 2813/27430 | 1291/9829 | 1.28 | 1.81E-29 |
| GO:0044464 | cell part | 5024/27430 | 2264/9829 | 1.26 | 8.17E-49 |
| GO:0003824 | catalytic activity | 1991/27430 | 919/9829 | 1.29 | 2.99E-21 |
| GO:0005488 | binding | 3393/27430 | 1534/9829 | 1.26 | 1.07E-31 |
| GO:0009987 | cellular process | 3860/27430 | 1739/9829 | 1.26 | 1.49E-35 |
| GO:0005215 | transporter activity | 400/27430 | 180/9829 | 1.26 | 4.84E-3 |
| GO:0032502 | developmental process | 896/27430 | 409/9829 | 1.27 | 2.85E-08 |
| GO:0002376 | immune system process | 390/27430 | 175/9829 | 1.25 | 7.19E-3 |
| GO:0008283 | cell proliferation | 292/27430 | 142/9829 | 1.36 | 2.41E-4 |
| GO:0005576 | extracellular region | 697/27430 | 320/9829 | 1.28 | 1.06E-06 |
| GO:0023052 | signaling | 904/27430 | 424/9829 | 1.31 | 1.34E-10 |
| GO:0048518 | positive regulation of biological process | 785/27430 | 359/9829 | 1.28 | 2.51E-07 |
| GO:0032501 | multicellular organismal process | 1124/27430 | 494/9829 | 1.23 | 3.43E-07 |
| GO:0051704 | multi-organism process | 189/27430 | 92/9829 | 1.36 | 0.0102 |
| GO:0043226 | organelle | 3372/27430 | 1509/9829 | 1.25 | 1.65E-28 |
| GO:0060089 | molecular transducer activity | 309/27430 | 151/9829 | 1.36 | 8.61E-05 |
| GO:0050789 | regulation of biological process | 1900/27430 | 871/9829 | 1.28 | 5.19E-19 |
| GO:0004872 | receptor activity | 347/27430 | 156/9829 | 1.25 | 0.0144 |
| GO:0030234 | enzyme regulator activity | 261/27430 | 122/9829 | 1.30 | 9.57E-3 |
| GO:0040011 | locomotion | 245/27430 | 115/9829 | 1.31 | 0.012 |
| GO:0051179 | localization | 1269/27430 | 575/9829 | 1.26 | 4.01E-11 |
| GO:0040007 | growth | 165/27430 | 81/9829 | 1.37 | 0.0170 |
| GO:0071840 | cellular component organization or biogenesis | 1015/27430 | 474/9829 | 1.30 | 1.57E-11 |
| GO:0065007 | biological regulation | 2029/27430 | 927/9829 | 1.28 | 8.67E-20 |
This table describes second GO terms significantly enriched in core regions based on GeneMerge1.2 software. 36 terms are detected to be significant here. Pop-frec describes the frequency of genes in the population with this term, and CR-frec describes the frequency of genes in the core regions with this term. Ratio is calculated by the comparison of a term within the core regions to that in genome wide. P value here is a Bonferroni corrected P value.
Pathway Enrichment Analyses
We detected over-represented pathways relevant to genes within detected core regions. Accordingly, 9 out of 213 (4.23%) pathways were found to be significant in the core regions (Table 5), e.g., 471 genes were contributed in the significant ‘metabolic pathways’ (bta01100) which could plausibly be involved in the processes of milk production; The term ‘chemokine signaling pathway’ (bta04062) showed significant over-representation involved in the processes of inflammatory immune response in mammary gland containing 89 genes in all. Other over-represented pathways also reflected some important biological processes, such as: ‘focal adhesion’ (bta04510) participated in cell motility and cell proliferation, ‘spliceosome’ (bta03040) assembled on the mRNA precursor with the function of folding the precursor into a conformation to allow transesterification to proceed.
