Abstract
This study characterized electrophilic and radical products derived from metabolism of capsaicin by cytochrome P450 and peroxidase enzymes. Multiple glutathione and β-mercaptoethanol conjugates (a.k.a., adducts), derived from trapping of quinone methide and quinone intermediates of capsaicin, its analogue nonivamide, and O-demethylated and aromatic hydroxylated metabolites thereof, were produced by human liver microsomes and individual recombinant human P450 enzymes. Conjugates derived from concomitant dehydrogenation of the alkyl terminus of capsaicin, were also characterized. Modifications to the 4-OH substituent of the vanilloid ring of capsaicinoids largely prevented the formation of electrophilic intermediates, consistent with the proposed structures and mechanisms of formation for the various conjugates. 5,5’-Dicapsaicin, presumably arising from bi-molecular coupling of free radical intermediates, was also characterized. Finally, the analysis of hepatic glutathione conjugates and urinary N-acetylcysteine conjugates from mice dosed with capsaicin confirmed the formation of glutathione conjugates of O-demethylated, quinone methide, and 5-OH-capsaicin in vivo. These data demonstrated that capsaicin and structurally similar analogues are converted to reactive intermediates by certain P450 enzymes, which may partially explain conflicting reports related to the cytotoxic, pro-carcinogenic, and chemoprotective effects of capsaicinoids in different cells and/or organ systems.
Keywords: Capsaicin, nonivamide, cytochrome P450, electrophile, glutathione conjugate
Introduction
Capsaicinoids are the pungent compounds in “hot” peppers. Capsaicin (trans-8-methyl-N-vanillyl-6-nonenamide) represents ~60–70% of all capsaicinoids in natural sources.(1) Humans are exposed to capsaicinoids through the consumption of peppers, and capsaicinoids (including cis- and trans- isomers of capsaicin) are also used in an array of consumer products, including: topical (e.g., Capzasin-HP and Qutenza/NGX-4010) and injectable (Adlea/ALGRX-4975) analgesics; nasal aerosols to treat allergic rhinitis(2) and cluster headache (Civamide; Winston Laboratories)(3,4); oral supplements for weight loss(5); and self-defense pepper spray products.(1) More recently, capsaicinoids have also been investigated as chemopreventive agents.(6–12)
Pharmacokinetic studies in rodents and humans show that topical and oral administration of capsaicinoids produce high local concentrations and moderate circulating concentrations, depending upon the dose and route of delivery. Using the NGX-4010 skin patch, a 1–1.5 h topical treatment of capsaicin at ~10 mg/kg in humans produced peak plasma concentrations of ~0.5–18 ng/mL (1.6–58 nM) in the roughly 3–30% of patients where quantifiable capsaicin levels occurred.(13) Capsaicin was cleared in most patients within 4.5 h and the estimated T1/2 was 1.6 h with AUC values of ~4.4 and 7.7 ng*h/mL (at ~1.5 h) and Cmax values of 1.4 and 2.9 ng/mL for 1 and 1.5 h patch applications. While detection of P450 metabolites of capsaicin were not reported, substantially higher concentrations of capsaicin and the detection of hydrolysis products and P450 metabolites arising from ω- and ω-1 hydroxylation and terminal dehydrogenation were reported at sites of patch application in a similar study.(14) Orally administered capsaicinoids are also efficiently (85–95% total dose) absorbed from the gastrointestinal lumen, with minor metabolism occurring in the intestine (mainly hydrolysis), but near complete decomposition in the liver.(15–17) In one study, a 5 g oral dose of capsicum (~26.6 mg capsaicin or ~0.4 mg/kg) in humans yielded peak plasma concentrations of capsaicin of 2.5 ng/mL (~8.2 nM), with Tmax, T1/2, and AUC values of ~0.78 h, 0.42 h, and ~1.7 ng*h/mL respectively.(18) It was suggested that the relatively low Cmax, short T1/2, and low AUC were the result of extensive metabolic decomposition, consistent with prior studies.(15,16) In a second study, oral dosing of rats (30 mg/kg; ~5× average daily intake of capsaicinoids by individuals in India, but where “beneficial effects have been observed”) demonstrated 94% absorption from the intestine with a peak serum concentration of 6.2 µM.(19) In this same study, substantially higher amounts were found in the liver (~45 mg/tissue), kidney (~7 mg/tissue), and intestine (~1,060 mg/tissue). However, the formation of P450-derived metabolites was not evaluated in either of these studies. Given reports that the average capsaicin intake for humans is 0.5–4 mg/kg/day (EC Scientific Committee on Food, 2002, http://ec.europa.eu/food/fs/sc/scf/out120_en.pdf), and that commonly used products containing capsaicinoids can produce substantial local and circulating levels of capsaicinoids, there is a critical need to understand how capsaicinoids interact with drug metabolizing enzymes, including the formation of reactive intermediates that may have extensive effects on biological systems.
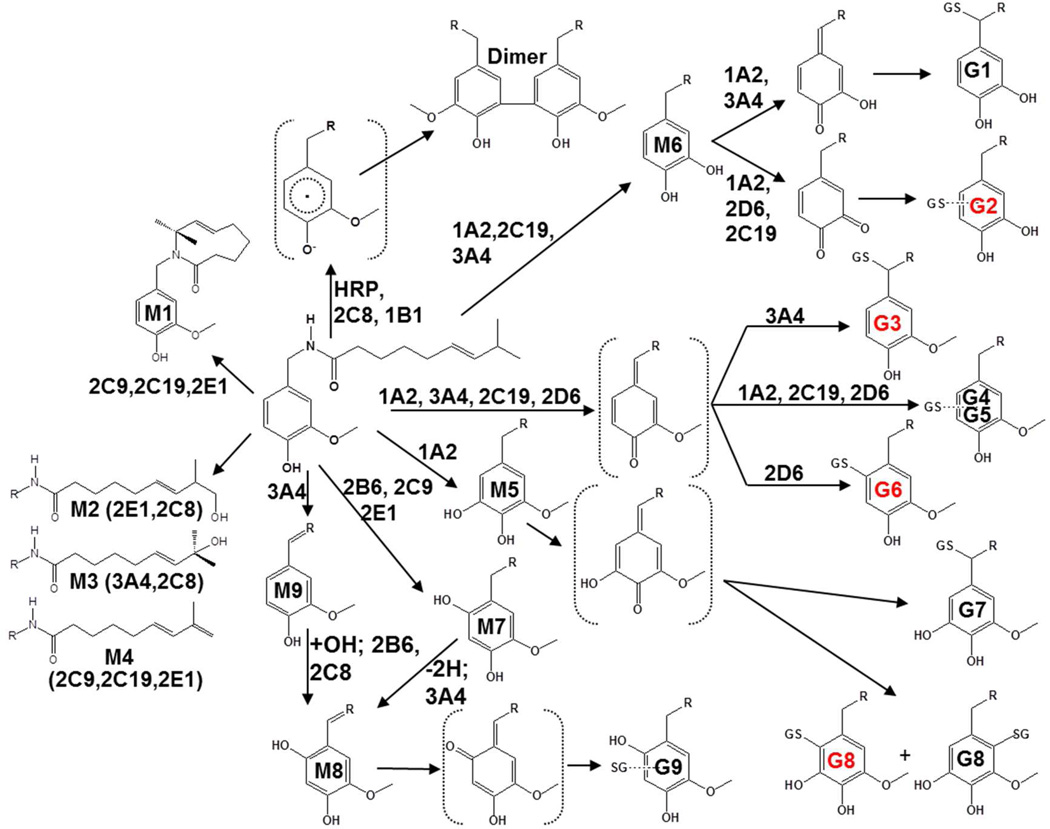
Prior studies of the metabolism of capsaicinoids by P450 enzymes characterized a macrocycle metabolite where the ω-1 carbon became covalently bound to the amide nitrogen (M1), ω- and ω-1 alcohols (M2 and M3), an alkyl terminal diene (M4), two aromatic hydroxylated products (M5 and M7), an O-demethylated catechol (M6), a unique oxygenated imide (M8), and an imide (M9)(20,21) (See Figure 7 for structures). Additionally, three glutathione (GSH) conjugates presumably derived from O-demethylation (M6), aromatic hydroxylation (M5 and M7), and the oxygenated imide (M8) were observed,(20) but the origins and characteristics of these GSH-conjugated metabolites were not thoroughly investigated.
Figure 7.
Proposed metabolic scheme for the formation of major GSH conjugates and 5,5’-dicapsaicin of capsaicinoids by P450 and peroxidase enzymes. Structures with a red label indicate that the product was detected in vivo in mouse liver, serum, or urine, as reported in Table 2.
The current study characterized the biochemical pathways leading to the formation of reactive intermediates from capsaicinoids by P450 enzymes, in an effort to better understand the relationship between P450-dependent metabolism and the effects of capsaicinoids on humans and animals. These studies also provide new insights into the likely metabolic fate of structurally related chemicals including olvanil, arvanil, linvanil, eugenol, capsiates, and gingerols.
Materials and Methods
Chemicals
Capsaicin (97%), nonivamide (N-vanillylnonanamide), NADPH, reduced glutathione (GSH), β-mercaptoethanol (BME), silver oxide, horseradish peroxidase (HRP; EC 1.11.1.7), hydrogen peroxide, H218O, octanoyl chloride, nonanoyl chloride, and CDCl3 were purchased from Sigma-Aldrich (St. Louis, MO). 2-chloronicotinoyl chloride was purchased from Alfa Aesar (Ward Hill, MA). HRP concentration was determined spectrophotometrically using the extinction coefficient ε402=1.02×105 M−1cm−1 and H2O2 using the extinction coefficient ε240 = 39.4 M−1cm−1. N-benzylnonanamide, N-(3-hydroxy-4-methoxybenzyl)nonanamide, N-(3,4-dihydroxybenzyl)nonanamide, N-(3-methoxybenzyl)nonanamide, N-(3,4-dimethoxybenzyl)nonanamide, and N-(4-hydroxybenzyl)nonanamide were synthesized as previously described.(22)
Metabolic Incubations
Pooled human liver microsomes and individual recombinant P450 enzymes were purchased from BD Biosciences (Franklin Lakes, NJ). Incubations (250 µL) in phosphate-buffered saline, pH 7.2 contained ~50 pmol P450 (human liver microsomes) or 25 pmol recombinant P450 enzyme, 2.5 mM NADPH, 2.5 mM GSH (or 1 mM BME when used in place of GSH), and capsaicin or capsaicinoid analogue, typically at 25 or 100 µM. Reactions were performed at 37°C and P450 turnover was initiated by addition of NADPH (2.5 mM). Reactions were terminated by adding 150 µL methanol containing 0.1 ng/µL internal standard; nonivamide was the internal standard for incubations containing capsaicin, and capsaicin was the internal standard for incubations containing nonivamide and the various synthetic analogues. Samples were clarified by centrifugation at 21,000 xg for 5 min, the supernatant was transferred to a clean tube, dried in a vacuum centrifuge, and stored frozen until assayed by LC/MS, as described below. In separate experiments utilizing HRP (25 nM) as the enzymatic catalyst for reactive intermediate formation, samples were prepared in 50 mM sodium acetate, pH 5.5 at room temperature and initiated by addition of 500 µM H2O2. Reactions using HRP were processed as described above. For determination of Km and the relative proportions of conjugate formation (Table 1), samples were prepared using substrate concentrations of 0.5, 1, 2.5, 10, 25, 50, 100, 250, and 500 µM. A 5 min incubation time was used for P450 studies and 1 min for HRP. Conjugate abundances, relative to the internal standard, were plotted versus concentration of substrate and fit with the Michaelis-Menten equation. For determination of relative quantities of conjugates, the concentration response curve was integrated and the area under the curve for each individual conjugate divided by the total AUC for all conjugates. Note: Differences in ionization efficiency could not be accounted for because analytical standards are not available for the GSH conjugates. Thus, data are relative values.