Table 5. Enriched pathway terms when comparing candidate genes to the whole genome using GeneMerge1.2.
| Pathway term | Description | Pop-frec | CR-frec | ratio | P value |
| bta04510 | Focal adhesion | 192/27430 | 93/9829 | 1.35 | 0.0464 |
| bta03040 | Spliceosome | 126/27430 | 67/9829 | 1.48 | 0.0107 |
| bta04270 | Vascular smooth muscle contraction | 122/27430 | 66/9829 | 1.51 | 5.94E-3 |
| bta05322 | Systemic lupus erythematosus | 189/27430 | 92/9829 | 1.36 | 0.0402 |
| bta04080 | Neuroactive ligand-receptor interaction | 318/27430 | 155/9829 | 1.36 | 3.00E-4 |
| bta04010 | MAPK signaling pathway | 266/27430 | 131/9829 | 1.37 | 9.80E-4 |
| bta01100 | Metabolic pathways | 1081/27430 | 471/9829 | 1.22 | 1.00E-5 |
This table describes significant pathway terms over-represented in core regions based on GeneMerge1.2 software. Pop-frec describes the frequency of genes in the population with this pathway, and CR-frec describes the frequency of genes in the core regions with this pathway. Ratio is calculated by the comparison of a term within the core regions to that in genome wide. P value here is a Bonferroni corrected P value.
Discussion
Due to serving as models for basic studies of some important pathways, cattle play a special role in biology (Bovine Genome Sequencing Initiative http://www.genome.gov/Pages/Research/Sequencing/SeqProposals/BovineSEQ). In the past few years, many studies were performed in the field of cattle genetics. For instance, Kaupe et al. [18] genotyped 1748 DNA samples of 38 different Bos taurus and Bos indicus cattle breeds to examine the DGAT1 polymorphism in cattle breeds; another research used Bos taurus and Bos indicus cattle to detect the utilization of low-quality roughage [19]. So far searching for genomic evidence of positive selection in cattle recently has been widely considered as an attractive strategy for identifying functional polymorphisms and providing important insight into the mechanism of evolution [20]. In this study, we carried out EHH tests to detect population-specific selection signatures by using the high density SNP chips in Chinese Holstein and then performed various systems biology analyses to confirm the biological significance for the identified core regions.
Totally, there were 125 core regions been detected potentially harboring selective signals at the significance level of p = 0.01. As Qanbari et al. (2010) said, by comparing with shorter extent of homozygositiy of other haplotypes presented in these regions, it can demonstrate the longer extent of homozygosity of some haplotypes were not simply because of low recombination rates but due to the strong recent selection. In a recent study of detecting selective signatures in German Holstein cattle based on EHH tests, a total of 161 regions were identified to exhibit the signals of positive selection. Annotation for these regions also revealed a list of functional candidate genes like SPATA17, DGAT1, FABP3 and ABCE1 [9]. Another study carried out by Flori et al. (2009) in three major French dairy cattle breeds, showed 13 highly significant regions under the recent selection. Some of them contained genes that with strong effects on milk production traits, such as GHR, LAP3 and ABCG2. These two researches revealed that most of genes targeted by artificial selection in dairy cattle mainly tended to improve milk production [21]. So, for further probing into the relationships between the identified selection signatures and important traits under selection in this study, we compared these 125 core regions with findings in our previous GWAS studies as well as a candidate gene database for economic traits in dairy cattle. A suite of genes in the identified core regions have been suggested relevant to milk production traits and mastitis. For instance, two core regions overlapped with the region harboring polycystic kidney disease 2 (PKD2) gene, which functioned as a calcium permeable cation channel and had been identified significantly associated with milk protein percentage [12]; G protein-coupled receptor 20 (GPR20) gene located in an overlapped area of 3 extended core regions on chromosome 14, and it is related to milk fat percentage traits. On BTA 6, a region with a haplotype showed the lowest P value (P = 0.0022) hiding the HECT domain and RLD 3 (HERC3) and polycystic kidney disease 2 (PKD2) gene, HERC3 gene involved in the pathway of ubiquitination mediated proteolysis and it mainly functions as a signal for 26S proteasome dependent protein degradation. Another signal on chromosome 6, which harboring ATP-binding cassette sub-family G member 2 (ABCG2) has been proved as a target of selection [22], [23]. These results indicated that these core regions presented biologic signals associated with milk production performance.
It was notable that the core region harboring DGAT1 gene merely showed a marginal significance (p = 0.057), although DGAT1 gene has been suggested as being under selection and been reported for many times. This could be due to the higher initial frequency of some beneficial alleles leading to less polymorphism in haplotypes [24]. Another possible reason could be the limitation of SNP density.