Table 1.
Summary of LC/MS2 properties, kinetics parameters of metabolite formation, and relative proportions of GSH conjugates and 5,5’-dicapsaicin in in vitro incubations using human liver microsomes.
| GSH Conjugate | Time (min) | [M+H]+ and SRM ion (m/z) |
Km (µM)† | % Total‡ |
|---|---|---|---|---|
| G1 | 14.4/15.9 | 597→308 | 20 ± 9 | 0.001 |
| G1* | 8.7 | 595→308 | --- | N.D. |
| G2 | 17.7/19.0 | 597→468 | 40 ± 3 | 71.4 |
| G2* | 11.1 | 595→466 | atypical, ≤1 | 0.7 |
| G3 | 20.1/21.9 | 611→308 | 7 ± 1 | 5.0 |
| G3* | 12.4/13.3 | 609→308 | atypical, ≤1 | 0.3 |
| G4 | 16.8 | 611→464 | 40 ± 15 | 0.4 |
| G4* | 10.5 | 609→462 | --- | N.D. |
| G5 | 19.5 | 611→464 | 24 ± 6 | 0.1 |
| G5* | 11.9 | 609→462 | --- | N.D. |
| G6 | 23.4 | 611→482 | 25 ± 5 | 1.0 |
| G6* | 14.2 | 609→480 | atypical, ≤1 | 0.02 |
| G7 | 14.5/16.0 | 627→308 | 4 ± 2 | .03 |
| G7* | 9.3/10.2 | 625→308 | --- | N.D. |
| G8 | 19.1/21.5 | 627→498 | 18 ± 3 | 3.4 |
| G8* | 11.8/13.1 | 625→496 | atypical, ≤1 | 0.3 |
| G9 | 25.6/26.7 | 627→498 | 10 ± 3 | 17.3 |
| G9* | 15.4/16.2 | 625→496 | atypical, ≤1 | 0.3 |
| Dimer | 46.5 | 609→440 | 260 ± 50 | 0.2 |
Refers to the G* (i.e., alkyl dehydrogenated forms) of the GSH conjugates.
Apparent Km values were generated from concentration metabolite abundance curves, as described in the methods section. --- Indicates that the Km was not calculated. N.D. indicates not detected. The designation “atypical” refers to the fact that formation of conjugate did not exhibit a normal Michaelis-Menten dose-response; that is maximum rates of formation occurred at lower concentrations and at higher concentrations the rates were diminished, presumably due to competition between capsaicin and dehydrogenated capsaicin for the enzyme active site.
The % total value was derived from dividing the area under the concentration-normalized peak area curve for each conjugate by the total AUC for all conjugates.
General LC/MS Methods
Sample residues were re-suspended in 70 µL 70% methanol:30% dH2O, clarified by centrifugation, and transferred to auto-sampler vials for analysis. GSH conjugates were assayed by LC/MS2 using a Thermo LCQ Advantage MAX (Thermo-Fisher, San Jose, CA) ion trap mass spectrometer interfaced with a Surveyor LC system. Samples (15 µL) were injected onto an Inertsil ODS-4 150 X 2.1 mm, 3µ C18 reverse-phase HPLC column (GL Sciences, Inc. Tokyo, Japan). The column was equilibrated at 40°C with a mobile phase comprised of 50% methanol and 50% 0.1% (v/v) formic acid at 0.2 mL/min. GSH conjugates were eluted using the following LC gradient: 50% methanol from 0–20 min.; 50→55% methanol from 20.1–23.5 min.; 55→95% methanol from 23.6–25 m in.; 95% methanol from 25→30 min.; and 50% methanol from 30.1–40 min. For BME conjugates the LC gradient was as follows: 45→60% methanol from 0–30 min.; 60% methanol from 30→45 min.; and 45% methanol from 45→60 min. The mass spectrometer was operated in positive electrospray ionization mode using parameters optimized to detect capsaicin. The sheath gas pressure (N2) was 50 u, the capillary temperature was 275°C and the capillary voltage was 5 kV. A collision offset value of 30% was used for MS2 and either selected-ion monitoring as outlined in Table 1, or full-scan product ion spectra were collected.
GSH and BME Conjugate LC/MS Analysis Parameters
Initial studies of GSH conjugate formation utilized data-dependent neutral-loss scanning for analytes that liberated pyroglutamate (−129 amu) or GSH (m/z 308) during MS2 analysis, as described by Baillie et al.(23) The m/z values for GSH conjugates produced from capsaicin using human liver microsomes were 595, 597, 609, 611, 625, and 627. Subsequent targeted MS2 analysis of GSH conjugates utilized these MH+ ions and collection of full scan (m/z 180→650) data for structural characterization, or selected reaction monitoring (SRM), using the ions indicated in the various figures herein. Analysis of GSH conjugates of nonivamide and other capsaicinoid analogues were adjusted to account for differences in the mass of the substrate (e.g., −12 amu for nonivamide vs. capsaicin). Conjugates of capsaicin with BME were assayed using the following MH+ values, based on the results for GSH conjugate formation: m/z 366, 368, 380, 382, 396, and 398. Full-scan MS2 data (m/z 100→400) were also collected for conjugate identification.
Interpretation of MSn Spectra
Interpretation of MS2 and MS3 spectra obtained for the GSH and BME conjugates of capsaicin, for purposes of structural elucidation, was facilitated by Mass Frontier 4.0 Software (Thermo, San Jose, CA). All software predictions were performed using default parameters.
18O Incorporation Studies
Differentiation of hydration of quinone methide intermediates versus P450-catalyzed hydroxyl rebound mechanisms for the formation of hydroxylated GSH conjugates was achieved by measuring 16O versus 18O incorporation into the conjugates from water. Fractional incorporation of 18O was calculated using Brauman’s least-squares method and 2-chloronicotinoyl chloride as a positive control, as previously described.(24) Incubations were performed in 50% v/v H218O.
Synthesis and NMR Analysis of Conjugate B6 (G6)
Capsaicin (2 mg) was dissolved in 2 mL dry acetonitrile and heated to 60°C. Silver oxide powder (150 mg) was added to the solution and the reaction was constantly mixed at 60°C for 7.5 minutes. The mixture was passed through a paper filter to remove the silver oxide and collected in a tube containing 200 µL BME. The solution was mixed, dried under N2, reconstituted in 200 µL 70% methanol:water, and the resulting BME B6-conjugate enriched by HPLC using the conditions described above for capsaicin BME conjugate analysis. [1H]-NMR of B6 was obtained at 600 MHz on a Varian INOVA equipped with a cryogenically cooled [1H] channel in 600 µL of CDCl3 (99.96% D).
Preparation and NMR Analysis of 5,5’-Dicapsaicin
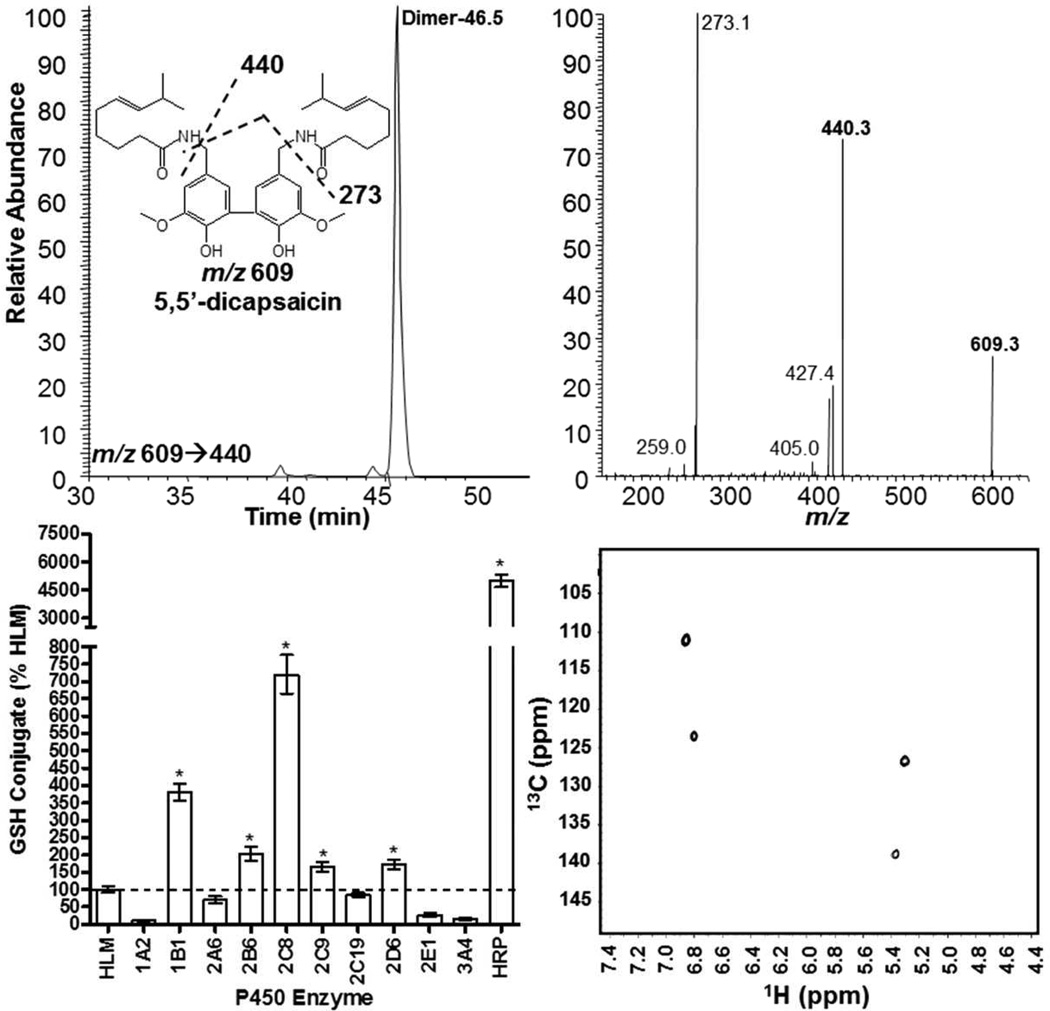
Capsaicin (500 µM) was incubated with HRP (500 nM) and H2O2 (1 mM) in 250 µL 50 mM sodium acetate, pH 5.5 for 10 minutes at room temperature. Reaction products from a 10 mL reaction were extracted into 10 mL n-butyl chloride two times, the organic extracts pooled, and dried under N2. The dried residue containing the dimer product was reconstituted in 200 µL 70% methanol:water, and enriched by HPLC by collecting the peak eluting at ~46.5 min (Figure 6) using the HPLC conditions described above for GSH conjugate analysis. [1H] and 13[C] NMR were obtained at 600 MHz on a Varian INOVA equipped with a cryogenically cooled [1H] channel in 100 µL of CDCl3 (99.96% D) in a 5 mm Shigemi NMR tube matched to CDCl3. [1H]-NMR data are shown in Supplemental Figure 1. Gradient heteronuclear single quantum coherence (gHSQC) NMR spectra were also acquired to ascertain the structure of the dimer product. Likewise, esterification of the dimer using nonanoyl chloride was used to confirm the dimer structure. This was performed by reacting excess nonanoyl chloride with the enriched dimer product in 100 µL 0.1 N NaOH (30 min at RT) followed by extraction of the product into n-butyl chloride, drying under N2, and subsequent MS2 and MS3 analysis of the doubly esterified product (Supplemental Figure 2).
Figure 6.
Chromatographic, spectral, and enzyme-specific production of 5,5’-dicapsaicin by human P450 enzymes and horseradish peroxidase. The top panel shows an extracted ion LC/MS2 chromatogram and the corresponding MS2 spectrum with illustrated fragmentation pathways for 5,5’-dicapsaicin (m/z 609). The bottom left panel shows quantitative results for the formation of 5,5’-dicapsaicin by individual human P450 enzymes and horseradish peroxidase. The bottom right panel shows the gHSQC spectrum used to confirm 5,5’-dicapsaicin. *Represents dimer production in excess of the quantity produced by human liver microsomes using ANOVA and p<0.05. Reactions were prepared as described under the methods section.
Synthesis of N-(Trimethoxybenzyl)nonanamide Analogues
For elucidation of the sites of aromatic hydroxylation for capsaicinoids (i.e., M5 and M7) and the corresponding GSH conjugates (G7, G8, and G9), N-(3,4,5-trimethoxybenzyl)nonanamide, N-(2,3,4-trimethoxybenzyl)nonanamide, and N-(2,4,5-trimethoxybenzyl)nonanamide were synthesized. 3,4,5-trimethoxybenzylamine (Sigma), 2,3,4-trimethoxybenzylamine (Matrix Scientific, Columbia, SC), and 2,4,5-trimethoxybenzylamine (Matrix Scientific) were reacted with an equimolar quantity of nonanoyl chloride and purified as previously described for the other nonivamide analogues (22). [1H]-NMR spectra of the products are shown in Supplemental Figures 3–5.
Analysis of GSH Conjugates in Mouse Liver
TRPV1−/− mice (~10 weeks; n=3) were treated i.p. with 10.0 mg/kg capsaicin (in 50% ethanol). Mice were sacrificed 30 min after treatment by lethal injection (100 mg/kg pentobarbital) and their livers were excised. The livers were washed, homogenized in ~5 volumes of 50% methanol:water, centrifuged at 2,500 xg for 10 min and again at 100,000 xg for 60 minutes. The supernatant was then lyophilized. The residue was reconstituted with 50% methanol:water and again clarified by centrifugation at 20,000 xg for 5 minutes prior to analysis for GSH conjugates using the following precursor-to-product ion transitions: m/z 597→468, 611→308, 611→482, and 627→498.
Analysis of N-Acetylcysteine Conjugates in Mouse Urine
Female CD-1 mice were treated p.o. or i.v. with 2.5 mg/kg capsaicin and urine was collected at 24 (n=5), 48 (n=2), and 96 h (n=2) and pooled for each time point. Urine (1.5 mL) was fortified with internal standard (nonivamide) at 100 ng/mL, diluted 2× in phosphate-buffered saline, pH 7.2, clarified by centrifugation, and loaded onto a 3 mL Hypersep C18 solid-phase extraction column (Thermo Scientific, Bellefonte, PA). The columns were washed with dH2O and the analytes eluted using methanol. The extracts were dried under N2 and reconstituted in mobile phase for LC/MS analysis. Conjugates previously identified through in vitro P450 incubations using N-acetylcysteine as the nucleophilic trapping agent were assayed using a GL Sciences C18 HPLC column (150×2.1 mm) using a mobile phase of 57.5% methanol:42.5% aqueous formic acid at 0.2 mL/min at 40°C (Supplemental Figure 6). The following precursor-to-product ion transitions were monitored using positive electrospray ionization: m/z 453→284, 467→298+304, 482→314, 306→137, and 294→137 for conjugates of O-demethylated, unmodified, and hydroxylated capsaicin, free capsaicin, and nonivamide.
Results
GSH Conjugates
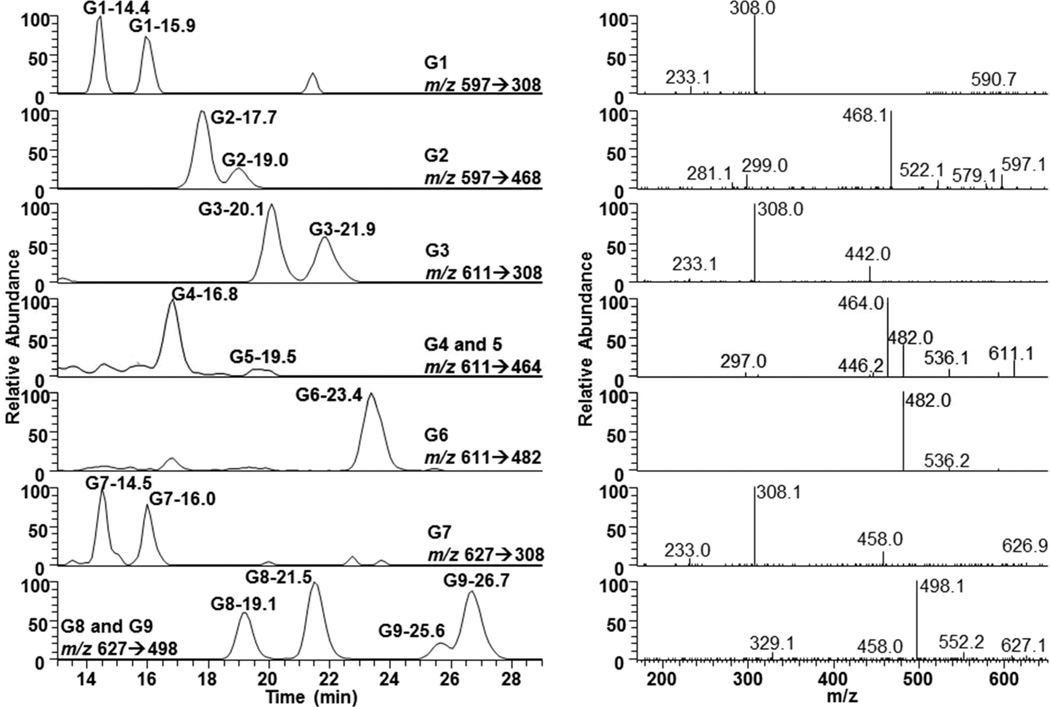
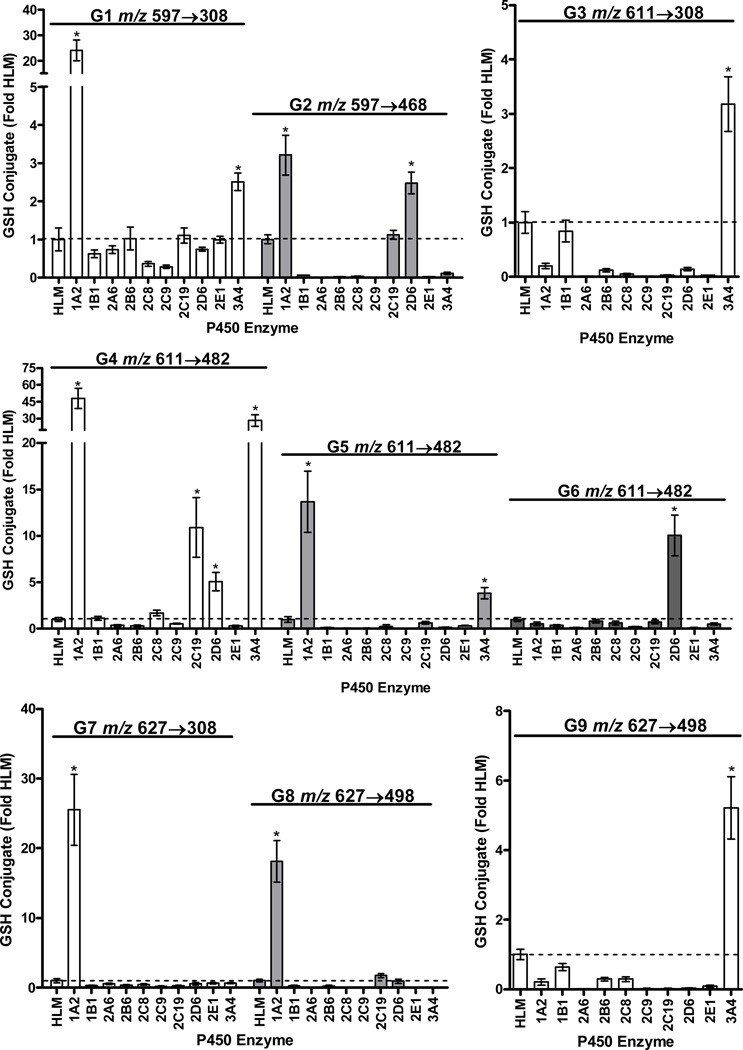
Representative chromatograms and the corresponding MS2 spectra of nine GSH conjugates produced from capsaicin by human liver P450 enzymes are shown in Figure 1. The structures of specific diagnostic fragment ions for the various GSH conjugates are shown in Figure 2 and Supplemental Figures 7–9. Additional characteristics are summarized in Table 1. Data comparing the production of the various GSH conjugates by individual recombinant P450 enzymes are in Figure 3 (25 µM substrate concentration) and Supplemental Figure 10 (100 µM substrate concentration). Chromatograms and spectra for minor “G*” conjugates (Table 1), derived from trapping of identical electrophiles of an alkyl dehydrogenated metabolite of capsaicin (i.e., M1 or M4) (20,21), are shown in Supplemental Figure 11.