In our study, we conducted enrichment analyses for the second GO terms and pathways, which makes use of specifically meaningful annotations rather than uninformative ones and provides insight into regional genomic function [25]. This has been proved to successfully identify over-represented terms for general annotation analyses such as post-genomic analysis, DNA chips and gene networks analysis, and QTL global meta-analysis [26]. In the analyses, we firstly carried out enrichment GO analyses on 418 candidate genes in the database [16]. There were 57 (32.76%) over-represented terms have been found, which also provided a strong indication that genes may be clustered in a manner that reflects their functional association with particular traits. Subsequently, enrichment analyses on genes in the identified core regions were performed and 36 second level GO terms (64.3%) were found to be significantly over-represented in core regions including a total of 2740 functional genes, of which 34 significant terms were coincide with the results of the analyses for 418 candidate genes. These results were consistent with our previous hypothesis that some functional genes associated with milk production traits, as reflected by GO terms could be clustered in core regions compared against the entire genome regions.
We finally analyzed the enriched pathways with the purpose to confirm the biological associations between core regions and quantitative traits. The results of the enriched pathway analyses showed a total of 9 out of 213 (4.23%) pathways were found to be significant in core regions, of which 6 (66.67%) significant pathways were included in the results of candidate genes, confirming that genes involved in functional biological processes. As expected, milk production processes were enriched in core regions, which revealed proofs of multiple genes in a same pathway presenting selective signatures. These results were consistent with the results reported by Qanbari et al. (2010) that the identified candidate genes reflected a panel of pathways such as steroid metabolism and transportation. All these demonstrated the feasibility of EHH test as well as the biological significance of the identified cores based on this method. Findings in our study could help detect functional candidate genes under positive selection for further genetic and breeding studies in Chinese Holstein.
In addition, it was not ignorable that the length of these significant core regions identified by EHH tests was generally large and contained many genes, thus reducing the accuracy of this method; On the other hand, the method of EHH test may lack sensitive for identifying lower-frequency selected alleles [10], resulting in some low-frequency functional genes not included in our results. As for the enriched GO term analyses to detect the distribution of functional genes, we found that the proportion of significant terms were limited. This could be because many of the bovine gene models remain un-annotated. Hence it only roughly reflects our hypothesis that core regions displaying signatures are suggested targets of recent selection. New powerful methods are needed to enhance the power for localizing the source of selection. Due to each single test for selective signatures can provide partial information, we could possibly combine them in a composite likelihood statistic. This idea has already been proved effectively in Human population [10]. Besides, as the differences of allele frequency between populations may be more palpable in the regions harboring causal variants, we can combine with the tests for selective signatures between populations to help us narrow core regions for further accurately positioning the causal variants.
In all, this study detected 125 core regions over the whole genome in Chinese Holstein using the EHH test associated with some bioinformatics analyses. Most of the genes included in these core regions were consistent with reported candidate genes for milk production and mastitis, of which some functional genes classified by GO terms or involved in important pathways of milk production traits were enriched in these identified regions. Our results provided evidence for the exploitability of the core regions identified by EHH tests, and eventually lay a substrate for further studies for targeting causal mutations underlying some important economic traits in Chinese Holstein.
Acknowledgments
We would like to give thanks to the two reviewers and the editor for their helpful comments and constructive suggestions that greatly improved our manuscript. We also thank Beijing Dairy Cattle Center for providing blood and semen samples.
Funding Statement
The authors appreciate the financial support provided by the National High Technology Research and Development Program of China (863 Program 2011AA100302), the National Natural Science Foundations of China (31272419, 30972092), the Natural Science Foundations of Beijing (6102016), New-Century Training Program Foundation for the Talents by the State Education Commission of China (NETC-10-0783), Scientific Research Foundation for the Returned Overseas Chinese Scholars of State Education Ministry. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Tian D, Araki H, Stahl E, Bergelson J, Kreitman M (2002) Signature of balancing selection in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America 99: 11525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang Z, Wang B, Tang K, Lee EJD, Chong SS, et al. (2005) A functional polymorphism within the MRP1 gene locus identified through its genomic signature of positive selection. Human molecular genetics 14: 2075. [DOI] [PubMed] [Google Scholar]
- 3. Andersson L, Georges M (2004) Domestic-animal genomics: deciphering the genetics of complex traits. Nature Reviews Genetics 5: 202–212. [DOI] [PubMed] [Google Scholar]
- 4. Akey JM, Zhang G, Zhang K, Jin L, Shriver MD (2002) Interrogating a high-density SNP map for signatures of natural selection. Genome research 12: 1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Voight BF, Kudaravalli S, Wen X, Pritchard JK (2006) A map of recent positive selection in the human genome. PLoS biology 4: e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sabeti PC, Reich DE, Higgins JM, Levine HZP, Richter DJ, et al. (2002) Detecting recent positive selection in the human genome from haplotype structure. Nature 419: 832–837. [DOI] [PubMed] [Google Scholar]
- 7. Lewontin R, Krakauer J (1973) Distribution of gene frequency as a test of the theory of the selective neutrality of polymorphisms. Genetics 74: 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tishkoff SA, Varkonyi R, Cahinhinan N, Abbes S, Argyropoulos G, et al. (2001) Haplotype diversity and linkage disequilibrium at human G6PD: recent origin of alleles that confer malarial resistance. Science 293: 455–462. [DOI] [PubMed] [Google Scholar]
- 9. Qanbari S, Pimentel E, Tetens J, Thaller G, Lichtner P, et al. (2010) A genome-wide scan for signatures of recent selection in Holstein cattle. Animal genetics 41: 377–389. [DOI] [PubMed] [Google Scholar]
- 10. Grossman SR, Shylakhter I, Karlsson EK, Byrne EH, Morales S, et al. (2010) A composite of multiple signals distinguishes causal variants in regions of positive selection. Science 327: 883–886. [DOI] [PubMed] [Google Scholar]
- 11. Bamshad M, Wooding SP (2003) Signatures of natural selection in the human genome. Nature Reviews Genetics 4: 99–111. [DOI] [PubMed] [Google Scholar]
- 12. Jiang L, Liu J, Sun D, Ma P, Ding X, et al. (2010) Genome wide association studies for milk production traits in Chinese Holstein population. PLoS One 5: e13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Browning SR, Browning BL (2007) Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. American journal of human genetics 81: 1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McKay SD, Schnabel RD, Murdoch BM, Matukumalli LK, Aerts J, et al. (2007) Whole genome linkage disequilibrium maps in cattle. BMC genetics 8: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tenesa A, Navarro P, Hayes BJ, Duffy DL, Clarke GM, et al. (2007) Recent human effective population size estimated from linkage disequilibrium. Genome research 17: 520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ogorevc J, Kunej T, Razpet A, Dovc P (2009) Database of cattle candidate genes and genetic markers for milk production and mastitis. Animal Genetics 40: 832–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Castillo-Davis CI, Hartl DL (2003) GeneMerge-post-genomic analysis, data mining, and hypothesis testing. Bioinformatics 19: 891. [DOI] [PubMed] [Google Scholar]
- 18. Kaupe B, Winter A, Fries R, Erhardt G (2004) DGAT1 polymorphism in Bos indicus and Bos taurus cattle breeds. Journal of dairy research 71: 182–187. [DOI] [PubMed] [Google Scholar]
- 19. Hunter R, Siebert B (1985) Utilization of low-quality roughage by Bos taurus and Bos indicus cattle. British Journal of Nutrition 53: 637–648. [DOI] [PubMed] [Google Scholar]
- 20. Beja-Pereira A, Luikart G, England PR, Bradley DG, Jann OC, et al. (2003) Gene-culture coevolution between cattle milk protein genes and human lactase genes. Nature Genetics 35: 311–313. [DOI] [PubMed] [Google Scholar]
- 21. Flori L, Fritz S, Jaffrézic F, Boussaha M, Gut I, et al. (2009) The genome response to artificial selection: a case study in dairy cattle. PLoS One 4: e6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Harder B, Bennewitz J, Reinsch N, Thaller G, Thomsen H, et al. (2006) Mapping of quantitative trait loci for lactation persistency traits in German Holstein dairy cattle. Journal of Animal Breeding and Genetics 123: 89–96. [DOI] [PubMed] [Google Scholar]
- 23. Gibbs RA, Taylor JF, Van Tassell CP, Barendse W, Eversole KA, et al. (2009) Genome-wide survey of SNP variation uncovers the genetic structure of cattle breeds. Science (New York, NY) 324: 528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Innan H, Kim Y (2004) Pattern of polymorphism after strong artificial selection in a domestication event. Proceedings of the National Academy of Sciences of the United States of America 101: 10667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hanni S, David A (2009) QTL global meta-analysis: are trait determining genes clustered? BMC Genomics 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xie H, Wasserman A, Levine Z, Novik A, Grebinskiy V, et al. (2002) Large-scale protein annotation through gene ontology. Genome research 12: 785. [DOI] [PMC free article] [PubMed] [Google Scholar]