Figure 1.
LC/MS2 chromatograms and spectra for the major GSH conjugates of capsaicin. Reactions were performed as outlined in the methods section using human liver microsomes and a concentration of 100 µM capsaicin. The identities of the conjugates and the specific MS2 criteria used for the analysis of each conjugate are provided for each chromatogram. All chromatograms are full scale for the precursor-to-product ion transition listed in the figure.
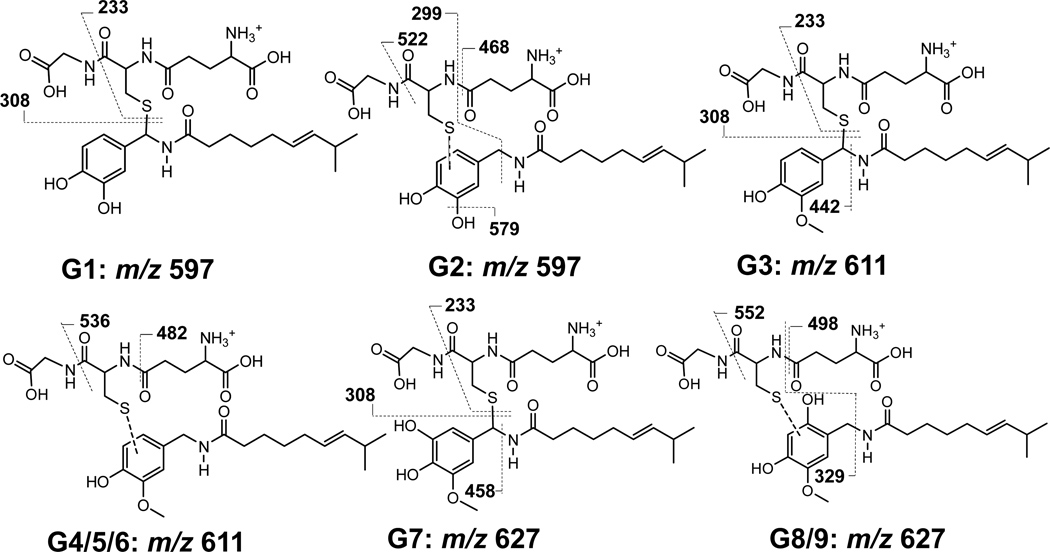
Figure 2.
Illustration of the diagnostic precursor-to-product ions and structures of the major GSH conjugates of capsaicin. Additional fragmentation ion data are provided in Supplemental Figures 7–9. Similar data for the corresponding BME conjugates of capsaicin are also provided in Supplemental Figures 12–14.
Figure 3.
Production of the various GSH conjugates of capsaicin by individual recombinant P450 enzymes. Reactions were performed as outlined in the methods section with a concentration of 25 µM capsaicin. Data using a 100 µM concentration and additional P450 enzymes are presented as Supplemental Figure 10. The identities of the conjugates and the specific MS2 analysis parameters used for metabolite detection are provided in the figures above the corresponding results from the enzyme screen. All data are relative to the amount of conjugate observed using human liver microsomes (n=3). *Represents a statistical difference in the amount of conjugate produced by a specific P450 enzyme relative to human liver microsomes using ANOVA and p<0.05.
BME Conjugates
In parallel studies, BME was used as the nucleophile to facilitate metabolite identification from MS2 spectra that were not dominated by fragmentation of GSH, as shown in Figure 1. Chromatograms and spectra of the BME conjugates B1-9, corresponding to G1-G9, are presented in Supplemental Figure 12 and the structures of diagnostic fragment ions for B1-9 are shown in Supplemental Figures 13 and 14.
G1 and G2
MS2 analysis of the minor G1 conjugate pair (m/z 597→308 at 14.4/15.9 min) and the major G2 conjugate pair m/z 597→468 at 17.7 and 19.0 min in Figure 1 indicated trapping of O-demethylated capsaicin (291 g/mol). The major fragment ion for G1 was m/z 308, representing GSH liberated from the conjugate (Figure 2, Supplemental Figure 7, top panel). This fragmentation pattern is characteristic of allylic GSH conjugates,(23) indicating that G1 was diasteriomers with GSH attached to the C7 benzylic carbon of O-demethylated capsaicin (M6). The proposed mechanism of G1 formation is presented in Figure 7.
The diagnostic fragment ions for G2 were m/z 597, m/z 522, and m/z 468 (Figures 1 and 2). These fragmentation properties are characteristic of loss of glycine (−75 amu) and pyroglutamate (−129) from GSH, and occur when GSH is attached to an aromatic carbon.(23) Thus, G2 formed from addition of GSH to the aromatic vanilloid ring of O-demethylated capsaicin (M6) at either the C2, C5, or C6 carbons (Figure 7). Consistent with the proposed mechanisms of formation and structural assignments in Figure 7, formation of equivalent conjugates from capsaicinoid analogues lacking the 4-OH vanilloid ring constituent (i.e., N-benzylnonanamide, N-(3-methoxybenzyl)nonanamide, and N-(3,4-dimethoxybenzyl)nonanamide) was diminished; was unchanged for analogues with either the exact or similar vanilloid ring structure (i.e., nonivamide and N-(3-hydroxy-4-methoxybenzyl)nonanamide); or was increased for N-(3,4-dihydroxybenzyl)nonanmide and N-(4-hydroxybenzyl)nonanamide (Supplemental Figure 15).
Characteristics of G1 and G2 Production
O-demethylation of capsaicin was previously shown to be catalyzed by CYP1A2 and 2C19 and to a lesser extent by CYP3A4 and 2D6.(20) Here, G1 and G2 were produced by CYP1A2, 2C19, 3A4, and 2D6 (Figure 3 and Supplemental Figure 10). Trapping of O-demethylated capsaicin that was dehydrogenated on the terminal alkyl position (i.e., G1* and G2*) was also observed (Supplemental Figure 11). G1* (8.7 min.) and G2* (11.1 min.) had molecular ions of m/z 595, consistent with trapping of an electrophile with a mass of 290 g/mol. No change in the m/z 308 fragment was observed for G1*, but the presence of m/z 428 for G2* indicated neutral loss of a dehydrogenated alkyl side chain (−167), versus −169 for capsaicin (20). G2 was the most abundant GSH conjugate (~71% total) and the calculated Km for formation was ~40 µM (Table 1); G1, G1*, and G2* were all <1% of the total GSH conjugates.
B1 and B2
B1 (corresponding to G1) and B2 (corresponding to G2) conjugates with an m/z value of 368 were observed at 7.6/10.3 min and 28.1/30.4 min (Supplemental Figure 12). The m/z 368 ion of the B1 and B2 conjugates was consistent with the addition of BME (+76 amu) to O-demethylated capsaicin (m/z 292). B1 uniquely fragmented to m/z 322 which was attributed to the loss of C2H6O (−46 amu) from BME attached to the C7 carbon, with additional loss of H2O to form m/z 304 (Supplemental Figure 13, top panel). Additionally, m/z 169 in the spectrum for B1 was consistent with an O-demethylated ring structure with -SCH2 attached to the C7 benzylic carbon. The MS2 spectrum of B1 also lacked diagnostic fragment ions corresponding to the alkyl side chain of capsaicin at m/z 170 and 153.(20)
B2 fragmented to m/z 199, 196, and 181, consistent with BME attached to the aromatic O-demethylated vanilloid ring minus the alkyl side chain of capsaicin (Supplemental Figures 12 and 13). Consistent with this structural assignment, scission of the amide bond also yielded m/z 170, 153, and 135 which correspond to the unmodified alkyl fragments of capsaicin, as previously reported.(20) Using nonivamide as the substrate, the fragment ion pattern for B1 was unchanged, despite a −12 amu shift in the molecular ion m/z 368. However, for B2 of nonivamide, the alkyl fragment ions m/z 170, 153, and 135 observed with capsaicin were appropriately shifted by −12 amu, further confirming the structures shown for B1/G1 and B2/G2 in Figure 7.
Identification of G3/G3* and Characteristics of Formation
The MS2 spectra of the two major analyte peaks for G3 in Figure 1, m/z 611→308 at 20.1/21.9 min, were indistinguishable. The MH+ ion of m/z 611 was consistent with GSH addition to unmodified capsaicin (305 g/mol). Like G1, the spectrum of G3 indicated liberation of GSH (m/z 308). Additional diagnostic fragment ions for G3 included m/z 442 attributable to GSH attached to the unmodified vanilloid ring as a result of neutral loss of 169 amu (i.e., the alkyl side chain of capsaicin) (Figures 1 and 2, Supplemental Figure 7, bottom panel). From these data, it was concluded that the G3 was diasteriomers derived from addition of GSH to the C7 benzylic carbon of a quinone methide intermediate (Figure 7). Similar to G1 and 2, a minor G3* conjugate arising from either bioactivation of the vanilloid ring of M1 or M4 was observed (Supplemental Figure 11). G3* liberated m/z 308, but had an MH+ ion of m/z 609, consistent with alkyl dehydrogenation of capsaicin prior to vanilloid ring metabolism to an electrophilic quinone methide intermediate. G3 was produced mainly by CYP3A4 and 1B1 (Figure 3 and Supplemental Figure 10) and was roughly the third most abundant GSH conjugate detected (5%), with a Km of ~40 µM (Table 1). Formation of G3 from nonivamide was comparable to capsaicin (Supplemental Figure 16), but absent for all analogues lacking a 4-OH moiety (i.e., N-benzylnonanamide, N-(3-hydroxy-4-methoxybenzyl)nonanamide, N-(3,4-dihydroxybenzyl)nonanamide, N-(3-methoxybenzyl)nonanamide, and N-(3,4-dimethoxybenzyl)nonanamide). Conversely, G3 formation from N-(4-hydroxybenzyl)nonanamide was greater, consistent with a quinone methide intermediate being the source of G3. The apparent lack of G3 formation from N-(3,4-dihydroxybenzyl)nonanamide, but extensive formation of G2 and G6 (Supplemental Data Figures 15 and 16) suggests that this analogue readily forms a 3,4-quinone intermediate rather than a quinone methide.
Analysis of B3
A BME conjugate pair, B3, corresponding to G3, (m/z 382→213) eluted at 24.5/26.4 min, was observed (Supplemental Figure 12). B3 fragmented to m/z 213 corresponding to the vanilloid ring+BME (i.e., m/z 137+76 amu; Supplemental Figure 13, middle panel), m/z 195 corresponding to m/z 213-H2O, and the characteristic alkyl chain ions of capsaicin at m/z 170 and 153,(20) consistent with the proposed structural assignment and mechanism of formation for G3 (Figure 7).
Identification of G4-6 and Characteristics of Formation
G4 and 5 (16.8 and 19.5 min; m/z 611→464) and G6 (m/z 611→482; 23.4 min) in Figure 1, were spectrally similar. Unlike G3, the MS2 spectra for these relatively minor conjugates were characterized by neutral loss of glycine (m/z 536), pyroglutamate (m/z 482), and pyroglutamate plus H2O (m/z 464), consistent with GSH being attached to the aromatic vanilloid ring of an intermediate quinine methide (Figure 7). CYP1A2, 3A4, 2C19, and 2D6 produced G4-6, with 2D6 being the primary catalyst for G6 (Figure 3 and Supplemental Figure 10). Corresponding G4-6* conjugates were also observed (Supplemental Figure 11), and like the other G* conjugates, only the fragment ions containing the dehydrogenated alkyl chain were decreased −2 amu. G4 and G5 formation was minimal for all of the capsaicinoid variants except nonivamide, which was roughly equivalent to capsaicin. However, G6 formation was increased for N-(3,4-dihydroxybenxyl)nonanamide (Supplemental Figure 16), suggesting that the origin of G6/B6 may be an intermediate 3,4-quinone with GSH/BME attached to the aromatic ring at either the C2, C5, or C6 positions.
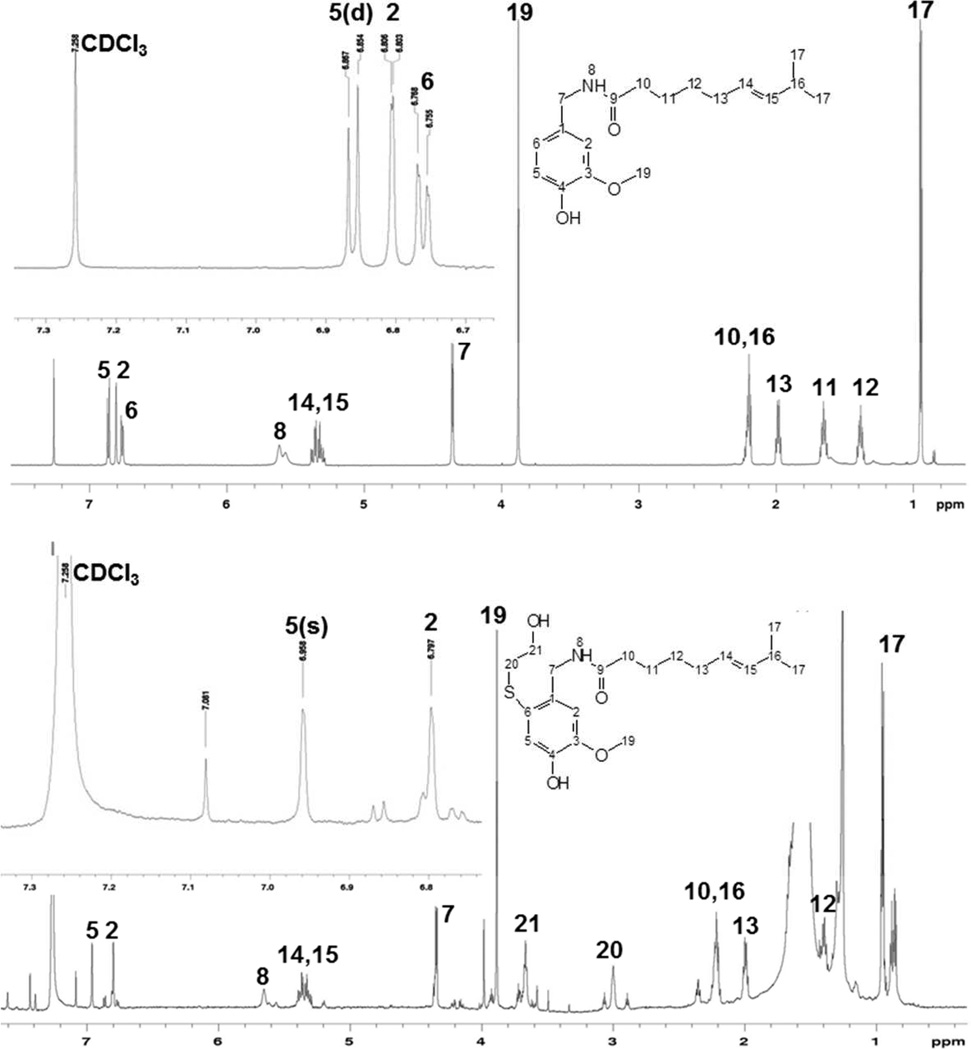
NMR Analysis of B6 (G6)
Because the identity of B6 (G6) was not fully elucidated through MSn analysis, and it was a major conjugate, the structure was pursued using NMR. The [1H]-NMR spectrum of synthetic B6 (Figure 4) was characterized by a loss of the doublet at 6.76 ppm attributable to the proton on C6, a shift in the resonance for the C5 proton from 6.86 to 6.96 ppm, loss of coupling (i.e., conversion to a singlet) between the C5 and C6 protons at 6.86 ppm for the proton on C5, loss of long-range coupling between the C2 and C6 protons, and the appearance of two new resonances at ~3.0 and 3.65 ppm, attributable to the two methylene carbons of BME. Thus, the identity of the B6 (G6) was concluded to be capsaicin with the thiol trapping agent attached to the C6 carbon, presumably derived from a quinone methide intermediate.
Figure 4.
600 MHz [1H]-NMR spectrum for capsaicin and B6, the BME equivalent of G6. Top spectrum is for capsaicin and the bottom spectrum is for B6. Insets provide an expansion of the aromatic proton resonances for capsaicin and B6. Data were collected as outlined in the methods section.
Identification of G7-G9
MS2 analysis of the G7 conjugate pair (m/z 627→308 at 14.5/16 min.), the G8 conjugate pair (m/z 627→498 at 19.1/21.5), and the G9 conjugates (m/z 627→498 at 25.6/26.7 min) (Figures 1 and 2) indicated trapping of an electrophile derived from hydroxylated capsaicin (321 g/mol) (i.e., a catechol), presumably M5, M7, or M8.(20) The diagnostic fragment ions for G7 were hydroxylated capsaicin plus GSH (m/z 627) and liberation of GSH (m/z 308) and liberation of the unmodified alkyl chain (m/z 458). G7 was characterized by essentially symmetrical peaks, consistent with diasteriomers arising from attachment of GSH to the C7 benzylic carbon of an intermediate quinone methide produced after hydroxylation of the vanilloid ring (Figure 7). Conversely, G8 and G9 exhibited neutral loss of glycine (−75) and pyroglutamate (−129), to yield m/z 552 and 498, respectively, consistent with GSH addition to the aromatic ring and two different carbon atoms.
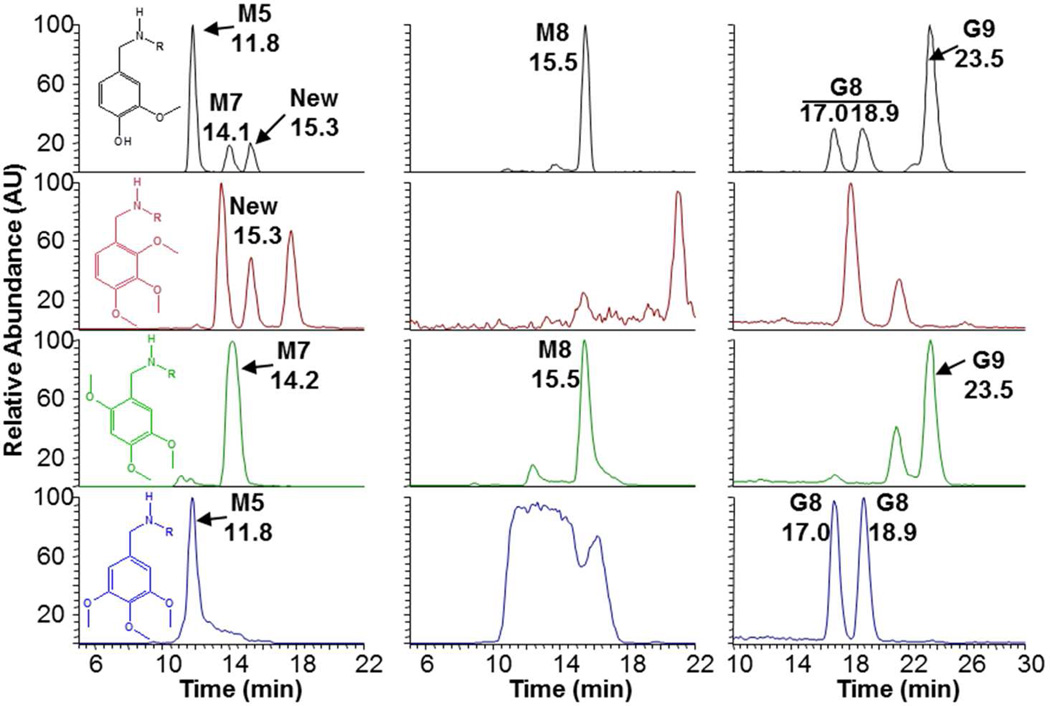
Aromatic hydroxylation of capsaicin was previously shown to be catalyzed by CYP1A2 and 2C19 (M5), and 2B6, 2C8, and 2E1 (M7), while M8 production occured with multiple enzymes including CYP3A4, 1A1, 2E1, 2C8, 2D6, and 2B6.(20) G7 and G8 were produced mainly by CYP1A2, suggesting these conjugates were derived from M5. Metabolism of N-(3,4,5-trimethoxy)nonanamide by human liver microsomes yielded an aromatic hydroxylated metabolite exhibiting an identical precursor-to-product ion transition (m/z 310→153; blue chromatograms) and HPLC retention time as nonivamide (black chromatograms), but neither N-(2,3,4-trimethoxy)nonanamide (red chromatograms) nor N-(2,4,5-trimethoxy)nonanamide (green chromatograms) produced this metabolite (Figure 5, bottom left panel). From these data it was concluded that M5 of capsaicin and nonivamide was the result of hydroxylation at the C5 position. Metabolism of N-(3,4,5-trimethoxy)nonanamide in the presence of GSH also produced conjugates with identical LC/MS characteristics to G7 (not shown) and G8 (Figure 5, bottom right panel, blue chromatogram), indicating that G7 and G8 are presumably formed by an intermediate quinone methide produced from 5-OH-capsaicin/nonivamide with addition of GSH at either the C7 benzylic position to yield diasteriomers (G7), or at the C3 or C6 positions to yield G8 (Figure 7). Using N-(2,3,4-trimethoxy)nonanamide as a substrate, neither M5, M7, M8, G7, G8, nor G9 were observed (Figure 5, red chromatogram, left side). Rather, a peak corresponding to a previously unidentified aromatic hydroxylated metabolite eluting at 15.3 minutes was observed, consistent with 3-OH-capsaicin/nonivamide. However, none of the GSH conjugates from this substrate corresponded with those of capsaicin or nonivamide. Finally, using N-(2,4,5-trimethoxy)nonanamide as a substrate, it was demonstrated that M7 was 6-OH-capsaicin (Figure 6, green chromatogram, left side). Additionally, this substrate uniquely yielded M8 (Figure 5 green chromatogram, center panel), indicating that M8 involved C6 oxygenation, contrary to the structure previously proposed.(20) N-(2,4,5-trimethoxy)nonanamide also uniquely produced G9 (Figure 5, green chromatogram, right side), demonstrating that G9 was formed from M7, presumably via dehydrogenation of M7 at the C7-NH bond and tautamerization to an electrophile (Figure 7), based on the relative production of M8 and G9 by CYP3A4 and 1A1, which are the dominant enzymes for catalyzing M8 formation and N-dehydrogenation of capsaicinoids.(20) Assays using all the other analogues (Supplemental Figures 17 and 18) were generally uninformative.
Figure 5.
Formation of aromatic alcohols M5 and M7, M8 and GSH conjugates G8 and 9 from N-(trimethoxybenzyl)nonanamide analogues. (Left) Formation of aromatic alcohols M5 and M7 (m/z 310→153), (center) M8 (m/z 308→168), and (right) G8 and G9 (m/z 615→486) from nonivamide (black), N-(2,3,4-trimethoxybenzyl)nonanamide (red), N-(2,4,5-trimethoxybenzyl)nonanamide (green), or N-(3,4,5-trimethoxybenzyl)nonanamide (blue) in incubations with human liver microsomes plus or minus GSH.
Characteristics of G7-9 Formation
For G7-9, hydroxylation occurred via direct P450-catalyzed oxygenation since incorporation of 18O from H2 18O was not observed (not shown). G* conjugates of G7-9 were also observed (Supplemental Figure 11). The MH+ ion for G7*(9.3 and 10.2 min.), G8* (11.8 and 13.1 min.), and G9* (15.4 and 16.2 min) was m/z 595, with no change in the diagnostic fragment ions of G7* relative to G7 (i.e., m/z 308), and −2amu shifts in the fragment ions for G8* and G9* relative to G8 and G9, consistent with alkyl dehydrogenation. G8 (~3%) and G9 (~17%) were the 4th and 2nd most abundant GSH conjugates produced (Table 1), while G7 was minor. The Km values for G7, G8, and G9 were 4, 18, and 10 µM, respectively (Table 1).
Analysis of B7-B9
B7 (corresponding to G7), B8 (corresponding to G8), and B9 (corresponding to G9) conjugates were observed with m/z values of 398 at 23.4/25.4 min., 30.2/34.1 min., and 37.5 min. (Supplemental Figure 12). The MH+ ion of 398 was consistent with the addition of BME (76 amu) to hydroxylated capsaicin (m/z 322; presumably M5). B7 fragmentation to a unique m/z 246 ion was predicted to arise from the addition of BME to the vanilloid ring at the C7 position (Supplemental Figure 14, top panel). B7 was also produced by CYP1A2, similar to G7 (not shown). B8 and B9 fragmented to m/z 229 and 211, also consistent with retention of BME on the hydroxylated vanilloid ring, where m/z 211 was consistent with the loss of H2O from m/z 229, and m/z 170 and 153 corresponded to the unmodified alkyl fragments derived from capsaicin.(20)
Identification of a Capsaicin Dimer
Two capsaicin dimers are known to be produced by peroxidases(25,26) by coupling of phenoxyl and/or carbon-centered radical intermediates.(27) Figure 6 illustrates that a capsaicin dimer was also produced by human liver microsomes and several individual P450 enzymes. The capsaicin dimer eluted at ~46.5 min and exhibited an m/z value of 609 with major fragment ions at m/z 440 and 273 (Figure 6). These ions corresponded to the loss of 1 and 2 alkyl chains (−169) from the dimer. The structure of the dimer was confirmed as 5,5’-dicapsaicin by 1H and 2D-gHSQC NMR analysis of the HPLC purified product. The 1H-NMR spectrum demonstrated two aromatic proton resonances at 6.8–6.9 ppm corresponding to the protons at C2 and C6 positions that lacked ortho-coupling (Supplemental Figure 1), consistent with a symmetrical dimer formed between C5 of two capsaicin molecules. Additionally, the 2D gHSQC spectrum showed only two correlations, further indicating a symmetrical dimer (Figure 6). Finally, acetylation of the dimer using octanoyl chloride confirmed the existence of two free hydroxyl groups on the dimer ring structure. The major product of the reaction was m/z 889 which fragmented to yield m/z 749, the monoacetylated ion, which further fragmented during MS3 to yield m/z 609, 440, and 273 (i.e., the original dimmer product) (Supplemental Figure 2). 5,5’-Dicapsaicin was produced primarily by CYP2D6, 2C9, and 2C8, but substantially less than that of HRP. Dimer formation was not affected by addition of GSH, indicating low reactivity of the precursor intermediates. For human liver microsomes, the Km for dimer formation was ~260 µM (Table 1). The dimer was a minor metabolite representing ~0.3% of the total conjugates formed (Table 1).
Assessment of GSH Conjugate Formation In Vivo
Similar to the in vitro studies, the O-demethylated conjugate G2 was the most abundant (~47% total) conjugate detected in livers of capsaicin-treated mice (Table 3). The quinone methide-derived G3 and G6 conjugates were also detected in liver extracts, representing 24% and 23% of the total, respectively. G8 was detected at ~6% of the total, but no other conjugates or the dimer were observed. Similarly, in mouse urine following either oral (2.5 mg/kg) or i.v. (2.5 mg/kg) capsaicin treatment, N-acetycysteine conjugates corresponding to O-demethylated capsaicin (G2), the quinone methide derived C7 conjugate (G3), the aromatic quinone methide conjugate (G6), and 5-OH-capsaicin (G8) were detected (Table 2).
Table 2.
Relative abundance of capsaicin, GSH conjugates, 5,5’-dicapsaicin, and N-acetylcysteine conjugates in mouse liver and urine.
|
Analyte |
% Total in Liver Extract* |
% Total in Mouse Urine Oral (2.5 mg/kg) / I.V. (2.5 mg/kg) doses† |
|||||
|---|---|---|---|---|---|---|---|
| NAC-Conjugate | Free Capsaicin | ||||||
| 24h | 48h | 96h | 24h | 48h | 96h | ||
| Capsaicin | ---- | ---- | ---- | ---- | 74/49 | 8/31 | 4/16 |
| G1 | N.D. | N.D. | N.D. | N.D. | ---- | ---- | ---- |
| G2 | 47 | 7/1 | 1/0.3 | 0.2/N.D. | ---- | ---- | ---- |
| G3 | 23 | 1/0.4 | 0.6/0.5 | 0.2/N.D. | ---- | ---- | ---- |
| G4 | N.D. | N.D. | N.D. | N.D. | ---- | ---- | ---- |
| G5 | N.D. | N.D. | N.D. | N.D. | ---- | ---- | ---- |
| G6 | 24 | 2/0.6 | 2/0.2 | 0.6/N.D. | ---- | ---- | ---- |
| G7 | N.D. | N.D. | N.D. | N.D. | ---- | ---- | ---- |
| G8 | 6 | 0.2/0.3 | N.D. | N.D. | ---- | ---- | ---- |
| G9 | N.D. | N.D. | N.D. | N.D. | ---- | ---- | ---- |
| Dimer | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
Values are percentages of total GSH conjugates detected in liver 30 min after i.p. injection with 10 mg/kg capsaicin. N.D. implies that the conjugate was not detected.
Values are the percentages of N-acetylcysteine conjugate and capsaicin in urine, determined using the sum of peak area ratios as the denominator and individual peak ratios as the numerator. Five livers were pooled and processed in parallel for control and treated mice. Pooled urine from 24h (n=5), 48h (n=2), and 96h (n=2) oral (2.5 mg/kg) or i.v. (2.5 mg/kg) treatments was used.
Discussion
Understanding the metabolism of capsaicinoids by P450 enzymes is important because humans are widely exposed to capsaicinoids and metabolism is a major determinant of their pharmacological and toxicological properties. Previous studies on capsaicinoid metabolism have suggested the formation of quinone, phenoxyl radical, and epoxide intermediates.(27,28) Additionally, P450 metabolism of capsaicin caused genotoxicity, (27,29) covalent binding of 3[H]-dihydrocapsaicin to hepatic microsomal proteins in vivo, and inhibition of P450 activity; including ethylmorphine deethylation by rat liver microsomes, prolongation of pentobarbital-induced sleep time,(28) inhibition of NNK bioactivation in vitro,(6) and inhibition of CYP2E1 in vitro by dihydrocapsaicin.(30) However, the origins and characteristics of the implied reactive metabolites were not determined.
This study provides new information on the formation of P450-generated reactive metabolites of capsaicin. Specifically, GSH and BME conjugates derived from quinone methide and quinone electrophiles of O-demethylated (M6), C5 (M5) and C6 (M7) aromatic hydroxylated and/or oxygenated imide (M8) metabolites of capsaicin, capsaicin itself, as well as the corresponding alkyl dehydrogenated forms of capsaicin (M1 and M4), were characterized. Although structurally similar alkyl phenols are known to form quinone methide intermediates as a result of P450 metabolism,(31–33) this metabolic process had not previously been demonstrated for capsaicinoids. The formation of a phenoxyl radical intermediate from capsaicin during P450 metabolism has also been proposed, but not shown. Studies of peroxidase-mediated capsaicinoid metabolism identified 5,5’-dicapsaicin and 4’-O-5-dicapsaicin ether metabolites.(25,26) Here, formation of the 5,5’-dicapsaicin dimer by several P450 enzymes and HRP was demonstrated indicating that P450 enzymes are also capable of oxidizing capsaicin to free radical intermediates. Finally, the formation of metabolites via sequential metabolism of capsaicin by P450 enzymes to produce alkyl dehydrogenated and vanilloid ring-modified products (i.e., the G* adducts) was novel. The identification of the G* conjugates, while minor, was interesting as it suggests that, while alkyl dehydrogenation of capsaicin (i.e., M1 and/or M4) diminishes activation of the capsaicin receptor (i.e., TRPV1) and TRPV1-mediated cytotoxicity,(20,34) these metabolic processes do not inhibit bioactivation of capsaicinoids to potentially toxic electrophiles. Similar conclusions can be made regarding O-demethylation (M6→G1 and G2) and aromatic hydroxylation (M5→G7 and G8 and M7/M8→G9), which either diminish or would be predicted from structure-activity studies to lessen activation of TRPV1,(20,22) but readily form electrophiles.
The majority of GSH and BME conjugates were produced by CYP1A2, 2C19, 2D6, and 3A4 at concentrations likely to occur at sites of exposure and/or absorption and metabolism (e.g., skin, intestine, liver) (Figure 3, Supplemental Figure 10). While there have been reports of capsaicinoid-induced inhibition of CYP enzyme function, most notably CYP2E1,(28,30,34) the formation of electrophilic intermediates by other CYPs is intriguing since a similar loss of function of CYP1A2, 2C19, 3A4 or 2D6 could have significant consequences that include a loss of, or change in the metabolic clearance mechanisms of capsaicinoids and other xenobiotics, potentially leading to unanticipated and undesirable effects. Accordingly, several recent studies indicated that capsaicinoids broadly inhibit CYP enzymes, including CYP1A2, 2C9, 2C19, and 3A4/5, via a competitive mechanism and that CYP3A4, 2B6, and 2D6 are potentially inhibited through a mechanism-based process similar to CYP2E1.(35,36) Finally, differential formation of G3 versus G6 by CYP3A4 and 2D6, respectively, was observed (Figure 3). Both conjugates appear to originate from a common quinone methide intermediate. However, CYP2D6 produced almost exclusively the C6 conjugate, G6, versus the more thermodynamically favorable C7 conjugate G3, as observed with CYP3A4 and other CYP enzymes (Figure 3). These data imply that specific enzyme:substrate interactions play vital roles in the initial bioactivation of the substrates and in determining how GSH interacts with electrophiles during the catalytic process.
The biological significance of reactive intermediate formation from capsaicinoids has not been thoroughly investigated. However, several studies have demonstrated that capsaicinoids may be chemopreventive.(6–10,37) Capsaicinoids reduce lung tumor formation and growth of transformed foci in animals treated with a mixture of the model carcinogens diethyl-, dibutylnitrosamine and dimethylnitrosourea.(38) Efficacy was also shown in models of prostate and pancreatic cancer(9–12,39,40) as well as in azoxymethane-induced gastric(41), colon(42) and 4-nitroquinoline-1-oxide-induced tongue(43) carcinogenesis models. Paradoxically, capsaicinoids were also reported to act as co-carcinogens in other organs in some of the same models,(38) and increased rates of liver and stomach cancer have been reported by epidemiological studies of populations that routinely consume high quantities of peppers.(37) These results are consistent with previous reports of genotoxicity for capsaicinoids following S9 bioactivation.(27,29)
Explanations for differences in promotion and/or prevention of cancer cell growth in different tissue and cell types are lacking. One hypothesis is that capsaicinoids attenuate procarcinogen bioactivation since capsaicinoids appear to inhibit P450 enzymes (including CYP2E1) involved in ethylmorphine deeythlation (CYP2D6 and 3A4),(28) pentobarbital clearance (CYP2B6)(28) NNK bioactivation (CYP2A6 and 2A13),(6–8) and benzo[a]pyrene bioactivation (CYP3A4).(44–46) Alternate hypotheses suggest that capsaicinoids are potent antioxidants, that capsaicinoids preferentially cause cell death in cancer cells that express TRPV1,(40,47) or selectively inhibit tumor NADH oxidase (tNOX or PMOR).(9,10) Unfortunately, all of these hypotheses do not fully address the incongruent effects of capsaicinoids in different tissue types using different tumor models. Based on the findings of this study and prior studies of TRPV1-mediated cytotoxicity of capsaicinoids, we posit that the cumulative degree of metabolic destruction, bioactivation, and TRPV1 activation balances the acute cytotoxic and chemopreventive effects versus the co-carcinogenic activity of capsaicinoids in different cell/tissue types. Specifically, cells/tissues with high P450 content, particularly CYP1A2 and 3A4, but low TRPV1 expression (e.g., liver) may be sensitized to carcinogens through additive macromolecule modification by electrophiles. Conversely, cells/tissues with high TRPV1 expression and low P450 expression (e.g., lungs, prostate, pancreas) may be more susceptible to acute cytotoxicity through TRPV1-mediated signaling. Partially supporting this hypothesis, we have shown that human lung adenocarcinoma (A549) cells express more TRPV1 and are more sensitive to cytotoxicity by capsaicin than human hepatoma (HepG2) cells.(34,48) Furthermore, inhibition of P450 function in both cell types using 1-aminobenzotriazole increased the cytotoxicity of capsaicin, with HepG2 cells exhibiting a much greater degree of sensitization, consistent with greater detoxification of capsaicinoids by P450 enzymes.(20,34,48)
In conclusion, capsaicinoids are highly amenable to biotransformation by P450 enzymes and peroxidases. These enzymes produce a variety of products including O-demethylated, oxygenated, dehydrogenated, electrophilic, and free radical metabolites. The biological significance of capsaicinoid bioactivation remains enigmatic, however the identification of electrophilic metabolites and the elucidation of specific enzymatic and chemical mechanisms of formation will facilitate future studies evaluating capsaicinoids as pharmaceutical and/or cytotoxic and chemopreventive agents.
Supplementary Material
Acknowledgements
Funding Support:
This work was supported by the following NIH grants: HL069813, HL013645, GM074249, and CA129038. The following NIH and NSF grants provided funding for the NMR instrumentation, RR13030, DBI 0002806.
The authors thank Dr. Alan R. Light and Ronald W. Hughen of the Department of Anesthesiology, University of Utah, for providing the TRPV1−/− mice, and Dr. Vy Tran (Department of Medicinal Chemistry) for assistance with NMR analysis of the nonivamide analogues.
Abbreviations
- NADPH
reduced nicotinamide adenine dinucleotide phosphate
- GSH
reduced glutathione
- BME
β-mercaptoethanol
- LC/MS2
liquid chromatography-tandem mass spectrometry
Footnotes
Supporting Information Available:
Supplementary data including NMR and LC/MS2 spectra, predicted fragment ions for the GSH and BME conjugates, enzyme-selective metabolite production, and quantification of conjugate formation from capsaicinoid analogues, is provided. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Reilly CA, Crouch DJ, Yost GS, Fatah AA. Determination of capsaicin, dihydrocapsaicin, and nonivamide in self-defense weapons by liquid chromatography-mass spectrometry and liquid chromatography-tandem mass spectrometry. J. Chromatogr. A. 2001;912:259–267. doi: 10.1016/s0021-9673(01)00574-x. [DOI] [PubMed] [Google Scholar]
- 2.Cheng J, Yang XN, Liu X, Zhang SP. Capsaicin for allergic rhinitis in adults. Cochrane Database Syst. Rev. 2006:CD004460. doi: 10.1002/14651858.CD004460.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fusco BM, Giacovazzo M. Peppers and pain. The promise of capsaicin. Drugs. 1997;53:909–914. doi: 10.2165/00003495-199753060-00001. [DOI] [PubMed] [Google Scholar]
- 4.Rapoport AM, Bigal ME, Tepper SJ, Sheftell FD. Intranasal medications for the treatment of migraine and cluster headache. CNS Drugs. 2004;18:671–685. doi: 10.2165/00023210-200418100-00004. [DOI] [PubMed] [Google Scholar]
- 5.Inoue N, Matsunaga Y, Satoh H, Takahashi M. Enhanced energy expenditure and fat oxidation in humans with high BMI scores by the ingestion of novel and non-pungent capsaicin analogues (capsinoids) Biosci. Biotechnol. Biochem. 2007;71:380–389. doi: 10.1271/bbb.60341. [DOI] [PubMed] [Google Scholar]
- 6.Miller CH, Zhang Z, Hamilton SM, Teel RW. Effects of capsaicin on liver microsomal metabolism of the tobacco-specific nitrosamine NNK. Cancer Lett. 1993;75:45–52. doi: 10.1016/0304-3835(93)90206-o. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Z, Hamilton SM, Stewart C, Strother A, Teel RW. Inhibition of liver microsomal cytochrome P450 activity and metabolism of the tobacco-specific nitrosamine NNK by capsaicin and ellagic acid. Anticancer Res. 1993;13:2341–2346. [PubMed] [Google Scholar]
- 8.Zhang Z, Huynh H, Teel RW. Effects of orally administered capsaicin, the principal component of capsicum fruits, on the in vitro metabolism of the tobaccospecific nitrosamine NNK in hamster lung and liver microsomes. Anticancer Res. 1997;17:1093–1098. [PubMed] [Google Scholar]
- 9.Mori A, Lehmann S, O'Kelly J, Kumagai T, Desmond JC, Pervan M, McBride WH, Kizaki M, Koeffler HP. Capsaicin, a Component of Red Peppers, Inhibits Growth of Androgen-Independent, p53 Mutant Prostate Cancer Cells. Cancer Res. 2006;66:3222–3229. doi: 10.1158/0008-5472.CAN-05-0087. [DOI] [PubMed] [Google Scholar]
- 10.Surh YJ. More than spice: capsaicin in hot chili peppers makes tumor cells commit suicide. Life Sci. 2002;94:1263–1265. doi: 10.1093/jnci/94.17.1263. [DOI] [PubMed] [Google Scholar]
- 11.Pramanik KC, Boreddy SR, Srivastava SK. Role of mitochondrial electron transport chain complexes in capsaicin mediated oxidative stress leading to apoptosis in pancreatic cancer cells. PLoS One. 2011;6:e20151. doi: 10.1371/journal.pone.0020151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pramanik KC, Srivastava SK. Apoptosis signal-regulating kinase 1- thioredoxin complex dissociation by capsaicin causes pancreatic tumor growth suppression by inducing apoptosis. Antioxid. Redox Signal. 2012;17:1417–1432. doi: 10.1089/ars.2011.4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Babbar S, Marier JF, Mouksassi MS, Beliveau M, Vanhove GF, Chanda S, Bley K. Pharmacokinetic analysis of capsaicin after topical administration of a high-concentration capsaicin patch to patients with peripheral neuropathic pain. Ther. Drug Monit. 2009;31:502–510. doi: 10.1097/FTD.0b013e3181a8b200. [DOI] [PubMed] [Google Scholar]
- 14.Chanda S, Bashir M, Babbar S, Koganti A, Bley K. In vitro hepatic and skin metabolism of capsaicin. Drug Metab. Dispos. 2008;36:670–675. doi: 10.1124/dmd.107.019240. [DOI] [PubMed] [Google Scholar]
- 15.Donnerer J, Amann R, Schuligoi R, Lembeck F. Absorption and metabolism of capsaicinoids following intragastric administration in rats. Naunyn Schmiedebergs Arch. Pharmacol. 1990;342:357–361. doi: 10.1007/BF00169449. [DOI] [PubMed] [Google Scholar]
- 16.Kawada T, Suzuki T, Takahashi M, Iwai K. Gastrointestinal absorption and metabolism of capsaicin and dihydrocapsaicin in rats. Toxicol. Appl. Pharmacol. 1984;72:449–456. doi: 10.1016/0041-008x(84)90121-2. [DOI] [PubMed] [Google Scholar]
- 17.Suresh D, Srinivasan K. Studies on the In Vitro Absorption of Spice Principles - Curcumin, Capsaicin and Piperine in Rat Intestines. Food and Chemical Toxicology. 2007;45:1437–1442. doi: 10.1016/j.fct.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 18.Chaiyasit K, Khovidhunkit W, Wittayalertpanya S. Pharmacokinetic and the effect of capsaicin in Capsicum frutescens on decreasing plasma glucose level. J. Med. Assoc. Thai. 2009;92:108–113. [PubMed] [Google Scholar]
- 19.Suresh D, Srinivasan K. Tissue Distribution and Elimination of Capsaicin, Piperine, and Curcumin Following Oral Intake in Rats. Indian J. Med. Res. 2010;131:682–691. [PubMed] [Google Scholar]
- 20.Reilly CA, Ehlhardt WJ, Jackson DA, Kulanthaivel P, Mutlib AE, Espina RJ, Moody DE, Crouch DJ, Yost GS. Metabolism of capsaicin by cytochrome P450 produces novel dehydrogenated metabolites and decreases cytotoxicity to lung and liver cells. Chem. Res. Toxicol. 2003;16:336–349. doi: 10.1021/tx025599q. [DOI] [PubMed] [Google Scholar]
- 21.Reilly CA, Yost GS. Structural and enzymatic parameters that determine alkyl dehydrogenation/hydroxylation of capsaicinoids by cytochrome p450 enzymes. Drug Metab. Dispos. 2005;33:530–536. doi: 10.1124/dmd.104.001214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas KC, Ethirajan M, Shahrokh K, Sun H, Lee J, Cheatham TE, 3rd, Yost GS, Reilly CA. Structure-activity relationship of capsaicin analogs and transient receptor potential vanilloid 1-mediated human lung epithelial cell toxicity. J. Pharmacol. Exp. Ther. 2011;337:400–410. doi: 10.1124/jpet.110.178491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baillie T, Davis MR. Mass Spectrometry in the Analysis of Glutathione Conjugates. Biol. Mass Spec. 1993;22:319–325. doi: 10.1002/bms.1200220602. [DOI] [PubMed] [Google Scholar]
- 24.Moore CD, Reilly CA, Yost GS. CYP3A4-Mediated oxygenation versus dehydrogenation of raloxifene. Biochemistry. 2010;49:4466–4475. doi: 10.1021/bi902213r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernal MAaRBA. 5,5'-Dicapsaicin, 4'-O-5-Dicapsaicin Ether, and Dehydrogenation Polymers with High Molecular Weights are the Main Products of the Oxidation of Capsaicin by Peroxidase from Hot Pepper. J. Agric. Food Chem. 1996;44:3085–3089. [Google Scholar]
- 26.Goodwin DC, Hertwig KM. Peroxidase-catalyzed oxidation of capsaicinoids: steady-state and transient-state kinetic studies. Arch. Biochem. Biophys. 2003;417:18–26. doi: 10.1016/s0003-9861(03)00321-7. [DOI] [PubMed] [Google Scholar]
- 27.Surh YJ, Lee SS. Capsaicin, a double-edged sword: toxicity, metabolism, and chemopreventive potential. Life Sci. 1995;56:1845–1855. doi: 10.1016/0024-3205(95)00159-4. [DOI] [PubMed] [Google Scholar]
- 28.Miller M, Brendel K, Burks TF, Sipes IG. Interaction of Capsaicinoids with Drug Metabolizing Systems. Biochem. Pharmacol. 1983;32:547–551. doi: 10.1016/0006-2952(83)90537-3. [DOI] [PubMed] [Google Scholar]
- 29.Lawson T, Gannett P. The mutagenicity of capsaicin and dihydrocapsaicin in V79 cells. Cancer Lett. 1989;48:109–113. doi: 10.1016/0304-3835(89)90045-1. [DOI] [PubMed] [Google Scholar]
- 30.Gannett P, Iversen P, Lawson T. The Mechanism of Inhibition of Cytochrome P450IIE1 by Dihydrocapsaicin. Biorg. Chem. 1990;15:185–198. [Google Scholar]
- 31.Iverson SL, Hu LQ, Vukomanovic V, Bolton JL. The influence of the p-alkyl substituent on the isomerization of o-quinones to p-quinone methides: potential bioactivation mechanism for catechols. Chem. Res. Toxicol. 1995;8:537–544. doi: 10.1021/tx00046a007. [DOI] [PubMed] [Google Scholar]
- 32.Thompson DC, Perera K, Krol ES, Bolton JL. o-Methoxy-4- alkylphenols that form quinone methides of intermediate reactivity are the most toxic in rat liver slices. Chem. Res. Toxicol. 1995;8:323–327. doi: 10.1021/tx00045a001. [DOI] [PubMed] [Google Scholar]
- 33.Bolton JL, Comeau E, Vukomanovic V. The influence of 4-alkyl substituents on the formation and reactivity of 2-methoxy-quinone methides: evidence that extended pi-conjugation dramatically stabilizes the quinone methide formed from eugenol. Chem. Biol. Interact. 1995;95:279–290. doi: 10.1016/0009-2797(94)03566-q. [DOI] [PubMed] [Google Scholar]
- 34.Reilly CA, Yost GS. Metabolism of capsaicinoids by P450 enzymes: a review of recent findings on reaction mechanisms, bio-activation, and detoxification processes. Drug Metab. Rev. 2006;38:685–706. doi: 10.1080/03602530600959557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Babbar S, Chanda S, Bley K. Inhibition and induction of human cytochrome P450 enzymes in vitro by capsaicin. Xenobiotica. 2010;40:807–816. doi: 10.3109/00498254.2010.520044. [DOI] [PubMed] [Google Scholar]
- 36.Takanohashi T, Isaka M, Ubukata K, Mihara R, Bernard BK. Studies of the toxicological potential of capsinoids, XIII: inhibitory effects of capsaicin and capsinoids on cytochrome P450 3A4 in human liver microsomes. Int. J. Toxicol. 2010;29:22S–26S. doi: 10.1177/1091581809360282. [DOI] [PubMed] [Google Scholar]
- 37.Archer VE, Jones DW. Capsaicin pepper, cancer and ethnicity. Med. Hypotheses. 2002;59:450–457. doi: 10.1016/s0306-9877(02)00152-4. [DOI] [PubMed] [Google Scholar]
- 38.Jang JJ, Cho KJ, Lee YS, Bae JH. Different modifying responses of capsaicin in a wide-spectrum initiation model of F344 rat. J. Korean Med. Sci. 1991;6:31–36. doi: 10.3346/jkms.1991.6.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanchez AM, Sanchez MG, Malagarie-Cazenave S, Olea N, Diaz-Laviada I. Induction of apoptosis in prostate tumor PC-3 cells and inhibition of xenograft prostate tumor growth by the vanilloid capsaicin. Apoptosis. 2006;11:89–99. doi: 10.1007/s10495-005-3275-z. [DOI] [PubMed] [Google Scholar]
- 40.Sanchez MG, Sanchez AM, Collado B, Malagarie-Cazenave S, Olea N, Carmena MJ, Prieto JC, Diaz-Laviada II. Expression of the transient receptor potential vanilloid 1 (TRPV1) in LNCaP and PC-3 prostate cancer cells and in human prostate tissue. Eur. J. Pharmacol. 2005;515:20–27. doi: 10.1016/j.ejphar.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 41.Kang JY, Alexander B, Barker F, Man WK, Williamson RC. The effect of chilli ingestion on gastrointestinal mucosal proliferation and azoxymethaneinduced cancer in the rat. J. Gastroenterol. Hepatol. 1992;7:194–198. doi: 10.1111/j.1440-1746.1992.tb00961.x. [DOI] [PubMed] [Google Scholar]
- 42.Yoshitani SI, Tanaka T, Kohno H, Takashima S. Chemoprevention of azoxymethane-induced rat colon carcinogenesis by dietary capsaicin and rotenone. Int. J. Oncol. 2001;19:929–939. doi: 10.3892/ijo.19.5.929. [DOI] [PubMed] [Google Scholar]
- 43.Tanaka T, Kohno H, Sakata K, Yamada Y, Hirose Y, Sugie S, Mori H. Modifying effects of dietary capsaicin and rotenone on 4-nitroquinoline 1-oxideinduced rat tongue carcinogenesis. Carcinogenesis. 2002;23:1361–1367. doi: 10.1093/carcin/23.8.1361. [DOI] [PubMed] [Google Scholar]
- 44.Anandakumar P, Kamaraj S, Jagan S, Ramakrishnan G, Vinodhkumar R, Devaki T. Capsaicin modulates pulmonary antioxidant defense system during benzo(a)pyrene-induced lung cancer in Swiss albino mice. Phytother. Res. 2008;22:529–533. doi: 10.1002/ptr.2393. [DOI] [PubMed] [Google Scholar]
- 45.Jang JJ, Kim SH, Yun TK. Inhibitory effect of capsaicin on mouse lung tumor development. In Vivo. 1989;3:49–53. [PubMed] [Google Scholar]
- 46.Modly CE, Das M, Don PS, Marcelo CL, Mukhtar H, Bickers DR. Capsaicin as an in vitro inhibitor of benzo(a)pyrene metabolism and its DNA binding in human and murine keratinocytes. Drug Metab. Dispos. 1986;14:413–416. [PubMed] [Google Scholar]
- 47.Sanchez AM, Martinez-Botas J, Malagarie-Cazenave S, Olea N, Vara D, Lasuncion MA, Diaz-Laviada I. Induction of the endoplasmic reticulum stress protein GADD153/CHOP by capsaicin in prostate PC-3 cells: a microarray study. Biochem. Biophys. Res. Commun. 2008;372:785–791. doi: 10.1016/j.bbrc.2008.05.138. [DOI] [PubMed] [Google Scholar]
- 48.Reilly CA, Taylor JL, Lanza DL, Carr BA, Crouch DJ, Yost GS. Capsaicinoids cause inflammation and epithelial cell death through activation of vanilloid receptors. Toxicol. Sci. 2003;73:170–181. doi: 10.1093/toxsci/kfg044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.